Abstract
Production of type I interferon (IFN; IFN-αβ) increases host susceptibility to Listeria monocytogenes, whereas type II IFN (IFN-γ) activates macrophages to resist infection. We show that these opposing immunological effects of IFN-αβ and IFN-γ occur because of cross talk between the respective signaling pathways. We found that cultured macrophages infected with L. monocytogenes were refractory to IFN-γ treatment as a result of down-regulation of the IFN-γ receptor (IFNGR). The soluble factor responsible for these effects was identified as host IFN-αβ. Accordingly, macrophages and dendritic cells (DCs) showed reduced IFNGR1 expression and reduced responsiveness to IFN-γ during systemic infection of IFN-αβ–responsive mice. Furthermore, the increased resistance of mice lacking the IFN-αβ receptor (IFNAR−/−) to L. monocytogenes correlated with increased expression of IFN-γ–dependent activation markers by macrophages and DCs and was reversed by depletion of IFN-γ. Thus, IFN-αβ produced in response to bacterial infection and other stimuli antagonizes the host response to IFN-γ by down-regulating the IFNGR. Such cross talk permits prioritization of IFN-αβ–type immune responses and may contribute to the beneficial effects of IFN-β in treatment of inflammatory diseases such as multiple sclerosis.
The innate immune system is the first line of defense against pathogenic microbes. Phagocytic cells of the innate immune system, including macrophages, DCs, and neutrophils, patrol host tissues and rapidly engulf any bacteria or particulate microbes they encounter. Once engulfed, most organisms are killed. However, several pathogens, like Listeria monocytogenes and Mycobacterium tuberculosis, have evolved strategies to replicate within resting macrophages and DCs (Pieters, 2008; Flannagan et al., 2009; Ray et al., 2009).
L. monocytogenes produces a hemolysin, listeriolysin O (LLO), which permits the bacterium to rupture phagosomes and escape into the cytosol of infected cells. Consequently, strains lacking expression of LLO (ΔHly) are avirulent. In addition, ΔHly L. monocytogenes fail to elicit the production of IFN-αβ by infected macrophages (O’Riordan et al., 2002). Production of IFN-αβ during L. monocytogenes infection is thought to be dependent on the detection of microbial products by a receptor present in the host cell cytosol (Leber et al., 2008). Although IFN-αβ elicits an antiviral state that promotes resistance to viral pathogens, IFN-αβ production increases the survival and replication of L. monocytogenes, M. tuberculosis, and several other pathogenic bacteria (Auerbuch et al., 2004; Carrero et al., 2004; O’Connell et al., 2004; Stanley et al., 2007; Qiu et al., 2008; Martin et al., 2009; Shahangian et al., 2009). Mechanisms for such probacterial effects of IFN-αβ have not been clearly defined, although previous work has correlated IFN-αβ production with increased cell death and differences in macrophage production of IL-10, IL-12, and TNF (Auerbuch et al., 2004; Carrero et al., 2004, 2006; O’Connell et al., 2004).
In contrast to IFN-αβ, IFN-γ is essential for host resistance to L monocytogenes and other intracellular pathogens (Buchmeier and Schreiber, 1985; Dalton et al., 1993). IFN-γ drives the differentiation of resting macrophages into an activated antimicrobial state (M1) that more efficiently restricts the growth of intracellular pathogens (Gordon, 2003). The effects of IFN-γ require its binding to the IFN-γ receptor (IFNGR) 1 subunit of a heterodimeric cell surface IFNGR. This binding triggers receptor clustering and activates a Janus kinase (JAK)–signal transducer and activator of transcription (STAT) signaling pathway that culminates in the binding of STAT1 to IFN-γ–activated sequence (GAS) elements in the DNA adjacent to IFN-γ–stimulated genes (Platanias, 2005). The expression of several IFN-γ–stimulated genes is up-regulated by IFN-γ, including those coding for class II MHC proteins (MHCII) and the transcriptional activator of MHCII, CIITA (Reith et al., 2005).
IFN-γ is produced in abundance by L. monocytogenes antigen-specific CD4+ and CD8+ T cells (Zenewicz and Shen, 2007; Harty and Badovinac, 2008). However, within the first few days of infection, the major sources of IFN-γ are NK cells of the innate immune system (Humann et al., 2007; Kang et al., 2008). This innate wave of IFN-γ production peaks around 24 h post infection (hpi) but fails to limit L. monocytogenes growth, which continues for the first 72 h after systemic infection. The continued bacterial growth in the face of innate IFN-γ suggests that the early production of IFN-γ is not sufficient to activate macrophage bactericidal activity.
In this paper, we present data indicating a mechanism by which L. monocytogenes prevents macrophage activation by innate IFN-γ. We find that both infected and bystander macrophages become refractory to stimulation by IFN-γ early after L. monocytogenes infection. This refractory state is the result of down-regulation of the IFNGR, which is induced by IFN-αβ released from L. monocytogenes–infected cells. IFN-αβ down-regulates cell surface IFNGR and attenuates macrophage activation during systemic L. monocytogenes infection only in mice expressing the receptor for type I IFN, IFN-αβ receptor (IFNAR). Mice lacking IFNAR expression consequently have increased expression of IFNGR and their reduced susceptibility to L. monocytogenes infection is dependent on IFN-γ. These studies reveal a mechanism by which IFN-αβ contributes to increased host susceptibility to bacterial infection and demonstrate a previously unappreciated mechanism of antagonistic cross talk between type I and II IFNs.
RESULTS AND DISCUSSION
L. monocytogenes infection inhibits macrophage responsiveness to IFN-γ
To test whether L. monocytogenes infection might suppress macrophage responses to IFN-γ, mouse BM-derived macrophages (BMMs) were subjected to a low multiplicity (multiplicity of infection [MOI] = 1) of WT L. monocytogenes (wt Lm) 2 h before treatment with IFN-γ. 20 h later, the infected and control BMMs were harvested and cell surface MHCII expression on live-gated cells was analyzed by flow cytometry (Fig. 1 A). Mock-infected BMM treated with IFN-γ showed 50–100× higher MHCII staining than BMM not treated with IFN-γ. However, nearly 95% of this IFN-γ–induced MHCII increase was blocked in BMM cultures that had been infected with wt Lm. These data suggest that the infection either specifically impaired expression of cell surface MHCII expression or more generally impaired macrophage responsiveness to IFN-γ.
Figure 1.
