Abstract
Our previous findings indicated that RSK2 plays a critical role in proliferation and cell transformation induced by tumor promoters, such as epidermal growth factor or 12-O-tetradecanoylphorbol-13-acetate, and that kaempferol, a natural compound found in edible plants, selectively inhibits RSK2 activity. However, the molecular mechanism for RSK2 activation is unclear. Herein, we provide evidence showing that NH2-terminal kinase domain (NTD) activation of RSK2 is required for the activation of the extracellular signal-regulated kinase–mediated COOH-terminal kinase domain (CTD). We also found that the NTD plays a key role in substrate phosphorylation and that kaempferol binds with the NTD but not the CTD in both the active and inactive forms. Homology modeling of the RSK2 NH2-terminal domain and small-molecule docking, validated by mutagenesis experiments, clearly showed that Val82 and Lys100 are critical amino acids for kaempferol binding and RSK2 activity. Furthermore, immunohistofluorescence and Western blot results indicated that the RSK2 protein level is markedly higher in cancer cell lines as well as cancer tissues compared with nonmalignant cell lines or normal tissues. In addition, kaempferol inhibited proliferation of malignant human cancer cell lines, including A431, SK-MEL-5 and SK-MEL-28, and HCT-116. These results indicate that targeting RSK2 with natural compounds, such as kaempferol, might be a good strategy for chemopreventive or chemotherapeutic application.
Introduction
The mitogen-activated protein kinases (MAPK) are important regulators of proliferation and oncogenesis (1, 2). The MAPK extracellular signal-regulated kinases (ERK) 1 and 2 (3, 4) mediate the phosphorylation of 90-kDa ribosomal S6 kinases (RSK), which are a family of widely expressed serine/threonine kinases that respond to many growth factors, peptide hormones, and neuro-transmitters (5, 6). RSK2 is a member of the p90RSK family and is activated by ERK1/2 and phosphoinositide-dependent kinase 1 (3, 4). When activated, RSK2 is translocated to the nucleus, where it can phosphorylate various nuclear proteins, including c-Fos, Elk-1, histones, cyclic AMP–responsive element binding (CREB) protein (7–11), activating transcription factor 4 (12), p53 (13), and NFAT3 (14). Based on its broad substrate specificity, the RSK2 protein is likely to mediate a variety of cellular processes, including proliferation and transformation as well as cell cycle.
The Ras/ERK pathway regulates cell proliferation, survival, growth and motility (15, 16), and tumorigenesis (17). RSK2 is a direct substrate kinase of ERKs and is located between ERKs and its own target transcription factors. Therefore, RSK2 is an important direct effector for transcriptional activation of down-stream target transcription factors. RSK2 phosphorylates and inactivates the proapoptotic protein BAD, a component of the cell death machinery, and RSK2 also up-regulates the transcription of antiapoptotic Bcl-2 through CREB phosphorylation and activation (18). Because apoptosis is presumed to have a protective function against carcinogenesis, these effects suggest that RSK2 may have a role in enhancing tumorigenesis. Furthermore, a recent report indicated that RSK2 is involved in prostate cancer cell proliferation (19) and in c-fos–dependent osteosarcoma development (20). Therefore, deciphering the molecular activation mechanism of RSK2 substrate phosphorylation and the mechanism of selective RSK2 inhibition is extremely important for understanding how to control RSK2 activity.
Recently, SL0101, the rhamnose form of kaempferol extracted from the tropical plant Forsteronia refracta, was shown to inhibit RSK2 activity and suppress proliferation of MCF-7 breast cancer cells or LNCaP prostate cancer cells (19, 21). We found that kaempferol selectively inhibited RSK2, but not RSK1 or RSK3, and this compound suppressed proliferation and cell transformation induced by epidermal growth factor (EGF) or 12-O-tetradecanoylphorbol-13-acetate (TPA; ref. 22). A homology model of the NH2-terminal kinase domain (NTD) RSK2 was the starting point for examining the docking of SL0101 or kaempferol in the protein ATP-binding pocket of RSK2 and suggested that both SL0101 and kaempferol could form good interactions with NTD RSK2 (23). Most recently, the inactive structure of NTD RSK1 at 1.9 Å resolution (24) was reported and the structure showed a 95% amino acid sequence identity with NTD RSK2. However, although accumulating results indicate that RSK2 plays an important role in cell proliferation and cancer development, the actual mechanism of activation and inhibition of RSK2 activity and the effect on its substrate(s) is not yet clearly understood.
