Abstract
We previously reported a Vietnamese-American family with isolated autosomal dominant occipital cephalocele. Upon further neuroimaging studies, we have recharacterized this condition as autosomal dominant Dandy-Walker with occipital cephalocele (ADDWOC). A similar ADDWOC family from Brazil was also recently described. To determine the genetic etiology of ADDWOC, we performed genome-wide linkage analysis on members of the Vietnamese-American and Brazilian pedigrees. Linkage analysis of the Vietnamese-American family identified the ADDWOC causative locus on chromosome 2q36.1 with a multipoint parametric LOD score of 3.3, while haplotype analysis refined the locus to 1.1 Mb. Sequencing of the five known genes in this locus did not identify any protein-altering mutations. However, a terminal deletion of chromosome 2 in a patient with an isolated case of Dandy-Walker malformation also encompassed the 2q36.1 chromosomal region. The Brazilian pedigree did not show linkage to this 2q36.1 region. Taken together, these results demonstrate a locus for ADDWOC on 2q36.1 and also suggest locus heterogeneity for ADDWOC.
Introduction
We recently reported a Vietnamese-American kindred with isolated autosomal dominant occipital cephalocele over three generations (Bassuk et al. 2004). While the original report of this kindred focused on the atretic cephalocele, our subsequent review of brain imaging in the proband and his affected second cousin showed Dandy-Walker malformation in addition to the cephalocele. Three other families with autosomal dominant Dandy-Walker malformation and occipital cephaloceles (ADDWOC) have been reported from Brazil, Spain, and China (Carvalho et al. 2006; Martinez-Lage et al. 1996; Zhao et al. 2007), signifying the worldwide occurrence of this heritable disorder. Here, we further characterize the phenotype of the original Vietnamese-American kindred, map the locus to 2q36.1 by genome-wide linkage analysis, and describe an overlapping phenotype in a child with deletion of this region. We also demonstrate locus heterogeneity by excluding the Brazilian family from this locus.
Materials and methods
Patient data and DNA gathering
The ADDWOC pedigrees comprised a 3-generation Vietnamese-American family with 16 and a Brazilian family with 6 affected individuals. Positive diagnosis was made by the presence of one or a combination of an occipital cephalocele or occult Dandy-Walker or cerebellar changes, as described in the results section. We tried to minimize any bias in this process by utilizing the opinions of multiple experts in the field in a blinded fashion. For the Vietnamese-American family, the attending neurologist contacted 14 family members, including 13 affected individual used in the affected-only linkage analysis, and administered informed consent with the approval of the local institutional review board. The Brazilian family was recruited separately and their clinical phenotype has previously been described (Carvalho et al. 2006). Genomic DNA was extracted from blood or buccal samples collected from family members using Gentra Autopure LS (Qiagen). A Siemens Trio 3 Tesla MRI Scanner at the Northwestern University Magnetic Resonance facility was used for imaging studies.
Microarray SNP analysis and DNA sequencing
The Affymetrix GeneChip Human Mapping 10 K Array (Xba 131) and the GeneChip Instrument System (Hybridization Oven 640, Fluidics Station 450, Scanner 3000) were used for single nucleotide polymorphism (SNP) genotyping at the Genomics Core Facility of Northwestern University Center for Genetic Medicine following the standard protocol supplied by the manufacturer. Affymetrix GCOS and GDAS software were used for scanning the arrays and assigning genotype calls. All array call rates were above 95%. Gene sequencing involved the exons and flanking 100 bp intronic or 200 bp promoter regions of the proband DNA amplified using the Platinum Pfx PCR kit (Invitrogen). Primer sequences are available upon request. Purified PCR amplification products were electrophoresed on ABI 3100 and 3730 DNA sequencers by the Genomics Core Facility at Northwestern University. Novel heterozygous SNPs found during gene sequencing of the proband DNA were resequenced using the same primers in all affected individuals.
Genome-wide linkage analysis
Of the 11,560 total SNPs, 662 were excluded from further analysis due to lack of positional information from Affymetrix or location on the X chromosome, as this condition does not follow an X-linked pattern of inheritance. Additionally, 52 SNPs with Mendelian inconsistencies were identified using PedCheck (O'Connell and Weeks 1998) and excluded from further analysis. The remaining 10,846 SNPs were subjected to parametric multipoint linkage analysis assuming a fully penetrant autosomal dominant mode of inheritance, equal SNP allele frequencies, and a disease allele frequency estimated at 1/10,000. easyLINKAGE Plus(Hoffmann and Lindner 2005) v5.02 was used to prepare the data for linkage computation using Allegro 2.0 f (Gudbjartsson et al. 2005). The haplotype diagram was prepared using HaploPainter (Thiele and Nurnberg 2005).
CGH and FISH analysis
DNA samples from the proband and his maternal grandfather were subjected to comparative genomic hybridization (CGH) on a 1-Mb resolution genome-wide array CGH platform (Spectral Genomics, Inc.) using manufacturer's recommendation (Iafrate et al. 2004). G-banded karyotype analysis and fluorescent in situ hybridization (FISH) on metaphase chromosomes was performed by the Genetic Services Laboratories at the University of Chicago. BAC clones from the RPCL-11 human library were selected across 2 q. Genomic DNA from BAC clones was isolated using a DNA isolation kit (Qiagen, Valencia, CA, USA) and directly labeled with Spectrum Orange-dUTP (Vysis/Abbot, Abbot Park, IL, USA) or Spectrum Green-dUTP (Vysis/Abbott) using a standard nick-translation kit (Vysis/Abbott). Slide, probe preparation, and hybridization were performed using standard methods. Additional information is available upon request.