L. monocytogenes infection suppresses macrophage responses to IFN-γ. (A) Cell surface MHC class II (MHCII) expression on live-gated C57BL/6 BMM treated with IFN-γ after mock infection (light shading) or infection with wt Lm (dark shading) at MOI = 1. The clear histogram depicts MHCII expression on mock-infected cells not treated with IFN-γ. (B) Semiquantitative RT-PCR was performed using complementary DNA template prepared from mock- or wt Lm–infected BMM at 10 hpi. Treatment with IFN-γ occurred 2 h after infection. (C) RAW264.7-CIITApIV reporter cells (Fortune et al., 2004) were mock or wt Lm infected. At 2 hpi, cells received fresh media containing 0 (none) or 100 U/ml IFN-γ. Luciferase reporter activity was measured 6 h later. (D) RAW264.7 cells were transfected with a GAS-luciferase reporter construct. A stable IFN-γ–responsive transfectant (RAW-GAS.6) was treated with IFN-γ at 2 hpi with wt Lm or a hemolysin-deficient L. monocytogenes strain (ΔHly Lm). Infected and control RAW-GAS.6 cells were lysed to assay luciferase activity at 10 hpi. (E) Immunoblotting for phospho-STAT1 after IFN-γ treatment of mock-infected or wt Lm–infected cells. Cells were treated with IFN-γ at the indicated times after infection and lysed 5, 15, or 30 min later. Similar results were seen using lysates prepared from four independent experiments. For C and D, bars show SE from three independent samples. Horizontal lines represent the level of expression on uninfected cells. Error bars indicate SEM. A Student’s t test was used to calculate p-values where indicated. Experiments in A–D were repeated at least three times.
Induction of MHCII transcription by IFN-γ requires the class II transactivator CIITA (Reith et al., 2005). To investigate the impact of wt Lm infection on IFN-γ–induced CIITA expression, we evaluated transcription of the CIITA-pIV isoform in control and infected BMM. IFN-γ treatment increased transcription of CIITA-pIV in mock-infected BMM as judged by semiquantitative RT-PCR (Fig. 1 B). However, no induction of CIITA-pIV transcripts was seen in IFN-γ–treated BMM previously infected with wt Lm. Similarly, wt Lm infection prevented IFN-γ–induced luciferase reporter activity in RAW-CIITApIV reporter macrophages (Fig. 1 C), which were derived from RAW264.7 macrophages by stable transfection with a CIITA-pIV-luciferase reporter construct (Fortune et al., 2004). Thus, infection of macrophages with L. monocytogenes suppressed induction of both CIITA and cell surface MHCII.
To further discern whether the suppressive effects of L. monocytogenes were specific to CIITA, we developed an additional set of reporter cell lines. RAW264.7 cells were stably transfected with pHTS-GAS, a reporter construct containing four GAS elements upstream of a luciferase open reading frame. Reporter activity in the resulting RAW-GAS reporter cells was strongly induced by IFN-γ (Fig. 1 D) and inhibited by pretreatment with the STAT1-inhibitory anticancer agent fludarabine (not depicted; Frank et al., 1999). When RAW-GAS.6 or additional independently derived GAS reporter cell lines were infected with wt Lm before IFN-γ treatment, the induction of reporter gene activity was reduced by ∼50% (Fig. 1 D). Infection with wt Lm failed to suppress luciferase reporter activity driven by an nfkb promoter (unpublished data). In contrast to wt Lm, infection with a live mutant L. monocytogenes strain lacking expression of the LLO hemolysin (ΔHly Lm) had no impact on IFN-γ–dependent reporter gene activity (Fig. 1 D). Likewise, heat-killed wt Lm failed to significantly suppress RAW-GAS or RAW-CIITApIV cell reporter activity in response to IFN-γ (unpublished data). Finally, we evaluated levels of phospho (Y701) STAT1 after treatment of mock- or wt Lm–infected macrophages with recombinant IFN-γ (Fig. 1 E). The results indicated that IFN-γ treatment elicits significantly less pSTAT1 in macrophages infected for 6 h. Conversely, the response to IFN-γ was comparable to that of mock-infected cells at 2 hpi. Thus, prolonged infection of macrophages with viable L. monocytogenes that is capable of accessing the macrophage cytosol results in impaired responsiveness of these cells to IFN-γ.
Down-regulation of IFNGR expression accounts for the suppression of macrophage responsiveness to IFN-γ
To investigate the mechanism by which L. monocytogenes suppressed IFN-γ responsiveness, we evaluated the impact of wt Lm infection on expression of several macrophage genes important for responsiveness to IFN-γ. Total RNA was harvested from mock- and wt Lm–infected BMM and used for Affymetrix genechip analysis. The normalized expression of stat1 and jak2 increased by nearly 10-fold with wt Lm infection, whereas jak1 and ifngr2 expression were not affected (Fig. 2 A). In contrast, expression of ifngr1 was reduced by nearly sevenfold in the wt Lm–infected macrophages. These data indicate that wt Lm infection dramatically affects the expression of genes involved in responses to IFN-γ. In particular, the suppressed transcription of ifngr1 might be expected to interfere with cell surface IFNGR expression and, thus, macrophage responsiveness to IFN-γ.
Figure 2.
Suppression of macrophage responsiveness to IFN-γ after L. monocytogenes infection reflects rapid down-regulation of the IFNGR. (A) Total RNA was isolated from mock- and wt Lm–infected BMM at 10 hpi and hybridized to Affymetrix genechips. The mean fold change in normalized expression between mock- and wt Lm–infected BMM is shown for the indicated genes encoding proteins involved in IFN-γ signaling. (B) Mock-infected and wt Lm–infected BMMs were harvested at the indicated hpi, stained with antibodies to IFNGR1, IFNGR2, or CD11b, and analyzed by flow cytometry. Mean channel fluorescence intensities (MFI) from three infected samples per group were determined and normalized to the mean MFI of three mock-infected samples using the following formula: relative surface staining = (MFI Lm infected)/(MFI mock infected). (C) C57BL/6 BMMs were mock or wt Lm infected. One half of each sample was stained for cell surface IFNGR1 and the other half was permeabilized with saponin and stained for total cellular IFNGR1. Shown are representative histograms of IFNGR1 staining for each BMM population. MFIs are indicated in parentheses. (D) Real-time RT-PCR was used to quantify the relative transcript abundance of IFNGR1 and IFNGR2 at the indicated hpi. Samples were from the experiment in B. (E) B6.IFNGR1−/− BMMs were mock or wt Lm infected and analyzed for surface expression of IFNGR2 and CD11b. Shown is relative surface staining on wt Lm–infected cells. Error bars indicate SEM for three samples per condition. Asterisks indicate significant (P < 0.05) variations between Lm-infected and mock-infected samples. ns indicates P > 0.05. Horizontal lines represent the level of expression on uninfected cells. Experiments in B–E were repeated at least three times.