In the present study, we found that the RSK2 protein level was obviously higher in malignant cancer cell lines and human cancer tissues compared with normal cells and tissues. Furthermore, we showed that the NTD RSK2 is a critical kinase domain for substrate phosphorylation, which requires ERK-mediated activation. We found that kaempferol binds and suppresses the activation of NTD RSK and that Val82 and Lys100 are critical amino acids for NTD RSK2 activity and inhibition by kaempferol resulted in decreased cell proliferation.
Materials and Methods
General Materials and Methods are included as Supplementary Materials and Methods.
In vitro kinase assay
A GST-NFAT3D4-261-365 protein was used as a substrate for an in vitro kinase assay with RSK2 (Upstate Biotechnology, Inc) and bacterial-expressed His-RSK2 was used for an in vitro kinase assay with active ERK1, ERK2, p38α, or p38β. Reactions were carried out at 30°C for 30 min in a mixture containing 50 μmol/L unlabeled ATP and 10 μCi [γ-32P]ATP and then were stopped by adding 6× SDS sample buffer. To test the effect of kaempferol on RSK2 activity, we used 10 μmol/L unlabeled ATP and 10 μCi [γ-32P]ATP. Samples were boiled and then separated by 12% SDS-PAGE and visualized by autoradiography, Western blotting, or Coomassie blue staining.
Pull-down assay with CNBr-kaempferol beads
Kaempferol was first coupled to a CNBr-activated Sepharose 4B matrix, and the binding between RSK2 and kaempferol was examined by affinity chromatography according to the manufacturer's suggested protocol. The active or bacterial-expressed RSK2 protein (100 ng) was tested for affinity binding with 30 μL of CNBr-kaempferol beads (50% slurry) by incubating for 2 h at 4°C. The beads were washed thrice and suspended in 20 μL of 1× SDS sample buffer. Bound proteins were resolved by 10% SDS-PAGE and visualized by Western blot using specific antibodies.
Tissue array
A human skin tissue array (SK801) was purchased from U.S. Biomax, Inc., and analysis was conducted according to the manufacturer's suggested protocols. The tissue array includes matched normal tissues (H1-H7), which were biopsied from the adjacent tissue of each cancer tissue (A1-A7) from seven individual patients. The slide was baked at 60°C for 2 h, deparaffinized, and rehydrated. Antigens were then unmasked by submerging the slide into boiling sodium citrate buffer (10 mmol/L; pH 6.0) for 10 min. The sample was blocked with 3% bovine serum albumin in 1× PBS/0.03% Triton X-100 in a humidified chamber for 1 h at room temperature, and then the RSK2 antibody (1:200 dilution in 1× PBS/0.03% Triton X-100) at 4°C in a humidified chamber overnight. The slide was washed and hybridized with the secondary antibody (anti-rabbit, donkey antibody) conjugated with Cy3 (1:1,000) for 1.5 h at room temperature in the dark.
Results
Abundance and activity of the RSK2 protein
Our previous studies indicated that RSK2 plays a critical role in skeletal muscle cell differentiation through its activation of NFAT3 in C2C12 myoblasts (14) and in cell proliferation and transformation induced by tumor promoters such as EGF or TPA through its phosphorylation of histone H3 at Ser10 (22). We also found that kaempferol, an abundant chemical compound present in edible plants, is a natural compound that specifically inhibits RSK2 activity, resulting in suppression of cell transformation as well as decreased histone H3 phosphorylation at Ser10 (22). However, the action and inhibitory mechanisms of kaempferol on RSK2 are not yet clear. To achieve a greater understanding, we constructed bacterial expression plasmids (pHis-RSK2-1-740, pHis-RSK2-1-373, pHis-RSK2-328-740, and pHis-RSK2-399-740) combined with the pET-46 Ek/LIC His fusion vector (Fig. 1A). The RSK2 proteins were purified from the BL21 bacterial strain using Ni-NTA agarose beads and confirmed by Coomassie blue R-250 staining or Western blotting using a RSK2 antibody or a His antibody (Fig. 1B). To examine whether the RSK2 proteins expressed in bacteria were active, we conducted an in vitro kinase assay with the NFAT3-261-365 protein and [γ-32P]ATP. The results indicated that none of the RSK2 proteins expressed in bacteria could phosphorylate NFAT3-261-365 (Fig. 1C). However, the positive control of commercially available active RSK2 strongly phosphorylated NFAT3-261-365 (Fig. 1C). These results indicated that full activation of RSK2 requires upstream kinase-mediated activation such as ERKs.
