Abstract
A population of cells that expresses the NK cell receptor NKp46 and produces interleukin (IL)-22 have recently attracted considerable attention. The identity of these cells is still the subject of speculation, being variably defined as a novel NK cell subset or as a population containing conventional NK (cNK) cell precursors. In this issue, two studies shed light on this conundrum, demonstrating that NKp46+ IL-22+ cells and cNK cells belong to distinct lineages.
NK cell receptors are expressed on a wide variety of lymphocytes, including CD1-restricted T cells (NKT), memory CTLs, γδ T cells, and lymphoid tissue initiator cells (Vivier and Anfossi, 2004; Veiga-Fernandes et al., 2007; Randall et al., 2008). Although NK cell receptors are generally defined by their functional capacity, their expression on different cell types may lead to different functional outputs caused by differential signal integration within each cell type. Therefore, defining cNK cells and NK cell precursors has been problematic, partly because of the disparity between NK cell receptor surface expression and NK function. The NK cell receptor NKp46 has been proposed as a phenotypic marker for NK cells, as it has been shown to be specifically expressed by NK1.1+ cNK cells across different mammalian species, but not on T cells (Walzer et al., 2007).
In this issue, two studies, one by Satoh-Takayama et al. and the other by Crellin et al., question the lineage of an NKp46+ population that also expresses the orphan nuclear hormone receptor RORc (also called RORγ) and secretes IL-22. Crellin et al. (2010) examine the phenotype and function of human NKp46+ cells at the clonal level, whereas Satoh-Takayama et al. (2010) use an elegant fate mapping strategy. Together, the studies show that NKp46+, RORc+ IL-22–producing cells are not cNK cell precursors, but rather a distinct lineage that is closely related to adult LTi-like cells.
Seeds of doubt in the NK family
Unlike T and B lymphocytes, which are defined by the expression of their canonical receptors TCR and BCR, respectively, the definition of other immune cell lineages requires careful use of both analytical and functional readouts because of the promiscuous expression of certain receptors on multiple cell types. This has led to significant controversy in the literature in assigning lineages to certain cell populations. Defining NK cell populations exemplifies this issue, as NK cell receptors are expressed on a large variety of hematopoietic cell types.
NK cells are a major component of the innate immune system, mediating killing of tumors and virus-infected cells. NK cells can also mount anamnestic responses, and thus presumably go beyond the conventional boundaries of innate and adaptive immunity (Cooper et al., 2009). Interestingly, IL7Rα+CD3−NK1.1+ cells have also been implicated in the neonatal phase of lymph node maturation (Coles et al., 2006).
NK cells are thought to differentiate from the common lymphoid progenitor that generates B and T lymphocytes. Interestingly, NK cell precursor activity has been found outside the bone marrow, suggesting that cNK cells may develop under the influence of local cues in the periphery. NK cell development is tightly controlled by IL-15, which is critical for their generation, differentiation, and survival (Kennedy et al., 2000). Defining cNK cells in the periphery is complicated because of their ability to differentiate into functionally distinct populations. In humans, CD56bright NK cells have an enhanced capacity for cytokine production, whereas CD56dim cells have more potential for cytotoxic killing. Similar populations can be found in mice based on their expression of CD27. Additionally, as seen with T helper subsets, NK cells can differentiate into NK1 and NK2 populations based on cytokine secretion profiles (Cooper et al., 2009). Despite the growing heterogeneity in NK populations, NKp46 is used to distinguish NK cells from other lymphoid populations across mammalian species (Walzer et al., 2007).
However, recent studies have described additional diversity within NKp46+ populations from the intestine and tonsils (Satoh-Takayama et al., 2008; Cella et al., 2009; Cupedo et al., 2009; Luci et al., 2009; Sanos et al., 2009). For example, mouse intestinal CD3−NKp46+ cells can be divided into two main subsets based on the expression of IL7Rα (CD127) and NK1.1 (Satoh-Takayama et al., 2008). Importantly, these subsets appear to mediate different functions because IL7Rα−NK1.1+ cells are similar to cNK cells, producing perforin and IFN-γ, whereas IL7Rα+NK1.1− cells lack these effector functions. Instead, they express IL-22 and RORc (Satoh-Takayama et al., 2008; Luci et al., 2009; Sanos et al., 2009). Interestingly, cells with similar characteristics to murine NKp46+IL7Rα+NK1.1− cells have also been described in human tonsils, intestinal lamina propria, and Peyer’s patches (Cella et al., 2009; Cupedo et al., 2009).