We thus evaluated cell surface staining for IFNGR1 and IFNGR2 subunits. IFNGR1 detection was highly specific using a two-step staining procedure (Fig. S1). Our results indicated that surface expression of IFNGR1 was rapidly reduced in the wt Lm–infected cells, with a maximal reduction of ∼60% at 8 hpi (Fig. 2 B). The reduction in IFNGR1 surface staining paralleled a reduction in total IFNGR1 protein levels, as determined by intracellular staining for IFNGR1 in saponin-permeabilized mock- and wt Lm–infected BMM (Fig. 2 C). Detection of IFNGR2 required a three-step staining procedure. Cell surface staining for IFNGR2 was also reduced by wt Lm infection with an extent and kinetics similar to that of IFNGR1 (Fig. 2 B). In contrast, cell surface staining for the integrin CD11b was not affected by wt Lm infection (Fig. 2 B; and Fig. S2, raw mean fluorescence intensity [MFI]). Thus, down-regulation of the IFNGR subunits is a specific consequence of infection. BMMs fail to produce IFN-γ in response to wt Lm (unpublished data), and IFN-γ−/− BMM retained the ability to down-regulate IFNGR in response to wt Lm infection (unpublished data). Thus, the specific loss of cell surface IFNGR expression was not attributable to ligand-induced IFNGR internalization.
The relative abundance of ifngr1 and ifngr2 transcripts was also evaluated in the mock- and wt Lm–infected BMM using quantitative RT-PCR. As predicted from the Affymetrix analysis, ifngr1 transcription was significantly reduced within 4 hpi (Fig. 2 D). However, we failed to observe significant reductions in ifngr2 transcription at any time point after the wt Lm infection. Given the contrasting behaviors of ifngr2 transcripts and IFNGR2 surface staining, we hypothesize that the stability or cell surface localization of IFNGR2 is tightly linked to that of IFNGR1 at a posttranscriptional level. Indeed, BMM from B6.IFNGR1−/− mice failed to down-regulate IFNGR2 when infected with wt Lm (Fig. 2 E).
Together, these findings demonstrated that wt Lm infection triggers a rapid decrease in cell surface expression of both IFNGR1 and IFNGR2 subunits of the IFNGR, albeit through distinct mechanisms. The reduced availability of the IFNGR provides a mechanistic basis for the reduction in responsiveness of wt Lm–infected BMM to IFN-γ.
IFNGR is selectively down-regulated on antigen presenting cell populations
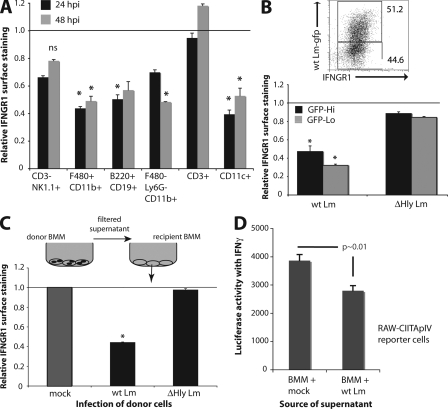
When C57BL/6 mice were infected i.v. with a sublethal dose of wt Lm (10,000 cfu), both splenic myeloid (CD11b+) and B lymphocyte (B220+CD19+) populations showed significant reductions in IFNGR1 staining from 24 to at least 48 hpi (Fig. 3 A). IFNGR1 staining remained low on CD11c+ gated DCs for at least 79 hpi (Fig. 3 A and see Fig. 5 A). Cell surface IFNGR1 staining was also slightly, but not significantly, reduced on NK1.1+CD3− NK cells (P > 0.05). However, no reduction was seen in IFNGR1 staining on gated CD3+ T cells. These results indicated that down-regulation of IFNGR1 selectively occurs on APC populations during the early stages of systemic infection with virulent L. monocytogenes. Furthermore, the results showed that IFNGR1 was down-regulated on nearly all APCs in L. monocytogenes–infected mice (Fig. 3 A), despite the fact that only a fraction of APCs are infected with live bacteria at the infection dose used (104). We thus hypothesized that a soluble factor released from L. monocytogenes-infected cells was responsible for IFNGR1 down-regulation.
Figure 3.
IFNGR down-regulation is restricted to APC populations and is a result of a suppressive soluble factor. (A) C57BL/6 mice (n = 3) were infected i.v. with ∼104 cfu wt Lm or given PBS alone. Spleens were harvested at indicated hpi and single cell suspensions were stained for IFNGR1 expression on the indicated cell populations by flow cytometry. Shown are mean + SEM IFNGR1 staining intensities from infected mice normalized to the same populations from mock-infected mice. Horizontal line represents the level of expression on uninfected cells. (B) BMMs were infected with GFP-tagged WT or ΔHly Lm stains at MOI = 5. At 5 hpi, cells were lifted and stained to evaluate surface IFNGR1 expression on the infected (GFPhi) and uninfected (GFPlo) populations. The bottom panel is a graphical depiction of the mean + SEM of normalized IFNGR1 expression levels for the GFPhi and GFPlo BMM populations. (C) Supernatants were harvested from WT or ΔHly infected donor BMMs at 8 hpi, filter sterilized, and transferred onto naive recipient BMMs. At 8 h after transfer, surface IFNGR1 expression was evaluated on the recipient BMMs by flow cytometry. Mean + SEM surface expression relative to mock-infected control samples is depicted. (D) RAW-CIITApIV reporter macrophage cells were treated for 6 h with sterile filtered supernatants from mock- or wt Lm–infected BMM, both of which were spiked with 100 U/ml IFN-γ. Supernatants were prepared as in C and luciferase activity was determined as in Fig. 1. Experiments were repeated at least three times. Error bars indicate SEM. *, P < 0.05.
Figure 5.
The resistance of IFNAR−/− APCs to IFNGR down-regulation correlates with their increased activation by IFN-γ and increased IFN-γ–dependent resistance of IFNAR−/− mice to systemic L. monocytogenes infection. (A) B6.IFNAR1−/− and congenic C57BL/6 mice were treated with PBS or 500 µg of anti–IFN-γ (XMG1.2) diluted in PBS 17 h before infection with a sublethal dose of wt Lm (9,000 cfu). Spleens and livers were harvested 72 h later. Gated monocytes and DCs were analyzed for cell surface MHCII expression. The MFI for each histogram is indicated in parentheses. Data are representative of results from three to four mice per group. (B) Livers from the infected mice were homogenized and dilution plated to determine bacterial burdens. Each point indicates an individual mouse. Bars indicate the mean values. The asterisk indicates a p-value of <0.05. Experiments were repeated twice.