Figure 1.
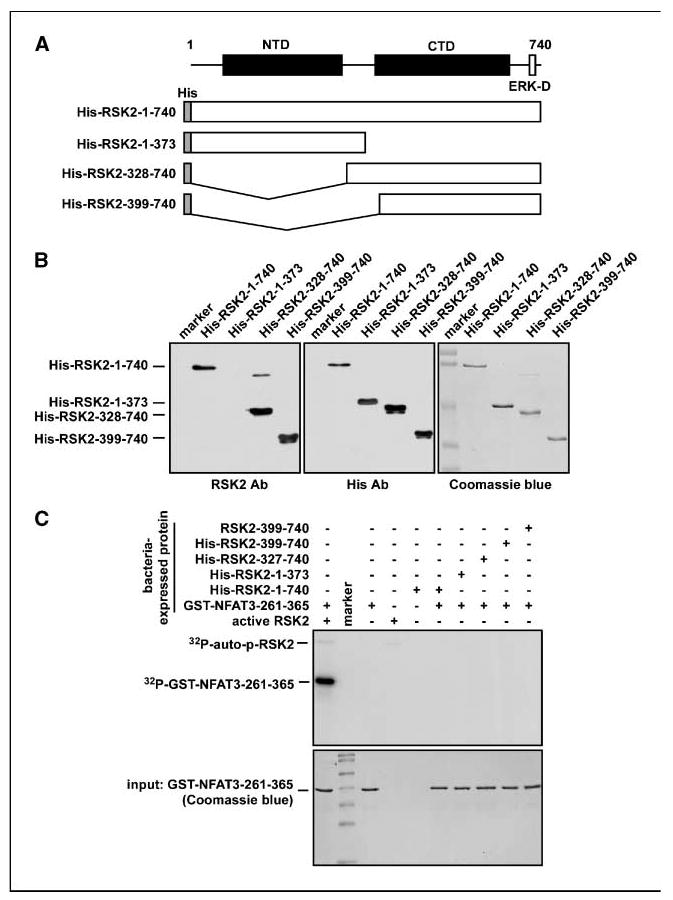
Expression and activities of full-length and truncated RSK2. A, schematic map of RSK2 constructs. RSK2-1-740, RSK2-1-373, RSK2-1-328-740, and RSK2-399-740 were each introduced into the pET-46 Ek/LIC His fusion vector. B, RSK2 proteins purified from BL21 were confirmed by Coomassie blue R-250 staining or Western blotting with an antibody against RSK2 (i.e., recognizes the CTD) or the His-tag. C, activity analysis of bacterial-expressed RSK2 proteins. The activity of purified RSK2 was analyzed by in vitro kinase assay using GST-NFAT3-261-365 and [γ-32P]ATP as described in Materials and Methods.
ERKs, but not p38, activate RSK2
Our previous study indicated that PD98059, a MAPK/ERK kinase (MEK) inhibitor, and SB202190, a p38 inhibitor, blocked histone H3 phosphorylation (25). RSK2 is an upstream kinase of histone H3 (Ser10; ref. 22). However, whether the only upstream kinase of RSK2 is ERK is unclear. To examine this idea, we conducted an in vitro kinase assay with bacterial-expressed His-RSK2-1-740 and identical units of active ERK1, ERK2, p38α, or p38β and [γ-32P]ATP. The results indicated that ERK1 or ERK2 strongly phosphorylated RSK2 (Fig. 2A) but not p38α or p38β (Fig. 2A). Furthermore, we determined that ERKs phosphorylated the linker region and the COOH-terminal region of RSK2 (Fig. 2B). Next, to determine RSK2 domain responsible for substrate phosphorylation, individual purified RSK2 proteins were subjected to a primary kinase reaction with active ERK2 and [γ-32P]ATP. One half of the reaction mixture (10 μL) was used to confirm phosphorylation of the RSK2 proteins by autoradiography (Fig. 2C). The remaining reaction mixture was divided into 2 μL aliquots and each was used sequentially in a secondary reaction with the substrate NFAT3-261-365. The autoradiography results indicated that NFAT3 phosphorylation was detected only with full-length His-RSK2-1-740 (Fig. 2D, lane 8) and not with His-RSK2-328-740 or His-RSK2-399-740 (Fig. 2D, lanes 9 and 10). Taken together with results shown in Figs. 1 and 2, these data indicate that substrate phosphorylation by RSK2 requires ERK-mediated preactivation and occurs through the NTD RSK2 but not the COOH-terminal kinase domain (CTD) RSK2.