Results
Brain imaging features in the expanded Vietnamese-American family
While the initial study of the Vietnamese-American family focused on the occipital cephalocele present in seven family members (Fig. 1a), we have now reviewed the brain imaging studies in the two most severely affected family members (III-3 and III-8 in Fig. 2) in more detail. In addition to the cephalocele, III-3 and III-8 demonstrate Dandy-Walker malformation characterized by a small and upwardly rotated cerebellar vermis, marked cystic enlargement of the fourth ventricle, and greatly enlarged posterior fossa (Fig. 1b).
Fig. 1.
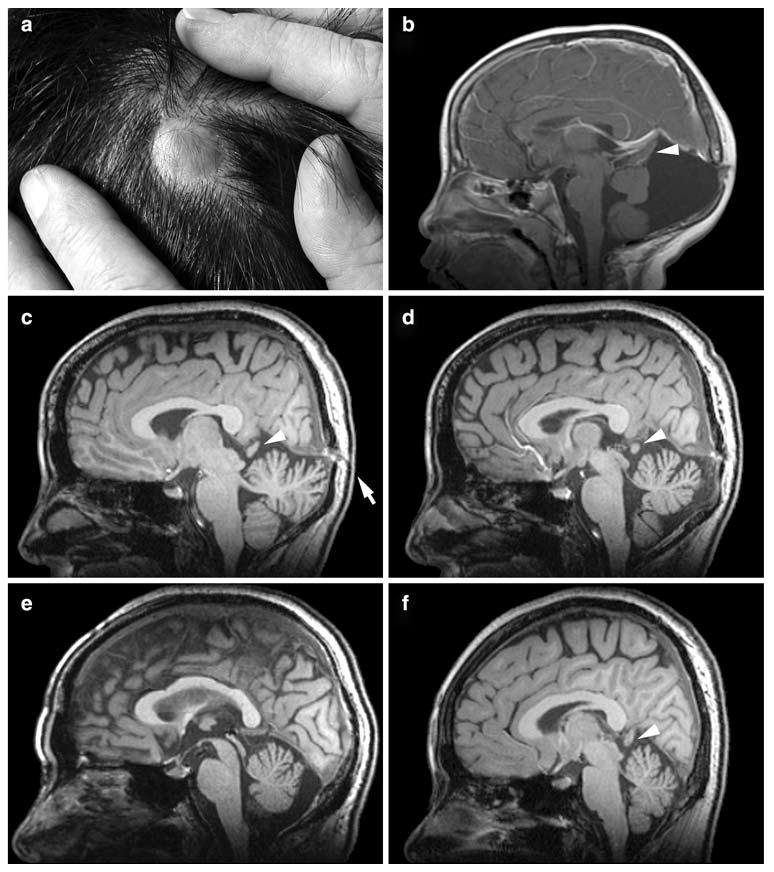
Representative occipital cephalocele (a) and MRI abnormalities (b–f) from the Vietnamese-American ADDWOC family. a Photograph of occipital cephalocele from individual II-8. b T1 post-contrast sagittal MRI of individual III-3. Note small and upwardly rotated cerebellar vermis and markedly enlarged fourth ventricle and posterior fossa. c Parasagittal T1 MRI of individual II-4, who was originally identified as an obligate carrier. Note skull defect and occult occipital cephalocele (arrow). d Mid-sagittal T1 MRI of individual II-4. Note cerebellar vermal hypoplasia and mildly enlarge fourth ventricle. e T1 sagittal MRI of individual I-2, who was originally identified as an obligate carrier. Note cerebellar vermal hypoplasia and mildly enlarged cisterna magna. f T1 sagittal MRI of individual II-2. Note mild cerebellar vermal hypoplasia. Arrowheads in b, c, d, and f point to the protrusion of occipital lobe through the tentorial notch
Fig. 2.
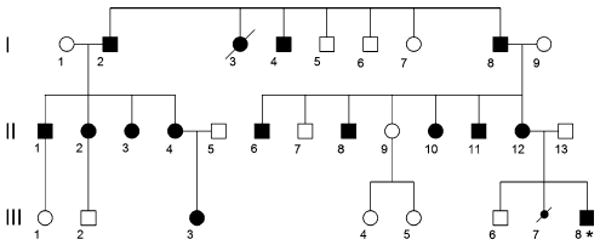
Pedigree of the Vietnamese-American ADDWOC family. Generation number is listed along the left margin, and individuals are numbered underneath each symbol. Thus, the proband (indicated by an asterisk) corresponds to individual III-8. Solid-shaded large symbols correspond to affected members. The smaller shaded symbol represents a spontaneously aborted fetus. Only the affected individuals (except I-3, I-4, and III-7) were included in the linkage analysis
We also obtained MRI studies in seven additional family members that lacked an obvious cephalocele including those we originally designated as obligate carriers for the cephalocele trait (I-2 and II-4). While II-4 lacked an obvious cephalocele, her MRI demonstrated an occult cephalocele in a parasagittal section (Fig. 1c) as well as cerebellar vermal hypoplasia with a mildly enlarged fourth ventricle in the mid-sagittal section (Fig. 1d). Similarly, I-2 demonstrated cerebellar vermal hypoplasia and a mildly enlarged cisterna magna (Fig. 1e). MRI studies of 4 other family members, II-1, II-2 (Fig. 1f), II-3, and II-6, also demonstrated mild to moderate cerebellar vermal hypoplasia. No anomalies were noted in the MRI study of II-7. While these family members did not display the full extent of posterior fossa developmental anomalies known as Dandy-Walker malformation, the cerebellar changes noted along with enlargement of the fluid spaces around the cerebellum are considered less severe forms of the same continuum of developmental anomalies. On the basis of these MRI findings in this family, we recharacterized this inherited condition as ADDWOC. Out of the total of 14 family members considered in this study, all except II-7 were included in our affected-only linkage analysis.
