Abstract
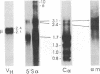
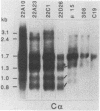
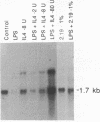
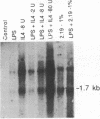
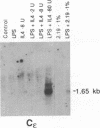
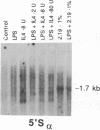
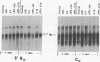
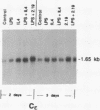
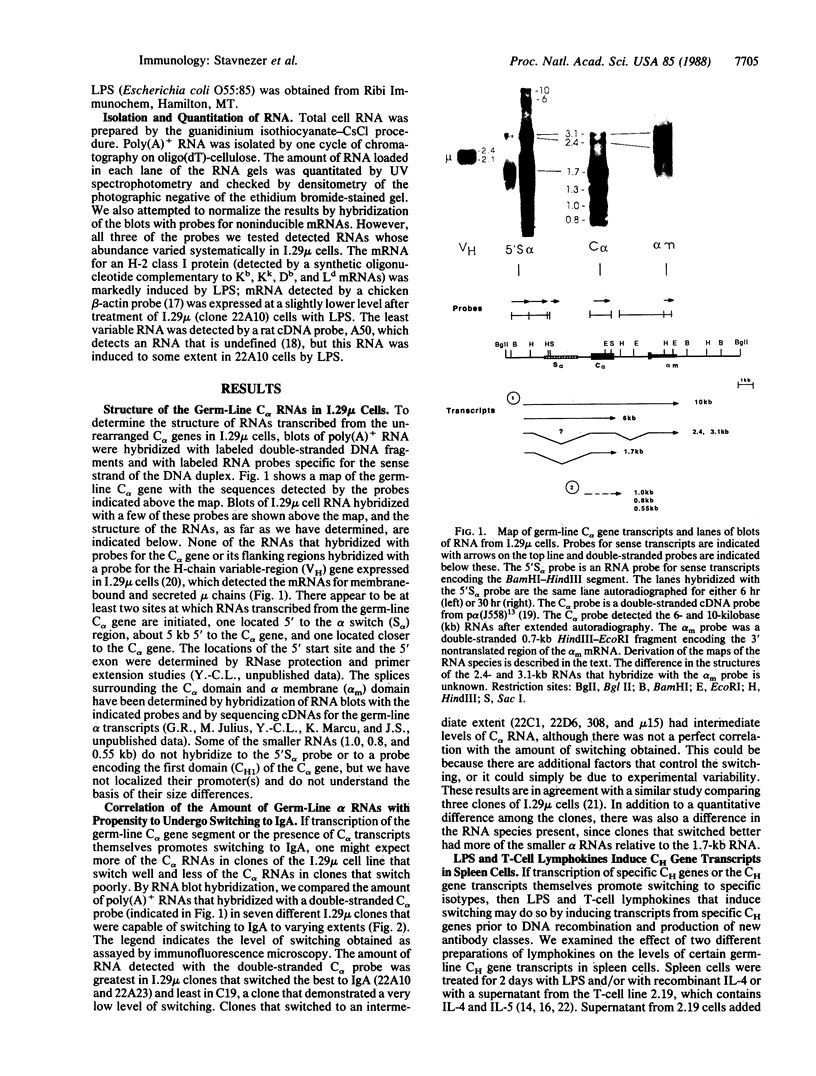
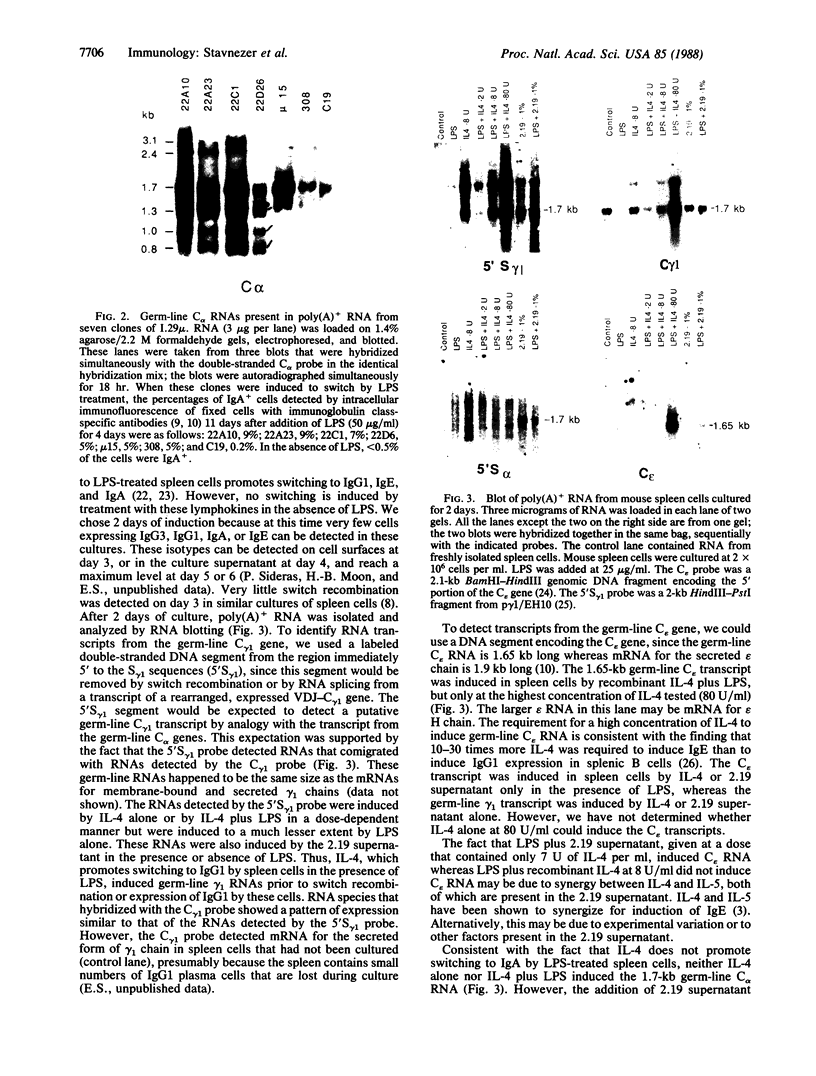
Immunoglobulin heavy-chain switching is effected by a DNA recombination event that replaces the C mu gene with one of the other heavy-chain constant-region (CH) genes located 3' to the C mu gene. How the specificity of this event is controlled is unknown. However, it has been shown that IgM+ cells capable of switching to specific isotypes have the corresponding unrearranged CH genes in an accessible or active chromatin state, as demonstrated by the fact that these specific CH genes are hypomethylated and are transcriptionally active. We now report that the RNAs transcribed from specific unrearranged CH genes are induced prior to switching under conditions that promote switching to these specific CH genes. For example, we find that bacterial lipopolysaccharide, which induces the IgM+ cell line I.29 mu to switch to IgA, induces transcripts from the germ-line C alpha gene(s) in I.29 mu cells prior to switch recombination. Two preparations of T-cell lymphokines (recombinant interleukin 4 and supernatant from the T-cell line 2.19, which contains interleukins 4 and 5) that promote switching to specific isotypes by lipopolysaccharide-treated spleen cells induce transcripts from the corresponding germ-line CH genes prior to expression of the new isotypes. For example, interleukin 4, which appears to be necessary for switching to IgE in vitro and in vivo, induces within 2 days large increases in germ-line C epsilon transcripts in lipopolysaccharide-treated spleen cells and in I.29 mu cells. The most straightforward interpretation of our data is that these lymphokines direct switching to specific isotypes by activating specific CH genes, making them accessible to the putative switch recombinase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberini C., Biassoni R., DeAmbrosis S., Vismara D., Sitia R. Differentiation in the murine B cell lymphoma I.29: individual mu + clones may be induced by lipopolysaccharide to both IgM secretion and isotype switching. Eur J Immunol. 1987 Apr;17(4):555–562. doi: 10.1002/eji.1830170419. [DOI] [PubMed] [Google Scholar]
- Bergstedt-Lindqvist S., Sideras P., MacDonald H. R., Severinson E. Regulation of Ig class secretion by soluble products of certain T-cell lines. Immunol Rev. 1984 Apr;78:25–50. doi: 10.1111/j.1600-065x.1984.tb00475.x. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Moore M. W., Yancopoulos G. D., Suh H., Lutzker S., Selsing E., Alt F. W. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986 Dec 11;324(6097):585–589. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Lopata M. A., MacDonald R. J., Cowan N. J., Rutter W. J., Kirschner M. W. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 1980 May;20(1):95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- Coffman R. L., Seymour B. W., Lebman D. A., Hiraki D. D., Christiansen J. A., Shrader B., Cherwinski H. M., Savelkoul H. F., Finkelman F. D., Bond M. W. The role of helper T cell products in mouse B cell differentiation and isotype regulation. Immunol Rev. 1988 Feb;102:5–28. doi: 10.1111/j.1600-065x.1988.tb00739.x. [DOI] [PubMed] [Google Scholar]
- Coffman R. L., Shrader B., Carty J., Mosmann T. R., Bond M. W. A mouse T cell product that preferentially enhances IgA production. I. Biologic characterization. J Immunol. 1987 Dec 1;139(11):3685–3690. [PubMed] [Google Scholar]
- Grabstein K., Eisenman J., Mochizuki D., Shanebeck K., Conlon P., Hopp T., March C., Gillis S. Purification to homogeneity of B cell stimulating factor. A molecule that stimulates proliferation of multiple lymphokine-dependent cell lines. J Exp Med. 1986 Jun 1;163(6):1405–1414. doi: 10.1084/jem.163.6.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harriman G. R., Kunimoto D. Y., Elliott J. F., Paetkau V., Strober W. The role of IL-5 in IgA B cell differentiation. J Immunol. 1988 May 1;140(9):3033–3039. [PubMed] [Google Scholar]
- Isakson P. C., Puré E., Vitetta E. S., Krammer P. H. T cell-derived B cell differentiation factor(s). Effect on the isotype switch of murine B cells. J Exp Med. 1982 Mar 1;155(3):734–748. doi: 10.1084/jem.155.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinashi T., Harada N., Severinson E., Tanabe T., Sideras P., Konishi M., Azuma C., Tominaga A., Bergstedt-Lindqvist S., Takahashi M. Cloning of complementary DNA encoding T-cell replacing factor and identity with B-cell growth factor II. Nature. 1986 Nov 6;324(6092):70–73. doi: 10.1038/324070a0. [DOI] [PubMed] [Google Scholar]
- Klein D., Nietupski J., Sirlin S., Stavnezer J. I.29 lymphoma cells express a nonmutated VH gene before and after H chain switch. J Immunol. 1988 Mar 1;140(5):1676–1684. [PubMed] [Google Scholar]
- Layton J. E., Vitetta E. S., Uhr J. W., Krammer P. H. Clonal analysis of B cells induced to secrete IgG by T cell-derived lymphokine(s). J Exp Med. 1984 Dec 1;160(6):1850–1863. doi: 10.1084/jem.160.6.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon G. G., Perry R. P. C mu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5'-nontranslatable exon. Nature. 1985 Dec 5;318(6045):475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- Lutzker S., Alt F. W. Structure and expression of germ line immunoglobulin gamma 2b transcripts. Mol Cell Biol. 1988 Apr;8(4):1849–1852. doi: 10.1128/mcb.8.4.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzker S., Rothman P., Pollock R., Coffman R., Alt F. W. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 1988 Apr 22;53(2):177–184. doi: 10.1016/0092-8674(88)90379-0. [DOI] [PubMed] [Google Scholar]