A soluble factor released from infected cells mediates IFNGR down-regulation and suppressed responses to IFN-γ
As a first step to directly evaluate whether a soluble factor mediated IFNGR down-regulation, BMMs were infected at a low multiplicity (MOI = 5) with a wt Lm strain expressing enhanced GFP. Both infected (GFPhi) and uninfected (GFPlo) BMM in these cultures down-regulated IFNGR1 expression (Fig. 3 B). In contrast, BMM infected with ΔHly Lm that expressed enhanced GFP failed to down-regulate IFNGR1 in either GFP+ or GFP− BMM. We next evaluated the ability of sterile filtered conditioned media from mock- or wt Lm–infected donor BMM to induce IFNGR down-regulation on uninfected recipient BMM. 8 h after transfer of the respective conditioned media, cell surface IFNGR1 was evaluated on recipient BMM. Recipient BMM treated with media from mock-infected donor BMM failed to down-regulate IFNGR1 (Fig. 3 C). In contrast, conditioned media from wt Lm–infected BMM induced a decrease in IFNGR1 staining that was similar to that seen with direct infection of the BMM by wt Lm. In addition, a soluble factor was sufficient to significantly suppress IFN-γ–dependent reporter gene activity in RAW-CIITApIV reporter cells, as shown by transfer of conditioned media from wt Lm– or mock-infected BMM (Fig. 3 D). Together, these results confirmed that a factor secreted from wt Lm–infected macrophages was sufficient to induce both IFNGR down-regulation and suppression of macrophage gene induction by IFN-γ.
IFN-αβ is responsible for IFNGR down-modulation
We asked whether other inflammatory stimuli might also induce macrophages to secrete factors that down-regulate the IFNGR. C57BL/6 and MyD88−/− BMMs were thus treated with TLR agonists or infected with wt Lm as a control. Similar IFNGR1 down-regulation was seen in both B6 and B6.MyD88−/− BMMs that were infected with wt Lm, indicating that MyD88-dependent TLR signaling was dispensable for induction of the soluble IFNGR down-regulating factor by live cytosolic L. monocytogenes (Fig. 4 A). Nonetheless, treatments with specific TLR agonists (including nonmethylated CpG oligodeoxynucleotides [ODNs], poly I:C, and, to a lesser extent, LPS) did induce significant IFNGR1 down-regulation (Fig. 4 A). In some cases, these treatments required MyD88 expression by the BMM. Scrambled control ODNs and the triacyl-lipopeptide Pam3Cys failed to elicit down-regulation of IFNGR1.
Figure 4.
Type I IFN mediates IFNGR down-regulation. (A) C57BL/6 (black) or MyD88−/− (gray) BMMs were infected with wt Lm at MOI = 5 or treated with the indicated TLR agonists as described in Materials and methods. After 8 h of infection or treatment, BMMs were lifted and surface IFNGR1 expression was assessed by flow cytometry. Shown is mean + SEM staining for IFNGR1 on each set of BMM relative to untreated BMM of the same genotype. (B) C57BL/6 or B6.IFNAR1−/− BMMs were infected with wt Lm at MOI = 5. At 8 hpi, surface expression of IFNGR1 and IFNGR2 was quantified by flow cytometry. Shown is the mean + SEM relative surface staining of each IFNGR subunit normalized to staining of mock-infected BMM. (C) Sterile filtered supernatants from mock- or wt Lm–infected donor BMM of the indicated genotype were transferred to BMM of the indicated recipient genotype as in Fig. 3. Shown is the relative surface expression of IFNGR1 in each cell population. (D) C57BL/6 BMMs were infected with wt Lm or in parallel treated with the indicated recombinant murine cytokines as described in Materials and methods. Wt Lm and IFN-β induced equivalent IFNGR1 down-regulation. Experiments were repeated at least three times. Horizontal lines represent the level of expression on uninfected cells. Error bars indicate SEM. *, P < 0.05.
Type I IFNs are produced by macrophages in response to cytosolic (but not ΔHly) L. monocytogenes infection, as well as by stimulation with CpG ODN, LPS, and pIC. To determine whether IFN-αβ might be the host factor responsible for IFNGR down-regulation, we evaluated IFNGR1 surface expression on macrophages from IFNAR1−/− mice after wt Lm infection. Strikingly, the infected IFNAR−/− macrophages failed to significantly down-regulate IFNGR1 or IFNGR2 (Fig. 4 B). We also used reciprocal transfers of sterile filtered supernatants from infected C57BL/6 or IFNAR1−/− donor BMMs to induce IFNGR down-regulation on uninfected recipient C57BL/6 or IFNAR1−/− BMMs. Staining for cell surface IFNGR1 on recipient BMM revealed that only those macrophages expressing the IFNAR were capable of significantly down-regulating IFNGR1 (Fig. 4 C). Thus, IFNAR signaling was necessary for the response to, but not the induction of, the factors that down-regulate the IFNGR.
To determine whether type I IFN was sufficient to mediate IFNGR down-regulation, we treated C57BL/6 BMM with a panel of recombinant commercial mouse cytokines. Down-regulation of IFNGR1 was not seen in macrophages treated with IL-6 or IL-10 (Fig. 4 D), two cytokines which are known to suppress IFN-γ signaling (Nagabhushanam et al., 2003; Dikopoulos et al., 2005; Carrero et al., 2006; Murray, 2007). In addition, IFNGR down-regulation was not seen in cells treated with recombinant IL-28/IFN-λ, a cytokine which shares signaling components with IL-10 and IFN-αβ (Donnelly et al., 2004). However, recombinant IFN-β induced a similar degree of IFNGR1 down-regulation as seen during wt Lm infection (Fig. 4 D). As expected, IFNGR1 down-regulation was not induced by IFN-β treatment of IFNAR1−/− BMM (unpublished data). These data indicate that IFNAR signaling is necessary and sufficient for down-regulation of IFNGR1.
Increased resistance of IFNAR1−/− mice to L. monocytogenes infection correlates with increased macrophage activation and requires IFN-γ
The results in the previous sections suggested that APC populations from IFNAR1−/− mice might respond better to IFN-γ and, thus, more efficiently clear in vivo bacterial infections. Indeed, IFNGR1 and MHCII cell surface staining were dramatically reduced on CD11c+CD3− DCs from wt Lm–infected B6 mice when compared with the same population from infected IFNAR1−/− or uninfected C57BL/6 mice (Fig. 5 A). Similar results were seen on gated Ly6G−CD11b+ inflammatory monocytes (unpublished data). It was previously reported that infection with wt Lm elicits similar serum concentrations of IFN-γ in IFNAR−/− and IFNAR+/+ mice (Auerbuch et al., 2004). Thus, the respective increases in MHCII expression seen in infected B6 and B6.IFNAR1−/− mice are not explained by differences in the amounts of IFN-γ produced in each mouse strain.