Interestingly, several family members (II-2, II-3, II-4, III-3) demonstrated mild protrusion of the occipital lobes through an apparently enlarged tentorial notch, which can be seen interposed between the splenium of the corpus callosum and the top of the vermis (arrowheads in Fig. 1). This anomaly resembles the dysplastic mesial occipital gyri seen in some patients with parietal foramina associated with mutations of ALX4 (Mavrogiannis et al. 2001; Reddy et al. 2000; Valente and Valente 2004) and suggests a subtle meningeal deficiency.
Genome-wide linkage analysis and sequencing
The genome-wide parametric LOD scores for this pedigree are shown in Fig. 3a. The most significant score was observed at a single locus spanning 1.5 cM on chromosome 2 (225.3–226.8 cM) with a LOD of 3.3. The maximum LOD score was obtained at rs951997 and rs1374536 (Fig. 3b). Haplotype analysis indicated that the co-segregating segment containing the ADDWOC locus is flanked proximally by rs724149 and distally by rs1371552 (Fig. 4), demarcating a 1.1 Mb region on chromosome 2q36.1 containing 5 known genes (Fig. 5). Sequencing the genes in this region revealed two previously reported SNPs, rs2166652 and rs2044537, and two novel polymorphisms, S1 and A1. The dbSNP annotated markers, rs2166652 and rs2044537, are located 100 and 173 bp upstream of FARSB exon 1. The novel S1 variation is a G to A transition at the third base position resulting in a synonymous change of the Ala36 residue of SGPP2, while the A1 variation is an A insertion/deletion in a polyA stretch 85 bp upstream of ACSL3 exon 4. Extending the linkage analysis to include the four additional polymorphisms discovered through sequencing did not further delineate the ADDWOC locus, though a LOD of 3.3 was extended to include A1 (Fig. 3b). No missense, nonsense, frameshift, or splice-site mutations were identified. Comparative genomic hybridization of I-8 and III-8 DNA on a 1 Mb resolution BAC array (Iafrate et al. 2004) failed to identify any significant gains or losses.
Fig. 3.
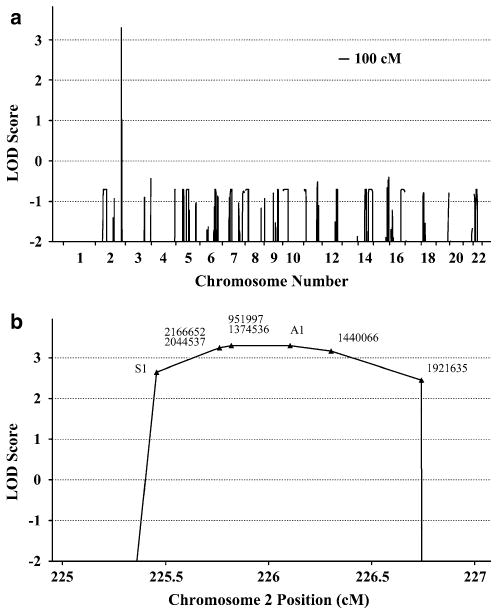
Genome-wide linkage analysis of the Vietnamese-American ADDWOC family. a Plot of multipoint parametric LOD scores across the autosomes demonstrates a single ADDWOC-linked locus on chromosome 2 with a significant LOD score of 3.30. b Higher magnification of the locus on chromosome 2 depicts the markers with significant LOD scores. Closed triangles are one or two nearby SNP markers with dbSNP rs numbers given next to them. S1 and A1 markers are novel SNPs in SGPP2 and ACSL3
Fig. 4.