- Marcu K. B., Banerji J., Penncavage N. A., Lang R., Arnheim N. 5' flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980 Nov;22(1 Pt 1):187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- Mowatt M. R., Dunnick W. A. DNA sequence of the murine gamma 1 switch segment reveals novel structural elements. J Immunol. 1986 Apr 1;136(7):2674–2683. [PubMed] [Google Scholar]
- Murray P. D., McKenzie D. T., Swain S. L., Kagnoff M. F. Interleukin 5 and interleukin 4 produced by Peyer's patch T cells selectively enhance immunoglobulin A expression. J Immunol. 1987 Oct 15;139(8):2669–2674. [PubMed] [Google Scholar]
- Nguyen H. T., Medford R. M., Nadal-Ginard B. Reversibility of muscle differentiation in the absence of commitment: analysis of a myogenic cell line temperature-sensitive for commitment. Cell. 1983 Aug;34(1):281–293. doi: 10.1016/0092-8674(83)90159-9. [DOI] [PubMed] [Google Scholar]
- Nishida Y., Kataoka T., Ishida N., Nakai S., Kishimoto T., Böttcher I., Honjo T. Cloning of mouse immunoglobulin epsilon gene and its location within the heavy chain gene cluster. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1581–1585. doi: 10.1073/pnas.78.3.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma Y., Sideras P., Naito T., Bergstedt-Lindquist S., Azuma C., Severinson E., Tanabe T., Kinashi T., Matsuda F., Yaoita Y. Cloning of cDNA encoding the murine IgG1 induction factor by a novel strategy using SP6 promoter. Nature. 1986 Feb 20;319(6055):640–646. doi: 10.1038/319640a0. [DOI] [PubMed] [Google Scholar]
- Sideras P., Bergstedt-Lindqvist S., MacDonald H. R., Severinson E. Secretion of IgG1 induction factor by T cell clones and hybridomas. Eur J Immunol. 1985 Jun;15(6):586–593. doi: 10.1002/eji.1830150611. [DOI] [PubMed] [Google Scholar]
- Snapper C. M., Finkelman F. D., Paul W. E. Differential regulation of IgG1 and IgE synthesis by interleukin 4. J Exp Med. 1988 Jan 1;167(1):183–196. doi: 10.1084/jem.167.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper C. M., Paul W. E. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987 May 22;236(4804):944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J., Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO J. 1986 Jan;5(1):95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Sirlin S., Abbott J. Induction of immunoglobulin isotype switching in cultured I.29 B lymphoma cells. Characterization of the accompanying rearrangements of heavy chain genes. J Exp Med. 1985 Mar 1;161(3):577–601. doi: 10.1084/jem.161.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter E., Krawinkel U., Radbruch A. Directed Ig class switch recombination in activated murine B cells. EMBO J. 1987 Jun;6(6):1663–1671. doi: 10.1002/j.1460-2075.1987.tb02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G. D., DePinho R. A., Zimmerman K. A., Lutzker S. G., Rosenberg N., Alt F. W. Secondary genomic rearrangement events in pre-B cells: VHDJH replacement by a LINE-1 sequence and directed class switching. EMBO J. 1986 Dec 1;5(12):3259–3266. doi: 10.1002/j.1460-2075.1986.tb04637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]