We further demonstrated that the differences in MHCII expression were the result of IFN-γ, rather than other factors, by evaluating staining on cells from B6 and B6.IFNAR1−/− mice given a neutralizing antibody to IFN-γ (XMG1.2) before wt Lm infection. The XMG1.2 treatment reduced MHCII expression on gated APCs to a similar basal level in both mouse strains (Fig. 5 A). Thus, although MHCII expression was increased by the infection in APCs from both IFNAR-expressing and IFNAR1-deficient mice, the response was more pronounced in the IFNAR1−/− animals.
Bacterial burdens present in the livers of infected B6, B6.IFNAR1−/−, and IFN-γ–depleted B6.IFNAR1−/− mice were also determined at 79 hpi with wt Lm. Organs from the control B6.IFNAR1−/− mice harbored ∼3–4 logs fewer bacteria when compared with C57BL/6 mice (Fig. 5 B), confirming the heightened resistance of IFNAR1−/− mice to wt Lm infection. However, this heightened resistance was completely abrogated by antibody-mediated depletion of IFN-γ in the B6.IFNAR1−/− mice pretreated with 500 µg neutralizing anti–IFN-γ antibody (XMG1.2). Indeed, the bacterial burdens in the IFN-γ–depleted IFNAR1−/− mice were not significantly different from those seen in control or IFN-γ–depleted C57BL/6 mice. Thus, the heightened responsiveness of IFNAR1−/− mice to IFN-γ accounts for their increased resistance to L. monocytogenes infection.
Concluding remarks
Our studies reveal that production of IFN-αβ early after L. monocytogenes infection down-regulates ifngr1 transcription and, hence, reduces surface expression of the IFNGR by ∼50–60%. Despite the partial nature of this reduction in IFNGR expression, the induction of IFN-γ–dependent gene expression by APCs is clearly affected both in vitro and in vivo. As shown in this paper, cells infected with L. monocytogenes respond poorly to IFN-γ, and supernatant from these cells impairs transcriptional and translational up-regulation of IFN-γ–inducible genes. To our knowledge, the ability of IFN-αβ to down-regulate IFNGR expression by APCs has not been previously reported. However, our findings do provide an explanation for the previously described ability of IFN-αβ to interfere with binding of IFN-γ to macrophages and B cells (Thompson et al., 1985; Yoshida et al., 1988). Our findings are also consistent with several older studies that showed that IFN-αβ treatment antagonizes the response of mouse and mature human macrophages to treatment with IFN-γ (Ling et al., 1985; Inaba et al., 1986; Yoshida et al., 1988).
We speculate that the ability of IFN-αβ to suppress IFNGR expression has evolved to permit the integration of coincident signals that occur during infection with agents that induce concurrent expression of both IFN-αβ and IFN-γ. By suppressing responsiveness of APCs to IFN-γ, IFN-αβ may prioritize the development of an antiviral IFN-αβ–type response to more effectively limit viral infections. The ability of IFN-αβ to suppress IFN-γ–type responses may also benefit the host by limiting collateral damage that might otherwise result from the rapid activation of macrophages and other APCs by IFN-γ. Indeed, IFN-β is commonly used to treat relapsing-remitting multiple sclerosis, an inflammatory autoimmune disease of the central nervous system (Hemmer et al., 2006; Borden et al., 2007). Likewise, IFN-αβ reduces disease severity in the murine multiple sclerosis model of experimental autoimmune encephalitis. Recent work shows the protective effect of IFN-αβ in experimental autoimmune encephalitis requires IFNAR1 expression on mouse myeloid cells (Prinz et al., 2008). In light of our findings, one may speculate that a key effect of IFN-αβ is to down-regulate IFNGR expression on myeloid cells, thereby reducing stimulation of autoimmune T cells and the consequences of IFN-γ produced by such T cells. Given that IFN-αβ does not reduce IFNGR expression in T cells, the integration of IFN-αβ and IFN-γ signals in T cells must entail distinct mechanisms.
IFNAR−/− mice have been shown to have heightened resistance to systemic L. monocytogenes infection, as judged by reduced bacterial burdens beginning within 3 or 4 d of systemic infection (Auerbuch et al., 2004; Carrero et al., 2004; O’Connell et al., 2004). L. monocytogenes–infected IFNAR−/− mice also produce lower amounts of IL-10 (perhaps because of increased splenocyte apoptosis) and increased production of IL-12 and TNF when compared with control animals (Auerbuch et al., 2004; Carrero et al., 2006). We propose that there may be a common mechanistic basis for such increased resistance. Given our data and previous results that IFN-γ enhances TNF production and suppresses IL-10 production by macrophages (Chomarat et al., 1993; Bundschuh et al., 1997; Déry and Bissonnette, 1999), we propose that the increased resistance of IFNAR−/− to L. monocytogenes is a result of their failure to down-regulate the IFNGR. Consistent with this model, we show that APCs are more highly activated after infection of IFNAR−/− mice and that these mice more efficiently limit bacterial replication at early times after infection. Indeed, both this increased APC activation and increased resistance to infection are completely abrogated by depletion of IFN-γ. A potential alternative explanation of our findings is that the increased IL-10 in the WT, but not IFNAR1−/−, mice suppresses the production of IFN-γ. Indeed, IL-10 suppresses IFN-γ production induced by treatment of cultured splenocytes from SCID mice with killed L. monocytogenes (Tripp et al., 1993). However, it was previously shown, and our findings confirm, that sera of both control and IFNAR−/− mice infected with live L. monocytogenes contain similar amounts of IFN-γ (Auerbuch et al., 2004). Thus, we favor the interpretation that IFN-αβ production in WT mice impairs responsiveness of APCs to IFN-γ and, thus, the host’s ability to limit bacterial replication and dissemination.
Recently, IFNAR−/− mice have also been shown to resist infection with several additional pathogenic bacteria (Stanley et al., 2007; Qiu et al., 2008; Martin et al., 2009; Shahangian et al., 2009). Some of these bacteria, such as M. tuberculosis and Chlamydia trachomatis, are known to suppress cellular responses to IFN-γ (Belland et al., 2003; Kincaid and Ernst, 2003; Pai et al., 2003). It thus appears likely that the mechanism for antagonistic cross talk between IFN-αβ and IFN-γ that we describe in this paper also impacts susceptibility to these other pathogenic bacteria. Additional understanding of the mechanisms regulating IFNGR down-regulation by IFN-αβ may lead to improved treatments for a variety of infectious and inflammatory diseases.