Figure 2.
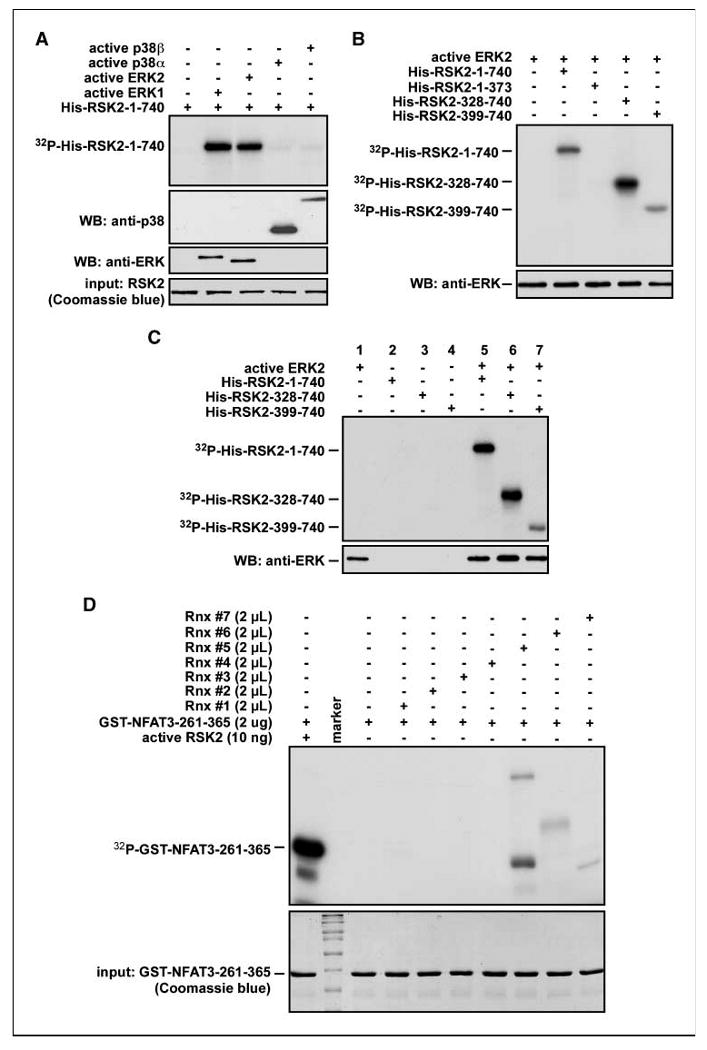
ERK-mediated RSK2 activation is required for substrate phosphorylation by NTD RSK2. A, ERK1 and ERK2, but not p38α or p38β, are RSK2 kinases. The commercially available active ERK1, ERK2, p38α, and p38β proteins (same units) were used for an in vitro kinase assay with bacterial-expressed RSK2-1-740 and [γ-32P]ATP as described in Materials and Methods. B, ERKs phosphorylated RSK2 at the linker region and CTD. Active ERK2 was used for an in vitro kinase assay with the indicated bacterial-expressed RSK2 proteins and [γ-32P]ATP as described in Materials and Methods. C and D, substrate phosphorylation by the NTD RSK2 requires ERK-mediated preactivation of RSK2. Bacterial-expressed RSK2 proteins were used for an in vitro kinase assay with active ERK2 and [γ-32P]ATP. A portion (10 μL) of the reaction mixture was used for autoradiography and 2 μL of reaction mixture were used for a secondary cascade kinase reaction with 2 μg of substrate, GST-NFAT3-261-365, and [γ-32P]ATP as described in Materials and Methods.