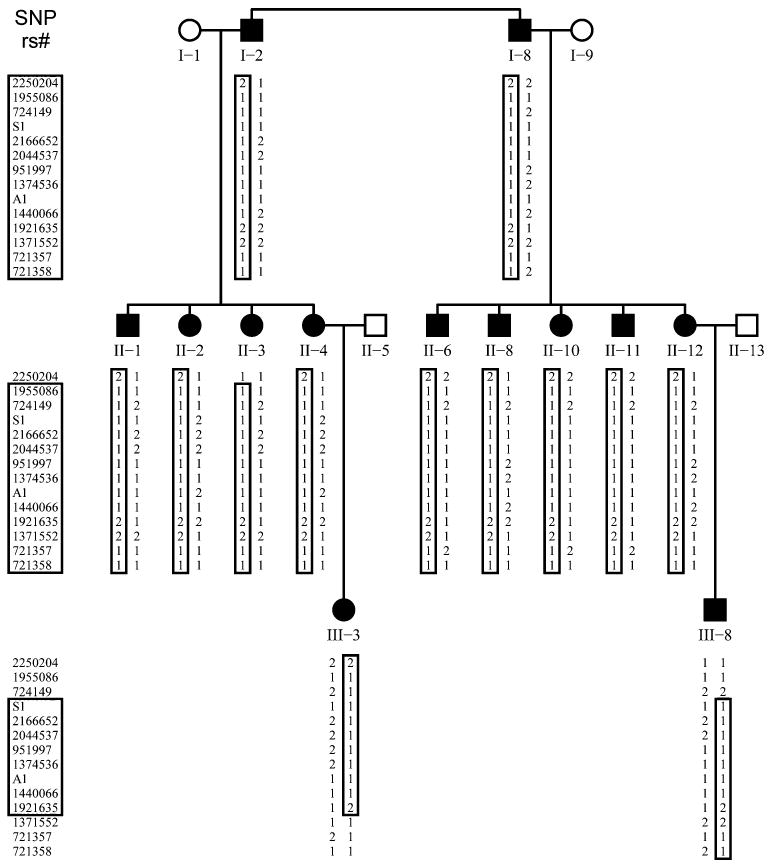
Haplotype analysis of the ADDWOC locus in the Vietnamese-American pedigree. SNP markers are listed along the left margin, and under each individual the corresponding alleles are listed. The two alleles for each SNP are represented by numbers 1 and 2. A1 allele 2 and S1 allele 2 are the newly discovered variants. The ancestral haplotype containing the ADDWOC affection locus is noted by the closed box. Chromosomal recombinations during meioses leading to individuals II-3, III-3, and III-8 narrowed the ADD-WOC haplotype, as noted by the smaller box in those individuals
Fig. 5.
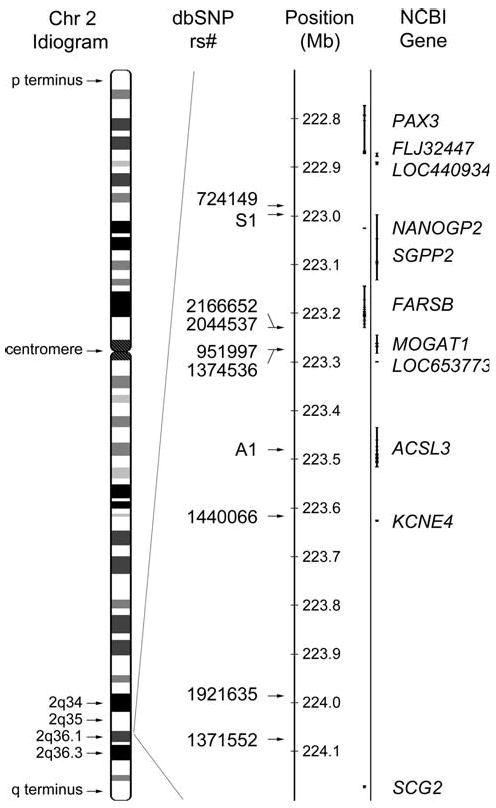
Features of the ADDWOC locus. Chromosome 2 idiogram is depicted on the left and the 2q36.1 region is magnified to its right. Positions of SNP markers and genes are marked along this region. There ADDWOC locus is limited by rs724149 proximally and rs1371552 distally. There are 5 identified genes and 2 pseudogenes (NANOGP2 and LOC653773) in the ADDWOC locus, and PAX3 is located 107 kb outside the locus. Positive and negative strand genes are depicted on the right and left, respectively
We also performed a genome-wide linkage analysis on the Brazilian family, which was genotyped using the same Affymetrix platform. No evidence for linkage was detected at the 2q36.1 locus (maximum LOD of −7.59 at rs1440066), nor at a single locus. Instead, multiple loci with LOD scores of 1.20 occurred across several chromosomes (data not shown). Taken together, the linkage results for the Vietnamese-American and Brazilian families demonstrate locus heterogeneity in ADDWOC.
Isolated Dandy-Walker patient with 2q36.1 deletion
We evaluated a 4-year-old boy with mild developmental delay who began walking at 14 months and uses about 50 words but no sentences at 4 years; formal intelligence testing has not been performed. His facial appearance was significant for a relatively prominent forehead, mildly downturned vermilion border of the upper lip, deep-set eyes and flat philtrum, and he has minimal high frequency hearing loss. His MRI shows mild cerebellar vermal hypoplasia and enlarged retrocerebellar fluid space (Fig. 6a). G-banded karyotype analysis revealed a small heterozygous deletion of distal 2q in 12 of 30 cells examined (Fig. 6b). FISH analysis confirmed a 20.5 Mb telomeric deletion encompassing the ADDWOC locus. The position of the deletion breakpoint was narrowed to a 0.2-Mb region of chromosome 2q36.1 between RP11-1080C21 and RP11-794I13 and 0.6 Mb centromeric to the ADDWOC locus. The karyotype is thus designated 46,XY,del(2)(q36.1→qter)[12]/46,XY[18]. Both parents had normal karyotypes.
Fig. 6.