MATERIALS AND METHODS
Mice.
IFNAR−/−, IL-6−/−, IFNGR−/−, and IFN-γ−/− mice were crossed to C57BL/6J (The Jackson Laboratory) for >10 generations. B6.IFNAR1−/− mice were originally obtained from DA. Portnoy (University of California, Berkeley, Berkeley, CA). STAT1−/− and isogenic 129/Sv mice were obtained from Taconic. Mice were housed in the National Jewish Health Biological Resource Center. All studies were approved by the National Jewish Health Institutional Animal Care and Use Committee.
Mouse infections.
Female mice between 8 and 10 wk of age were used for all in vivo experiments. Mice were infected (tail vein) with 0.5–2 × 104 cfu of log-phase mouse passaged L. monocytogenes strain 10403S. 24–96 h later, spleens and livers were harvested for analysis. Spleens were treated with 0.3% collagenase type IV (Worthington Biochemical Corporation) to release phagocytic and adherent cell populations then processed into single cell suspensions for staining and flow cytometry. Bacterial CFUs in infected tissues were determined by dilution plating as previously described (Humann et al., 2007).
Macrophages and cell lines.
To culture BMM, cells were flushed from both femurs of mice and cultured for 6 d in BM macrophage media (DMEM supplemented with 10% FBS, 1% sodium pyruvate, 1% L-glutamine, 1% penicillin/streptomycin, 2-mercaptoethanol, and 10% L-cell conditioned media). Fresh media was added at day 3 and BMMs were used for experiments on day 7. RAW264.7 macrophage cells stably transfected with a CIITApIV-luciferase construct (RAW-CIITApIV reporter cells) were provided by J. Ernst (New York University, New York, NY; Fortune et al., 2004). RAW.GAS6 reporter macrophages were generated in our laboratory by stable transfection with linearized pHTS-GAS (Biomyx Technology). RAW-CIITApIV and RAW-GAS.6 reporter cells were cultured with selection in 400 µg/ml neomycin or 100 µg/ml hygromycin, respectively.
Infection of cultured macrophages and immunoblotting.
BMM or RAW reporter cell lines were cultured overnight in antibiotic-free media and then infected with log-phase L. monocytogenes 10403S (wt Lm) or the isogenic ΔHly strain provided by D.A. Portnoy. Macrophages were infected at MOI = 1–5 for 30 min, washed three times in PBS, and given fresh media. At 1 h after infection, gentamicin was added to a concentration of 50 µg/ml to kill extracellular bacteria. For immunoblotting studies, control or infected macrophages were treated with 100 U/ml IFN-γ at 2 or 6 hpi. Cells were rinsed in PBS and lysed in SDS-PAGE buffer (62.5 mM Tris-HCL, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, and 0.01% bromophenol blue) supplemented with HALT phosphatase and protease inhibitors (Thermo Fisher Scientific). Lysates were separated by SDS-PAGE and immunoblotted with rabbit anti-pY701 STAT1, total STAT1, or mouse anti-actin using commercial antibodies (Cell Signaling Technology and Millipore) followed by secondary HRP-labeled anti–rabbit and anti–mouse (Thermo Fisher Scientific).
Luciferase assay.
Reporter cells were plated at 2 × 106 per well in 6-well plates and mock-infected or infected with WT or ΔHly L. monocytogenes. At 2 hpi, the culture media was replaced with fresh media containing 50 µg/ml gentamicin plus 0 or 100 U/ml of recombinant mouse IFN-γ (Invitrogen). Lysates were harvested at 10 hpi using lysis buffer from the Enhanced Luciferase Assay kit (BD) and frozen at −20°C. Luminescence was measured using injectors and kit reagents on a Synergy 2 reader with injectors (BioTek).
Flow cytometry.
Collagenase-treated splenocytes were incubated 1 min in ACK lysis buffer to lyse red blood cells then pelleted (Humann et al., 2007). BMMs were lifted from culture dishes with Nozyme (Specialty Media) and pelleted. Fc receptors were blocked before staining using supernatant from hybridoma 2.4G2 (rat anti-CD16/32). Surface staining used PBS/1% FCS/0.01% NaN3. Intracellular staining used Cytofix/Cytoperm solutions (BD). To detect MHCII and IFNGR1 expression, cells were stained with biotinylated antibodies to pan-MHCII (eBioscience; clone M5/114.15.2) or IFNGR1/CD119 (BD), followed by streptavidin-APC secondary antibody. Directly conjugated M5/114.15.2 (BioLegend) was used for some experiments. To detect IFNGR2, we used a three-step staining procedure with hamster anti–mouse IFNGR2 (Abcam; antibody 21570), followed by biotinylated goat anti–hamster IgG (eBioscience) and streptavidin-APC (eBioscience). To compare the effects of various treatments on receptor surface expression levels, the mean channel fluorescence intensities (MFIs) for each of three infected or treated samples per group were normalized to mean control MFI for the same receptor using the following formula: relative surface staining = (MFI treated)/(mean MFI control). All graphs depict the mean of these calculations plus SE. For statistical analyses, we compared the raw MFIs for each of at least three control and three treated samples. The following antibodies were used to identify splenocyte populations: NK1.1-PE (PK136), Ly6G-PE (1A8), CD8α-PE (53–6.7), CD4-FITC (RM4-5), and B220-PECy5 (TÜ116; BD); and F480-PECy5 (BM8), CD11b-PECy5 (M1/70), CD19-PE (MB19-1), CD3-PECy5 (4B12), and CD11c-PE or Pacific Blue (N418; eBioscience). Stained cells were run on FACSCalibur (BD) or DakoCytomation CyAn (Dako) machines and analyzed using FlowJo software (Tree Star, Inc.).
Affymetrix analysis and RT-PCR.