The NTD RSK2 is responsible for substrate phosphorylation ex vivo
To examine whether the NTD RSK2 is responsible for substrate phosphorylation under physiologic conditions, we constructed an NTD-deleted RSK2 (RSK2-NTDD) and a CTD-deleted RSK2 (RSK2-CTDD; Fig. 3A). The vectors, including the full-length RSK2 (RSK2 FL), were transfected into 293 cells and expression was confirmed by Western blotting (Fig. 3B). The proteins were immunoprecipitated with an Xpress-tagged antibody (Fig. 3C) and directly subjected to an in vitro kinase assay with NFAT3-261-365 and [γ-32P]ATP. The results showed that RSK2-FL, but not RSK2-NTDD or RSK2-CTDD, phosphorylated NFAT3-261-365 (Fig. 3D). These results showed that the NTD is responsible for substrate phosphorylation and NTD activation requires activation by the CTD.
Figure 3.
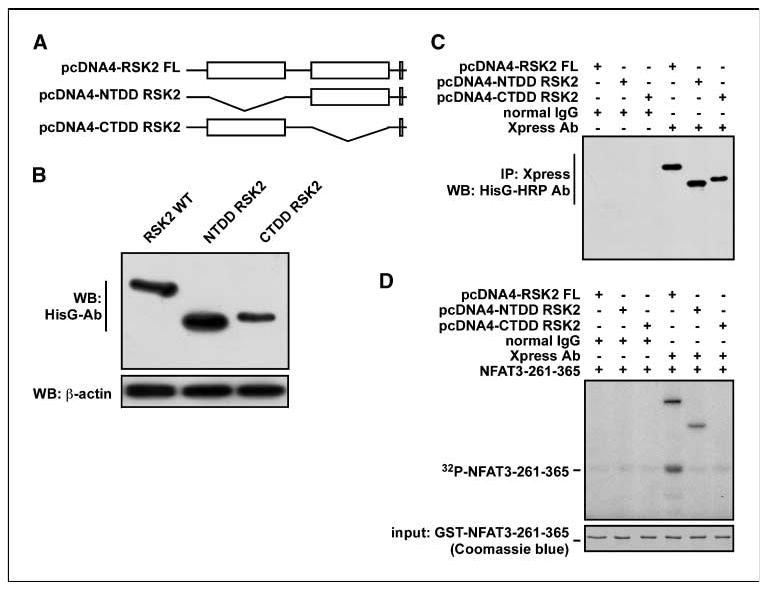
The NTD of RSK2 is responsible for substrate phosphorylation. A, schematic map of kinase domain deletion RSK2 constructs. B, the indicated RSK2 expression vectors were transiently introduced into 293 cells and protein expression was visualized by Western blotting with HisG-specific antibodies. C and D, immunoprecipitation/kinase assay of RSK2 with GST-NFAT3-261-365. Total cell lysates (200 μg) were used for immunoprecipitation with the Xpress antibody and agarose A/G beads. C, one half of immunoprecipitated RSK2 was used for confirmation of immunoprecipitation with HisG-horseradish peroxidase and visualized by enhanced chemiluminescence. D, the other half was used for an in vitro kination assay with 2 μg of GST-NFAT3-261-365 and [γ-32P]ATP as described in Materials and Methods.
Kaempferol binds with the NTD of RSK2
Our previous study showed that kaempferol inhibits RSK2 activity in vitro and ex vivo (22). However, the molecular mechanism of kaempferol inhibition of RSK2 has not been elucidated. To examine our hypothesis, we conjugated kaempferol with CNBr-Sepharose 4B beads and conducted an in vitro pull-down assay with an active RSK2 protein (100 ng; Fig. 4A). The results showed that the active RSK2 protein bound with CNBr-kaempferol beads but not with unconjugated CNBr-control beads (Fig. 4A). Furthermore, we found that kaempferol binds with either bacterial-expressed RSK2 (inactive) or active RSK2 (commercially available; Fig. 4B). Next, we conducted a pull-down assay with CNBr-kaempferol beads and each bacterial-expressed RSK2 protein (same molar ratio), including His-RSK2-1-740, His-RSK2-1-373, His-RSK2-328-740, and His-RSK2-399-740. Western blot results indicated that the RSK2 proteins that harbored the NTD, including His-RSK2-1-740 and His-RSK2-1-373, bound with CNBr-kaempferol beads (Fig. 4C). Furthermore, results from a pull-down assay using CNBr-kaempferol beads and lysates from cells that overexpressed RSK2 FL, NTD RSK2, or CTD RSK2 showed that RSK2 FL bound with kaempferol (Fig. 4D). Unexpectedly, the CTD RSK2 did not bind to CNBr-kaempferol beads, indicating that unknown factor(s) may interfere with kaempferol and CTD RSK2 binding.