The observation that NKp46+IL7Rα+NK1.1− also express RORc raises questions about their pedigree. RORc is involved in a variety of different processes, including thymocyte survival, differentiation of IL-17–producing T cells, and development of LTi cells, but it is dispensable for the development of cNK cells (Sun et al., 2000; Ivanov et al., 2006; Satoh-Takayama et al., 2008). Thus, these cells may express a putative universal NK marker, but they also express a factor shown to be essential for the development of different cell lineages. Given the promiscuity of surface molecule expression, accurate classification of NK lineages may need to rely more on transcription factor expression patterns. This strategy has been instrumental for dissecting T cell lineages. For example, differential expression of the transcription factors Foxp3, Gata-3, T-bet, and RORc has, rather reliably so far, identified T helper cell subsets that could not be distinguished through the expression of surface markers alone (Dong and Flavell, 2000).
A putative missing link
Although cNK cells have been studied for many years, the role of LTi cells in the immune system has been addressed only relatively recently. It is commonly accepted that development of secondary lymphoid tissues depends on productive interactions between hematopoietic lymphoid tissue inducer (LTi) cells and stroma lymphoid tissue organizer cells (Veiga-Fernandes et al., 2007; Randall et al., 2008). LTi cells were first characterized by Saxer in 1896 as wandering lymphocytes found in the early stages of developing lymph node anlagen (Sabin, 1913). The current common view is that LTi cells express RORc, IL-7Rα, lymphotoxin α1β2, and, in mice, CD4 (Randall et al., 2008). Despite the absence of lineage marker expression on these cells, experimental evidence has suggested that LTi cells may be developmentally related to conventional NK cells (Mebius et al., 1997). For instance, in the absence of helix–loop–helix inhibitor Id2, both LTi cells and NK cells fail to develop, as E2A-encoded E proteins repress the development of both cell types (Yokota et al., 1999; Boos et al., 2007). Furthermore, NK1.1+ cells can develop in in vitro cultures of fetal LTi cells (Mebius et al., 1997). However, despite being developmentally related, it is clear that LTi and conventional NK cells belong to distinct lineages. Thus, although RORc is required for LTi development, it is dispensable for the development of NK cells (Sun et al., 2000; Satoh-Takayama et al., 2008). Conversely, the transcription factor E4BP4 is absolutely required for the development of conventional NK cells, but LTi development is seemingly unaffected in E4BP4-deficient mice, as revealed by normal development of secondary lymphoid organs (Gascoyne et al., 2009). Finally, although cNK cells are highly dependent on IL-15, LTi development is not affected by the absence of this cytokine (Kennedy et al., 2000; Vosshenrich et al., 2005; Satoh-Takayama et al., 2008).
The discovery of RORc+NKp46+IL7Rα+NK1.1− cells and their human equivalents raised the possibility that this cell type could be the missing developmental link between LTi and NK cells. In fact, it was shown that human CD56− IL7Rα+ LTi-like cells develop into CD56+IL7Rα+ cells after in vitro culture (Cupedo et al., 2009). Given their immature phenotype, it is possible that the population of NKp46+IL7Rα+ cells could contain NK cell precursor activity, which can differentiate in a local environment and give rise to specialized NK subsets (Colonna, 2009).
Unmasking the unconventional NKp46 cells
In this issue, two studies address the question of whether mouse and human NKp46+ cells are NK precursor cells or a distinct lineage. Because Id2 is essential for LTi and NK cell development, the study by Satoh-Takayama et al. (2010) starts by revisiting the impact of Id2 ablation on enteric NKp46+ cells (Yokota et al., 1999). Analysis of Id2-deficient mice revealed that, indeed, normal development of NKp46+ subsets required Id2, strengthening the notion that, at least developmentally, these cells are related to LTi and cNK cells. To further explore the relationship between enteric NKp46+ cells and their putative relatives, LTi and cNK cells, the authors examined mice with altered expression of IL-15 or IL-7. Analysis of enteric NKp46+ cells in mice that overexpress IL-15 in the intestine revealed that although NKp46+IL7Rα+NK1.1− homeostasis was not perturbed, the numbers of NKp46+IL7Rα−NK1.1+ cells were significantly increased in these mice. Conversely, although the homeostasis of NKp46+IL7Rα+NK1.1− cells was strongly impaired by an absence of IL-7, NKp46+IL7Rα−NK1.1+ cNK cell numbers were not affected. Thus, NK1+IL7Rα− and NK1−IL7Rα+ subsets are seemingly two separate cell subsets with respect to IL-7 and IL-15 dependency (Fig. 1).