Total RNA was isolated from C57BL/6 BMM 10 h after mock or wt Lm infection using the RNeasy kit with DNase treatment (QIAGEN). For each of two to three independent infections, RNA was pooled from 3–6 wells of BMM and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 Arrays by the University of Colorado Cancer Center Gene Expression core. Intensity data from chips was normalized and probe sets given a p (present)-value by Genechip operating software were further analyzed. Raw microarray data are available from the National Center for Biotechnology Information Gene Expression Omnibus, Accession no. GSE19374. To identify differentially expressed genes (P < 0.05 by ANOVA), normalized hybridization data were further analyzed using GeneSifter software (Geospiza). For RT-PCR, complementary DNA synthesis was conducted with 1 µg of total RNA using Oligo(dT) primers (Promega). Semiquantitative RT-PCR was done as previously described (Humann et al., 2007) using oligonucleotide primers 5′-GAGACTGCATGCAGGCAGCA-3′ and 5′-GGTCGGCATCACTGTTAAGGA-3′ for C2ta-pIV. Real-time quantitative PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems) and an ABI PRISM 7700 Sequence detector. Conditions for amplification of the target sequences were the following: 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C. Efficiency of amplification with each primer set was confirmed in control experiments. All samples were run in triplicate. Commercial oligonucleotide primer sets from Applied Biosystems were used to quantify ifngr1 and ifngr2 transcripts. The ifngr1 or ifngr2 transcript abundance in each sample was normalized to gapdh. The fold change in expression was calculated using 2−ΔΔCt method.
Supernatant transfers.
Culture supernatants from mock- or L. monocytogenes–infected donor BMMs were harvested at 8 hpi and centrifuged at 500 g for 5 min. Supernatants were sterile filtered through a 0.2-µm syringe filter and frozen at −20°C. Donor BMMs were stained for IFNGR1 to confirm infection-induced down-regulation. Conditioned supernatants were thawed and 1 ml was used to replace media for 106 uninfected recipient BMM or RAW-CIITApIV macrophages in 12-well plates. Recipient cells were harvested for analysis of IFNGR surface expression at 8 h or luciferase activity at 6 h.
TLR and cytokine stimulations.
Uninfected BMMs were treated with the indicated TLR agonists and cytokines for 8 h before analysis. CpG, scrambled CpG, and ultrapure LPS (InvivoGen) were used at 1 µM (CpG) and 10 ng/ml (LPS). Poly I:C (GE Healthcare) was used at 10 µg/ml. Pam3Cys was a gift from R. Kedl (Colorado University, Denver, CO) and used at 1 µg/ml. Recombinant mouse IL-6, IL-10, and IL-28 (eBioscience) were used at respective final concentrations of 0.01, 0.3, and 0.2 ng/ml. Recombinant mouse IFN-β (R&D Systems) was used at 100 U/ml.
Statistical analysis.
All experiments were repeated at least three times. Asterisks (*) in the figures indicate differences deemed significant (P < 0.05) by a two-tailed Student’s t test or the Mann-Whitney test. All error bars in graphs indicate SEM for three samples per experimental group.
Online supplemental material.
Fig. S1 illustrates control stains for IFNGR1 on WT and IFNGR1−/− BMM, demonstrating the specificity of the staining procedures. Fig. S2 depicts the raw unmanipulated MFIs for IFNGR1, IFNGR2, and CD11b staining in a representative experiment. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091746/DC1.
Acknowledgments
The authors thank David Riches, Yosef Refaeli, Laurent Gapin, and Caroline Cole for scientific discussions and Caroline Cole and Terry Potter for reading of the manuscript.
This work was funded by National Institutes of Health grant AI-065638 and a developmental project through the Rocky Mountain RCE (AI-065357), both to L.L. Lenz. J. Humann received additional support from National Institutes of Health training grants AI075-05 and AI52066-06.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BMM
- BM-derived macrophages
- GAS
- IFN-γ–activated sequence
- hpi
- h post infection
- IFNAR
- IFN-αβ receptor
- IFNGR
- IFN-γ receptor
- JAK
- Janus kinase
- LLO
- listeriolysin O
- MFI
- mean fluorescence intensity
- MOI
- multiplicity of infection
- ODN
- oligodeoxynucleotide
- STAT
- signal transducer and activator of transcription
- wt Lm
- WT L. monocytogenes
References
- Auerbuch V., Brockstedt D.G., Meyer-Morse N., O’Riordan M., Portnoy D.A. 2004. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200:527–533 10.1084/jem.20040976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R.J., Nelson D.E., Virok D., Crane D.D., Hogan D., Sturdevant D., Beatty W.L., Caldwell H.D. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA. 100:15971–15976 10.1073/pnas.2535394100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden E.C., Sen G.C., Uze G., Silverman R.H., Ransohoff R.M., Foster G.R., Stark G.R. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975–990 10.1038/nrd2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier N.A., Schreiber R.D. 1985. Requirement of endogenous interferon-gamma production for resolution of Listeria monocytogenes infection. Proc. Natl. Acad. Sci. USA. 82:7404–7408 10.1073/pnas.82.21.7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundschuh D.S., Barsig J., Hartung T., Randow F., Döcke W.D., Volk H.D., Wendel A. 1997. Granulocyte-macrophage colony-stimulating factor and IFN-gamma restore the systemic TNF-alpha response to endotoxin in lipopolysaccharide-desensitized mice. J. Immunol. 158:2862–2871 [PubMed] [Google Scholar]
- Carrero J.A., Calderon B., Unanue E.R. 2004. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 200:535–540 10.1084/jem.20040769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero J.A., Calderon B., Unanue E.R. 2006. Lymphocytes are detrimental during the early innate immune response against Listeria monocytogenes. J. Exp. Med. 203:933–940 10.1084/jem.20060045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomarat P., Rissoan M.C., Banchereau J., Miossec P. 1993. Interferon γ inhibits interleukin 10 production by monocytes. J. Exp. Med. 177:523–527 10.1084/jem.177.2.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton D.K., Pitts-Meek S., Keshav S., Figari I.S., Bradley A., Stewart T.A. 1993. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 259:1739–1742 10.1126/science.8456300 [DOI] [PubMed] [Google Scholar]
- Déry R.E., Bissonnette E.Y. 1999. IFN-gamma potentiates the release of TNF-alpha and MIP-1alpha by alveolar macrophages during allergic reactions. Am. J. Respir. Cell Mol. Biol. 20:407–412 [DOI] [PubMed] [Google Scholar]