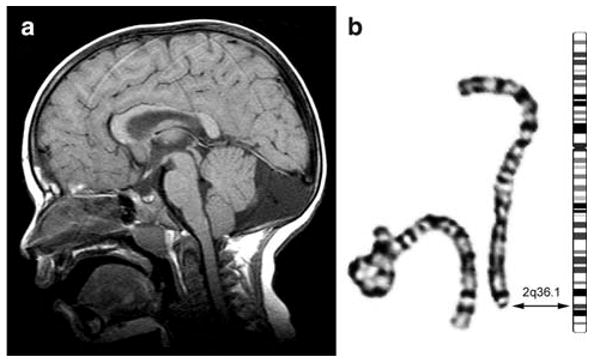
MRI of isolated Dandy-Walker patient with deletion overlapping ADDWOC locus. a T1 sagittal MRI demonstrates enlarged cisterna magna and cerebellar vermal hypoplasia. b G-banded partial karyotype of chromosome 2 homologs in this patient shows the normal homolog (left) and the deleted homolog (right) next to a chromosome 2 ideogram. The double arrow points to the deletion breakpoint in band 2q36.1 on both the idiogram and deleted chromosome 2. The formal karyotype is: 46,XY,del(2)(q36.1→qter) [12]/46,XY[18]
Discussion
The pathogenesis of Dandy-Walker malformation is unknown but certainly heterogeneous. Most patients are sporadic with an empiric sibling recurrence risk less than 5% for the isolated form of the disease (Murray et al. 1985). Families with autosomal dominant inheritance of typical Dandy-Walker malformation without an occipital cephalocele have not been reported, perhaps in part due to the occult nature of some of the weaker variants. In this paper; however, we cite several reports of families that feature a combination of Dandy-Walker malformation and occipital cephaloceles, which have recently been described.
The genetic causes of Dandy-Walker malformation and occipital cephaloceles are not well understood, although the last several years have seen some progress. The genes for several congenital anomaly syndromes associated with occipital cephaloceles have been reported; these include Knobloch, Joubert, Meckel-Gruber, Roberts, and Walker-Warburg syndromes (Beltran-Valero de Bernabe et al. 2002; Dixon-Salazar et al. 2004; Ferland et al. 2004; Kyttala et al. 2006; Parisi et al. 2004; Sertie et al. 2000; Smith et al. 2006; Valente et al. 2006; van Reeuwijk et al. 2005; Vega et al. 2005). Our studies in patients with Dandy-Walker malformation and overlapping chromosome deletions mapped the first two Dandy-Walker loci to 3q24 and 6p25, and identified ZIC1 and ZIC4 as the first causative genes for Dandy-Walker malformation (Descipio et al. 2005; Grinberg et al. 2004). However, no genes associated with the novel ADDWOC syndrome have been identified.
In the continuum of posterior fossa developmental anomalies, Dandy-Walker malformation is often associated with developmental delay, difficulty with balance, spasticity, poor fine-motor control, and seizures. Hearing and/or visual difficulties are also associated with poor intellectual development. Those presenting with milder variants in this continuum can, however, have mild or no symptoms, a normal neurological exam, and normal intellectual development (Hart et al. 1972; Tal et al. 1980). In the Vietnamese-American pedigree, only the third generation (III-3 and III-8) presented with Dandy-Walker malformation and the associated symptoms. The affected family members in the first and second generation, however, showed only milder posterior fossa variations while functioning normally as adults and with no associated signs or symptoms other than the presence of an occipital cephalocele. Although originally our diagnostic criteria were limited to the presence of an occipital cephalocele or obligate-carrier status, after recharacterization of this condition as ADDWOC we were able to use the neuroradiological features of Dandy-Walker malformation and milder anomalies in the cerebellum and its surrounding fluid spaces as a more sensitive means of designating the affection status in this pedigree.
A genome-wide linkage screen of ADDWOC in this three-generation Vietnamese-American family resulted in the most significant evidence for linkage on chromosome 2 with a LOD score of 3.30 at 225.3–226.8 cM. We believe, given our assumptions, the maximum theoretical LOD score achievable for this pedigree is 3.61. The obtained LOD score is equivalent to the threshold value of 3.3 for significant linkage of a Mendelian disorder in the presence of locus heterogeneity (Terwilliger and Ott 1994). Haplotype analysis confirmed that this relatively small 1.1 Mb locus co-segregated with the ADDWOC phenotype. Additionally, we identified an isolated Dandy-Walker patient with a 2q36.1→qter deletion encompassing the 2q36.1 ADDWOC locus. Another isolated patient with a 2q35→qter deletion was also reported to have a Dandy-Walker malformation (Waters et al. 1993), which is consistent with the presence of a Dandy-Walker locus in this region. A second genome-wide linkage screen of ADD-WOC in a four-generation Brazilian family failed to detect evidence for linkage at 2q36.1, providing evidence for locus heterogeneity of the disorder.
Sequencing the coding exons, immediately flanking intronic regions, and promoter regions of SGPP2, FARSB, MOGAT1, ACSL3, and KCNE4 in the 2q36.1 locus failed to identify the disease-causing variant in our Vietnamese-American ADDWOC family. Further, comparative genomic hybridization using a BAC array failed to identify any significant gains or losses. Nevertheless, a higher resolution BAC array or analysis using multiplex ligation-dependent probe amplification (MLPA) may be able to uncover deletions or other ploidy variations. It is also possible that a mutation in an uncharacterized gene in this locus or a mutation in a regulatory region affecting the expression of a gene in this locus is causative for the ADDWOC phenotype. Furthermore, genes outside of this locus may also have enhancers/repressors located within the 2q36.1 locus. Regulatory elements have been identified 1 Mb away from the gene whose expression they control (Bien-Willner et al. 2007; Jeong et al. 2006). In fact, point mutations in a SHH enhancer located greater than 1 Mb away from the SHH gene have been identified as the causative mutations in families with triphalangeal thumb and preaxial polydactyly (Gurnett et al. 2007). Therefore, it is possible that mutations of regulatory regions within the ADDWOC locus may affect the expression of genes located outside of this locus.