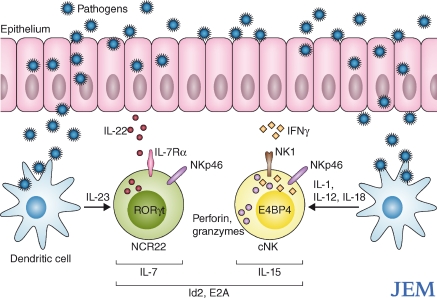
Figure 1.
NCR22 and conventional NK cells have different function and ontogeny. NCR22 and cNK cells home to mucosal sites and mucosal-associated lymphoid tissues. Upon interaction with microbial components, mucosal dendritic cells produce IL-23, which stimulates NCR22 cells to secrete IL-22, which is believed to protect the mucosa and control inflammation. In response to dendritic cell-derived cytokines such as IL-1, IL-12, and IL-18, cNK cells produce IFN-γ, perforin and granzymes, which target pathogen infected cells. In the absence of helix-loop-helix inhibitor Id2, both NCR22 and conventional NK cells fail to develop. However, the development of these cell types differs in its dependency on IL-7, IL-15, Rorγt and E4BP4.
These findings are consistent with the study by Crellin et al. (2010), which initially examined whether human LTi-like CD56−IL7Rα+ or CD56+IL7Rα+ cells convert to cNK cells upon in vitro expansion. Using bulk cultures and clonal analysis, the authors show that CD56−IL7Rα+ LTi-like cells differentiate in to CD56+IL7Rα+ cells, but importantly, both subsets maintain RORc expression and do not convert to cNK cells. Interestingly, in vitro–differentiated CD56+IL7Rα+ cells acquire poor cytolytic activity in comparison to cNK cells. These observations support the idea that LTi-like cells and CD56+IL7Rα+ cells belong to a lineage distinct from cNK cells.
Satoh-Takayama et al. (2010) provide further support for this concept with elegant in vivo fate mapping experiments. BAC transgenic mice that express Cre recombinase under control of the Rorc promoter were bred to mice that have the eYFP gene knocked into the ubiquitously expressed ROSA26 locus, which is preceded by a triple polyadenylation signal flanked by two loxP sites. In these mice, cells that express Rorc at some point during development maintain eYFP expression from that point onward. The authors found that although NKp46+IL7Rα+ cells in the intestine expressed eYFP, no eYFP expression was observed in NKp46+IL7Rα− enteric cells or cNK cells found in lymphoid organs.
Collectively, these two studies demonstrate that despite expressing NK receptors, NKp46+IL7Rα+ cells are a distinct lineage and not precursors of cNK cells. In general, the naming of cells and receptors brings with it welcome simplicity in an ever complex world of cell types, but it also brings a burden of expectation. Thus, the expression of NK receptors does not mean a cell has NK function. Satoh-Takayama et al. (2010) have termed these recently identified NKp46+RORc+IL-22+ cells “NCR22s,” succinctly highlighting the key functional features of these cells: natural cytotoxicity receptor expression, RORc expression, and IL-22 production, which differentiates them from cNK cells in both phenotype and function. (Fig. 1)
Future perspectives and unanswered questions
The development and maintenance of specialized lymphoid stromal cells is essential for the generation of protective immune responses. Although the function of LTi lineage in lymphoid organ development has been well established, their role in adult tissue maintenance is less clear. The discoveries that IL-22 production by NCR22 cells may mediate mucosal immune defense, and adult LTi-like cells contribute to stromal regeneration after LCMV infection, suggest a significant role for the adult LTi lineage in the maintenance and regeneration of adult tissues (Satoh-Takayama et al., 2008; Scandella et al., 2008). However, many interesting questions remain regarding the function of NCR22 cells and the role of IL-22 within specific tissue microenvironments in development, homeostasis, and disease.
Since Sykes and Ballas first showed expression of NK cell receptors on T cells 20 yr ago (Ballas and Rasmussen, 1990; Sykes, 1990), the presence of NK cell receptors on unconventional lymphocytes has identified exciting new cell populations. NCR22s are the latest incarnation of this phenomenon.