- Dikopoulos N., Bertoletti A., Kröger A., Hauser H., Schirmbeck R., Reimann J. 2005. Type I IFN negatively regulates CD8+ T cell responses through IL-10-producing CD4+ T regulatory 1 cells. J. Immunol. 174:99–109 [DOI] [PubMed] [Google Scholar]
- Donnelly R.P., Sheikh F., Kotenko S.V., Dickensheets H. 2004. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J. Leukoc. Biol. 76:314–321 10.1189/jlb.0204117 [DOI] [PubMed] [Google Scholar]
- Flannagan R.S., Cosío G., Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7:355–366 10.1038/nrmicro2128 [DOI] [PubMed] [Google Scholar]
- Fortune S.M., Solache A., Jaeger A., Hill P.J., Belisle J.T., Bloom B.R., Rubin E.J., Ernst J.D. 2004. Mycobacterium tuberculosis inhibits macrophage responses to IFN-gamma through myeloid differentiation factor 88-dependent and -independent mechanisms. J. Immunol. 172:6272–6280 [DOI] [PubMed] [Google Scholar]
- Frank D.A., Mahajan S., Ritz J. 1999. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat. Med. 5:444–447 10.1038/7445 [DOI] [PubMed] [Google Scholar]
- Gordon S. 2003. Alternative activation of macrophages. Nat. Rev. Immunol. 3:23–35 10.1038/nri978 [DOI] [PubMed] [Google Scholar]
- Harty J.T., Badovinac V.P. 2008. Shaping and reshaping CD8+ T-cell memory. Nat. Rev. Immunol. 8:107–119 10.1038/nri2251 [DOI] [PubMed] [Google Scholar]
- Hemmer B., Nessler S., Zhou D., Kieseier B., Hartung H.P. 2006. Immunopathogenesis and immunotherapy of multiple sclerosis. Nat. Clin. Pract. Neurol. 2:201–211 10.1038/ncpneuro0154 [DOI] [PubMed] [Google Scholar]
- Humann J., Bjordahl R., Andreasen K., Lenz L.L. 2007. Expression of the p60 autolysin enhances NK cell activation and is required for listeria monocytogenes expansion in IFN-gamma-responsive mice. J. Immunol. 178:2407–2414 [DOI] [PubMed] [Google Scholar]
- Inaba K., Kitaura M., Kato T., Watanabe Y., Kawade Y., Muramatsu S. 1986. Contrasting effect of α/β- and γ-interferons on expression of macrophage Ia antigens. J. Exp. Med. 163:1030–1035 10.1084/jem.163.4.1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.J., Liang H.E., Reizis B., Locksley R.M. 2008. Regulation of hierarchical clustering and activation of innate immune cells by dendritic cells. Immunity. 29:819–833 10.1016/j.immuni.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid E.Z., Ernst J.D. 2003. Mycobacterium tuberculosis exerts gene-selective inhibition of transcriptional responses to IFN-gamma without inhibiting STAT1 function. J. Immunol. 171:2042–2049 [DOI] [PubMed] [Google Scholar]
- Leber J.H., Crimmins G.T., Raghavan S., Meyer-Morse N.P., Cox J.S., Portnoy D.A. 2008. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 4:e6 10.1371/journal.ppat.0040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling P.D., Warren M.K., Vogel S.N. 1985. Antagonistic effect of interferon-beta on the interferon-gamma-induced expression of Ia antigen in murine macrophages. J. Immunol. 135:1857–1863 [PubMed] [Google Scholar]
- Martin F.J., Gomez M.I., Wetzel D.M., Memmi G., O’Seaghdha M., Soong G., Schindler C., Prince A. 2009. Staphylococcus aureus activates type I IFN signaling in mice and humans through the Xr repeated sequences of protein A. J. Clin. Invest. 119:1931–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. 2007. The JAK-STAT signaling pathway: input and output integration. J. Immunol. 178:2623–2629 [DOI] [PubMed] [Google Scholar]
- Nagabhushanam V., Solache A., Ting L.M., Escaron C.J., Zhang J.Y., Ernst J.D. 2003. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 171:4750–4757 [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Saha S.K., Vaidya S.A., Bruhn K.W., Miranda G.A., Zarnegar B., Perry A.K., Nguyen B.O., Lane T.F., Taniguchi T., et al. 2004. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J. Exp. Med. 200:437–445 10.1084/jem.20040712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan M., Yi C.H., Gonzales R., Lee K.D., Portnoy D.A. 2002. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc. Natl. Acad. Sci. USA. 99:13861–13866 10.1073/pnas.202476699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai R.K., Convery M., Hamilton T.A., Boom W.H., Harding C.V. 2003. Inhibition of IFN-gamma-induced class II transactivator expression by a 19-kDa lipoprotein from Mycobacterium tuberculosis: a potential mechanism for immune evasion. J. Immunol. 171:175–184 [DOI] [PubMed] [Google Scholar]
- Pieters J. 2008. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 3:399–407 10.1016/j.chom.2008.05.006 [DOI] [PubMed] [Google Scholar]
- Platanias L.C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375–386 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- Prinz M., Schmidt H., Mildner A., Knobeloch K.P., Hanisch U.K., Raasch J., Merkler D., Detje C., Gutcher I., Mages J., et al. 2008. Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity. 28:675–686 10.1016/j.immuni.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Qiu H., Fan Y., Joyee A.G., Wang S., Han X., Bai H., Jiao L., Van Rooijen N., Yang X. 2008. Type I IFNs enhance susceptibility to Chlamydia muridarum lung infection by enhancing apoptosis of local macrophages. J. Immunol. 181:2092–2102 [DOI] [PubMed] [Google Scholar]
- Ray K., Marteyn B., Sansonetti P.J., Tang C.M. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7:333–340 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- Reith W., LeibundGut-Landmann S., Waldburger J.M. 2005. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5:793–806 10.1038/nri1708 [DOI] [PubMed] [Google Scholar]
- Shahangian A., Chow E.K., Tian X., Kang J.R., Ghaffari A., Liu S.Y., Belperio J.A., Cheng G., Deng J.C. 2009. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119:1910–1920 10.1172/JCI35412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley S.A., Johndrow J.E., Manzanillo P., Cox J.S. 2007. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 178:3143–3152 [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Zhang Z., Fournier A., Tan Y.H. 1985. Characterization of human beta-interferon-binding sites on human cells. J. Biol. Chem. 260:563–567 [PubMed] [Google Scholar]
- Tripp C.S., Wolf S.F., Unanue E.R. 1993. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc. Natl. Acad. Sci. USA. 90:3725–3729 10.1073/pnas.90.8.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R., Murray H.W., Nathan C.F. 1988. Agonist and antagonist effects of interferon α and β on activation of human macrophages. Two classes of interferon gamma receptors and blockade of the high-affinity sites by interferon alpha or beta. J. Exp. Med. 167:1171–1185 10.1084/jem.167.3.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz L.A., Shen H. 2007. Innate and adaptive immune responses to Listeria monocytogenes: a short overview. Microbes Infect. 9:1208–1215 10.1016/j.micinf.2007.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]