The PAX3 transcriptional start site is located 107 kb centromeric to the ADDWOC 2q36.1 locus (Fig. 5) and proceeds from the negative strand, suggesting PAX3 cis-regulatory elements may lie within the ADDWOC locus. An association between PAX3 mutations and the ADDWOC clinical phenotype is suggested by observations in both murine Pax3 mutant lines, and in Waardenburg syndrome, which involves PAX3 mutations in humans (Tassabehji et al. 1993). However, it is not clear whether putative changes in PAX3 expression due to disruption of its regulatory elements can mimic the effects of exon mutations described in mice or humans. Genetic lesions in murine Pax3 lead to developmental abnormalities such as severe defects of the neural tube and neural crest derivatives (Franz and Kothary 1993; Mansouri et al. 1996, 1994; Moase and Trasler 1989) including absent cerebellum (Keller et al. 2004) and hindbrain cranioschisis (Robert 1954). Expression analysis of PAX3 in human fetuses by in situ hybridization detects transcripts in the ventricular zone at the mid–hindbrain boundary, and in the dorsal region of the ventricular zone and the roof plate of the medulla and the spinal cord (Terzic and Saraga-Babic 1999). This posterior neural tube expression pattern is consistent with a role for PAX3 in the development of posterior fossa structures, the structures primarily affected by ADDWOC. In addition, while individuals with Waardenburg syndrome typically present with abnormalities of eye and nose formation and deafness and pigmentation anomalies, several cases have been described with posterior neural tube defects (Hol et al. 1995; Kujat et al. 2007; Lu et al. 2007; Nye et al. 1998; Shim et al. 2004), providing an additional link between PAX3 and posterior neural tube structures. In silico analysis did not reveal any significant homology between PAX3 transcript and the sequence of the identified 2q36.1 locus, ruling out an undiscovered PAX3 homolog in this region.
We should finally point out that the appearance of the most severely affected members of the Vietnamese-American pedigree in the third generation (III-3 and III-8) suggests the phenomenon of anticipation in this family. The number and depth of pedigrees is too small to definitively conclude whether this condition is associated with anticipation. Additionally, we did not observe any nucleotide repeat expansion in the areas we sequenced (i.e., exons and immediate intronic or promoter regions). Further, the di-, tri-, tetra- and higher order nucleotide repeats in this locus, similarly to other parts of the genome, are too numerous to be sequenced entirely using the simple sequencing technique utilized in this study. Nevertheless, we cannot rule out that a nucleotide repeat expansion somewhere in this locus may play a role in this condition. Further sequencing in the conserved non-coding or nucleotide repeat regions of the 2q36.1 locus may help to uncover the genetic basis for ADDWOC.
Acknowledgments
We would like to thank Hoang Le and Dung Tri Pham, HoChiMinh City Hospital, HoChiMinh City, Vietnam, for their assistance in patient recruitment. A.J. is supported by NIH pre-doctoral NRSA grant F30-NS51962. K.A.A. is supported by NIH Pre-doctoral grant GM007839–26. A.G.B. is supported by NIH grant K08-NS48174.
Contributor Information
Ali Jalali, Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Kimberly A. Aldinger, Committee on Neurobiology, The University of Chicago, Chicago, IL, USA
Ajit Chary, Department of Pediatrics, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
David G. Mclone, Department of Neurosurgery, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Robin M. Bowman, Department of Neurosurgery, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Luan Cong Le, HoChiMinh City Hospital, HoChiMinh City, Vietnam.
Phillip Jardine, Department of Paediatrics, Bristol Royal Hospital for Children, Bristol, UK.
Ruth Newbury-Ecob, Department of Clinical Genetics, St Michael's Hospital, Bristol, UK.
Andrew Mallick, Department of Paediatrics, Yeovil Hospital, Somerset, UK.
Nadereh Jafari, Center for Genetic Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Eric J. Russell, Department of Radiology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
John Curran, Department of Radiology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Pam Nguyen, Department of Radiology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA.
Karim Ouahchi, Department of Pathology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, UK.
Charles Lee, Department of Pathology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, UK.