References
- Ballas Z.K., Rasmussen W. 1990. NK1.1+ thymocytes. Adult murine CD4-, CD8- thymocytes contain an NK1.1+, CD3+, CD5hi, CD44hi, TCR-V beta 8+ subset. J. Immunol. 145:1039–1045 [PubMed] [Google Scholar]
- Boos M.D., Yokota Y., Eberl G., Kee B.L. 2007. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204:1119–1130 10.1084/jem.20061959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K., Doherty J.M., Mills J.C., Colonna M. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles M.C., Veiga-Fernandes H., Foster K.E., Norton T., Pagakis S.N., Seddon B., Kioussis D. 2006. Role of T and NK cells and IL7/IL7r interactions during neonatal maturation of lymph nodes. Proc. Natl. Acad. Sci. USA. 103:13457–13462 10.1073/pnas.0604183103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M. 2009. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity. 31:15–23 10.1016/j.immuni.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Colonna M., Yokoyama W.M. 2009. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 10:1103–1110 10.1038/embor.2009.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crellin N.K., Trifari S., Kaplan C.D., Cupedo T., Spits H. 2010. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J. Exp. Med. 207:■■■–■■■ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T., Crellin N.K., Papazian N., Rombouts E.J., Weijer K., Grogan J.L., Fibbe W.E., Cornelissen J.J., Spits H. 2009. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat. Immunol. 10:66–74 10.1038/ni.1668 [DOI] [PubMed] [Google Scholar]
- Dong C., Flavell R.A. 2000. Control of T helper cell differentiation—in search of master genes. Sci. STKE. 2000:pe1 10.1126/stke.2000.49.pe1 [DOI] [PubMed] [Google Scholar]
- Gascoyne D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H.J. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Ivanov I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., Littman D.R. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Kennedy M.K., Glaccum M., Brown S.N., Butz E.A., Viney J.L., Embers M., Matsuki N., Charrier K., Sedger L., Willis C.R., et al. 2000. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15–deficient mice. J. Exp. Med. 191:771–780 10.1084/jem.191.5.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci C., Reynders A., Ivanov I.I., Cognet C., Chiche L., Chasson L., Hardwigsen J., Anguiano E., Banchereau J., Chaussabel D., et al. 2009. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10:75–82 10.1038/ni.1681 [DOI] [PubMed] [Google Scholar]
- Mebius R.E., Rennert P., Weissman I.L. 1997. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 7:493–504 10.1016/S1074-7613(00)80371-4 [DOI] [PubMed] [Google Scholar]
- Randall T.D., Carragher D.M., Rangel-Moreno J. 2008. Development of secondary lymphoid organs. Annu. Rev. Immunol. 26:627–650 10.1146/annurev.immunol.26.021607.090257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin F.R. 1913. The origin and development of the lymphatic system. In Hospital Reports Monographs New Series No. V The Johns Hopkins Press; p. 65–70 [Google Scholar]
- Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. 2009. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10:83–91 10.1038/ni.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C.A.J., Di Santo J.P. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD32NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207:■■■–■■■ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandella E., Bolinger B., Lattmann E., Miller S., Favre S., Littman D.R., Finke D., Luther S.A., Junt T., Ludewig B. 2008. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat. Immunol. 9:667–675 10.1038/ni.1605 [DOI] [PubMed] [Google Scholar]
- Sun Z., Unutmaz D., Zou Y.R., Sunshine M.J., Pierani A., Brenner-Morton S., Mebius R.E., Littman D.R. 2000. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 288:2369–2373 10.1126/science.288.5475.2369 [DOI] [PubMed] [Google Scholar]
- Sykes M. 1990. Unusual T cell populations in adult murine bone marrow. Prevalence of CD3+CD4-CD8- and alpha beta TCR+NK1.1+ cells. J. Immunol. 145:3209–3215 [PubMed] [Google Scholar]
- Veiga-Fernandes H., Coles M.C., Foster K.E., Patel A., Williams A., Natarajan D., Barlow A., Pachnis V., Kioussis D. 2007. Tyrosine kinase receptor RET is a key regulator of Peyer’s patch organogenesis. Nature. 446:547–551 10.1038/nature05597 [DOI] [PubMed] [Google Scholar]
- Vivier E., Anfossi N. 2004. Inhibitory NK-cell receptors on T cells: witness of the past, actors of the future. Nat. Rev. Immunol. 4:190–198 10.1038/nri1306 [DOI] [PubMed] [Google Scholar]
- Vosshenrich C.A., Ranson T., Samson S.I., Corcuff E., Colucci F., Rosmaraki E.E., Di Santo J.P. 2005. Roles for common cytokine receptor gamma-chain-dependent cytokines in the generation, differentiation, and maturation of NK cell precursors and peripheral NK cells in vivo. J. Immunol. 174:1213–1221 [DOI] [PubMed] [Google Scholar]
- Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. 2007. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 104:3384–3389 10.1073/pnas.0609692104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 397:702–706 10.1038/17812 [DOI] [PubMed] [Google Scholar]