William B. Dobyns, Departments of Human Genetics and Neurology, The University of Chicago, Chicago, IL, USA
Kathleen J. Millen, Departments of Human Genetics and Neurology, The University of Chicago, Chicago, IL, USA
Joao M. Pina-Neto, Department of Genetics, School of Medicine, Universidade de São Paolo, Ribeirão Preto, Brazil
John A. Kessler, Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
Alexander G. Bassuk, Email: alexander-bassuk@uiowa.edu, Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, IL, USA; Department of Pediatrics, Northwestern University Feinberg School of Medicine, Chicago, IL, USA
References
- Bassuk AG, McLone D, Bowman R, Kessler JA. Autosomal dominant occipital cephalocele. Neurology. 2004;62:1888–1890. doi: 10.1212/01.wnl.0000125255.90915.5c. [DOI] [PubMed] [Google Scholar]
- Beltran-Valero de Bernabe D, Currier S, Steinbrecher A, Celli J, van Beusekom E, van der Zwaag B, Kayserili H, Merlini L, Chitayat D, Dobyns WB, Cormand B, Lehesjoki AE, Cruces J, Voit T, Walsh CA, van Bokhoven H, Brunner HG. Mutations in the O-mannosyltransferase gene POMT1 give rise to the severe neuronal migration disorder Walker-Warburg syndrome. Am J Hum Genet. 2002;71:1033–1043. doi: 10.1086/342975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien-Willner GA, Stankiewicz P, Lupski JR. SOX9cre1, a cisacting regulatory element located 1.1 Mb upstream of SOX9, mediates its enhancement through the SHH pathway. Hum Mol Genet. 2007;16:1143–1156. doi: 10.1093/hmg/ddm061. [DOI] [PubMed] [Google Scholar]
- Carvalho DR, Giuliani LR, Simao GN, Santos AC, Pina-Neto JM. Autosomal dominant atretic cephalocele with phenotype variability: report of a Brazilian family with six affected in four generations. Am J Med Genet A. 2006;140:1458–1462. doi: 10.1002/ajmg.a.31255. [DOI] [PubMed] [Google Scholar]
- Descipio C, Schneider L, Young TL, Wasserman N, Yaeger D, Lu F, Wheeler PG, Williams MS, Bason L, Jukofsky L, Menon A, Geschwindt R, Chudley AE, Saraiva J, Schinzel AA, Guichet A, Dobyns WE, Toutain A, Spinner NB, Krantz ID. Subtelomeric deletions of chromosome 6p: molecular and cytogenetic characterization of three new cases with phenotypic overlap with Ritscher-Schinzel (3C) syndrome. Am J Med Genet A. 2005;134:3–11. doi: 10.1002/ajmg.a.30573. [DOI] [PubMed] [Google Scholar]
- Dixon-Salazar T, Silhavy JL, Marsh SE, Louie CM, Scott LC, Gururaj A, Al-Gazali L, Al-Tawari AA, Kayserili H, Sztriha L, Gleeson JG. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland RJ, Eyaid W, Collura RV, Tully LD, Hill RS, Al-Nouri D, Al-Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart YY, Ruvolo M, Walsh CA. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- Franz T, Kothary R. Characterization of the neural crest defect in Splotch (Sp1H) mutant mice using a lacZ transgene. Brain Res Dev Brain Res. 1993;72:99–105. doi: 10.1016/0165-3806(93)90163-5. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. doi: 10.1038/ng1420. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A. Allegro version 2. Nat Genet. 2005;37:1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- Gurnett CA, Bowcock AM, Dietz FR, Morcuende JA, Murray JC, Dobbs MB. Two novel point mutations in the long-range SHH enhancer in three families with triphalangeal thumb and preaxial polydactyly. Am J Med Genet A. 2007;143:27–32. doi: 10.1002/ajmg.a.31563. [DOI] [PubMed] [Google Scholar]
- Hart MN, Malamud N, Ellis WG. The Dandy-Walker syndrome. A clinicopathological study based on 28 cases. Neurology. 1972;22:771–780. doi: 10.1212/wnl.22.8.771. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus-automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- Hol FA, Hamel BC, Geurds MP, Mullaart RA, Barr FG, Macina RA, Mariman EC. A frameshift mutation in the gene for PAX3 in a girl with spina bifida and mild signs of Waardenburg syndrome. J Med Genet. 1995;32:52–56. doi: 10.1136/jmg.32.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 2004;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujat A, Veith VP, Faber R, Froster UG. Prenatal diagnosis and genetic counseling in a case of spina bifida in a family with Waardenburg syndrome type I. Fetal Diagn Ther. 2007;22:155–158. doi: 10.1159/000097117. [DOI] [PubMed] [Google Scholar]
- Kyttala M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestila M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- Lu W, Zhu H, Wen S, Laurent C, Shaw GM, Lammer EJ, Finnell RH. Screening for novel PAX3 polymorphisms and risks of spina bifida. Birth Defects Res A Clin Mol Teratol. 2007;79:45–49. doi: 10.1002/bdra.20322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Stoykova A, Gruss P. Pax genes in development. J Cell Sci Suppl. 1994;18:35–42. doi: 10.1242/jcs.1994.supplement_18.5. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- Martinez-Lage JF, Martinez Robledo A, Poza M, Sola J. Familial occurrence of atretic cephaloceles. Pediatr Neurosurg. 1996;25:260–264. doi: 10.1159/000121136. [DOI] [PubMed] [Google Scholar]
- Mavrogiannis LA, Antonopoulou I, Baxova A, Kutilek S, Kim CA, Sugayama SM, Salamanca A, Wall SA, Morriss-Kay GM, Wilkie AO. Haploinsufficiency of the human homeobox gene ALX4 causes skull ossification defects. Nat Genet. 2001;27:17–18. doi: 10.1038/83703. [DOI] [PubMed] [Google Scholar]
- Moase CE, Trasler DG. Spinal ganglia reduction in the splotch-delayed mouse neural tube defect mutant. Teratology. 1989;40:67–75. doi: 10.1002/tera.1420400109. [DOI] [PubMed] [Google Scholar]
- Murray JC, Johnson JA, Bird TD. Dandy-Walker malformation: etiologic heterogeneity and empiric recurrence risks. Clin Genet. 1985;28:272–283. doi: 10.1111/j.1399-0004.1985.tb00401.x. [DOI] [PubMed] [Google Scholar]
- Nye JS, Balkin N, Lucas H, Knepper PA, McLone DG, Charrow J. Myelomeningocele and Waardenburg syndrome (type 3) in patients with interstitial deletions of 2q35 and the PAX3 gene: possible digenic inheritance of a neural tube defect. Am J Med Genet. 1998;75:401–408. [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA, Bennett CL, Eckert ML, Dobyns WB, Gleeson JG, Shaw DW, McDonald R, Eddy A, Chance PF, Glass IA. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet. 2004;75:82–91. doi: 10.1086/421846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy AT, Hedlund GL, Percy AK. Enlarged parietal foramina: association with cerebral venous and cortical anomalies. Neurology. 2000;54:1175–1178. doi: 10.1212/wnl.54.5.1175. [DOI] [PubMed] [Google Scholar]
- Robert A. Analysis of the developmental effects of a lethal mutation in the house mouse. J Exp Zool. 1954;127:305–329. [Google Scholar]
- Sertie AL, Sossi V, Camargo AA, Zatz M, Brahe C, Passos-Bueno MR. Collagen XVIII, containing an endogenous inhibitor of angiogenesis and tumor growth, plays a critical role in the maintenance of retinal structure and in neural tube closure (Knobloch syndrome) Hum Mol Genet. 2000;9:2051–2058. doi: 10.1093/hmg/9.13.2051. [DOI] [PubMed] [Google Scholar]
- Shim SH, Wyandt HE, McDonald-McGinn DM, Zackai EZ, Milunsky A. Molecular cytogenetic characterization of multiple intrachromosomal rearrangements of chromosome 2q in a patient with Waardenburg's syndrome and other congenital defects. Clin Genet. 2004;66:46–52. doi: 10.1111/j.0009-9163.2004.00276.x. [DOI] [PubMed] [Google Scholar]
- Smith UM, Consugar M, Tee LJ, McKee BM, Maina EN, Whelan S, Morgan NV, Goranson E, Gissen P, Lilliquist S, Aligianis IA, Ward CJ, Pasha S, Punyashthiti R, Malik Sharif S, Batman PA, Bennett CP, Woods CG, McKeown C, Bucourt M, Miller CA, Cox P, Algazali L, Trembath RC, Torres VE, Attie-Bitach T, Kelly DA, Maher ER, Gattone VH, II, Harris PC, Johnson CA. The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat Genet. 2006;38:191–196. doi: 10.1038/ng1713. [DOI] [PubMed] [Google Scholar]
- Tal Y, Freigang B, Dunn HG, Durity FA, Moyes PD. Dandy-Walker syndrome: analysis of 21 cases. Dev Med Child Neurol. 1980;22:189–201. doi: 10.1111/j.1469-8749.1980.tb04327.x. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Read AP, Newton VE, Patton M, Gruss P, Harris R, Strachan T. Mutations in the PAX3 gene causing Waardenburg syndrome type 1 and type 2. Nat Genet. 1993;3:26–30. doi: 10.1038/ng0193-26. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J. Handbook of human genetic linkage. Johns Hopkins University Press; Baltimore: 1994. [Google Scholar]
- Terzic J, Saraga-Babic M. Expression pattern of PAX3 and PAX6 genes during human embryogenesis. Int J Dev Biol. 1999;43:501–508. [PubMed] [Google Scholar]
- Thiele H, Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, Fazzi E, Signorini S, Louie CM, Bellacchio E, Bertini E, Dallapiccola B, Gleeson JG. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- Valente KD, Valente M. Epilepsy in one family with parietal foramina: an incidental finding? J Neurol Neurosurg Psychiatry. 2004;75:1648–1649. doi: 10.1136/jnnp.2004.035733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Reeuwijk J, Janssen M, van den Elzen C, Beltran-Valero de Bernabe D, Sabatelli P, Merlini L, Boon M, Scheffer H, Brockington M, Muntoni F, Huynen MA, Verrips A, Walsh CA, Barth PG, Brunner HG, van Bokhoven H. POMT2 mutations cause alpha-dystroglycan hypoglycosylation and Walker-Warburg syndrome. J Med Genet. 2005;42:907–912. doi: 10.1136/jmg.2005.031963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega H, Waisfisz Q, Gordillo M, Sakai N, Yanagihara I, Yamada M, van Gosliga D, Kayserili H, Xu C, Ozono K, Jabs EW, Inui K, Joenje H. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- Waters BL, Allen EF, Gibson PC, Johnston T. Autopsy findings in a severely affected infant with a 2q terminal deletion. Am J Med Genet. 1993;47:1099–1103. doi: 10.1002/ajmg.1320470734. [DOI] [PubMed] [Google Scholar]
- Zhao X, Chi L, Zhao Y, Chi Z. A five-generation family with occipital encephalocele. Clin Neurol Neurosurg. 2007;109:81–84. doi: 10.1016/j.clineuro.2006.03.003. [DOI] [PubMed] [Google Scholar]