Figure 4.
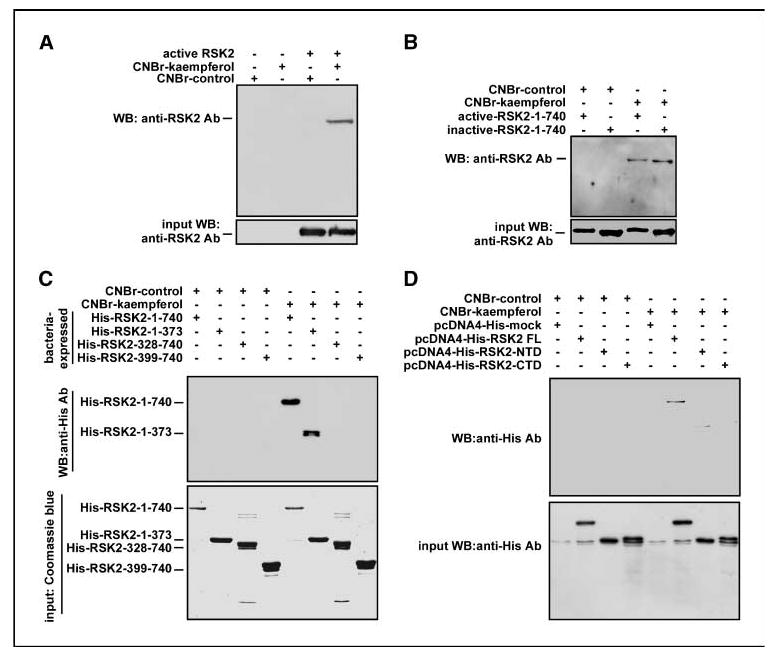
Kaempferol binds with the NTD of RSK2. A and B, RSK2 binds with kaempferol. Commercially available active RSK2 (100 ng) or bacterial-expressed RSK2 was mixed with 30 μL of CNBr-kaempferol beads (50% slurry) for 2 h at 4°C. Beads were then washed and RSK2 binding was visualized by Western blotting with an RSK2 antibody. C, kaempferol binds with the NTD of RSK2. To identify the RSK2 domain that binds with kaempferol, RSK2 proteins (200 ng) as indicated (same molar ratio) were mixed with 30 μL of CNBr-kaempferol beads (50% slurry) at 4°C for 2 h. The binding proteins were visualized by Western blotting with a His-specific antibody or an RSK2 antibody. D, RSK2 FL, but not NTD RSK2 or RSK2-CTD alone, binds with kaempferol. The indicated RSK2 expression vector was transiently introduced into 293 cells. Total cell lysates (200 μg) were used for CNBr-kaempferol binding and the bound proteins were visualized by Western blotting with a His-specific antibody.
Val82 and Lys100 are required for RSK2 activity
To better understand how small molecules such as kaempferol interact with the catalytic site of NTD RSK2 to modulate its activity, we modeled the NTD RSK2 (Fig. 5A) based on the crystal structure of the highly homologous phosphorylated protein kinase C (PKC) protein in complex with staurosporine (Protein Data Bank code: 1xjd). An NTD RSK2 homology model based on the complex between protein kinase A and ANP has already been reported (23). Using the modeled NTD RSK2, we docked kaempferol or SL0101 in silico to the ATP-binding pocket, allowing not only the ligand to be flexible but also the amino acids forming the protein binding site to achieve a more realistic view of the possible protein-ligand interactions (see Supplementary Materials and Methods). Both kaempferol and SL0101 showed the capability of forming numerous favorable connections within the NTD RSK2 ATP-binding pocket as indicated by a good docking score of −12.8 and −13.3 kcal/mol, respectively. The bound orientation of our predicted complexes was different from a model previously proposed (23). In our model, kaempferol seemed to form three hydrogen bonds with the backbone atoms of Asp148 and Leu150 in the hinge region of the NTD RSK2 (Fig. 5B). In contrast to the docking pose anticipated in ref. 23, a hydrogen bond might be formed with the amino group of the conserved Lys100 in the phosphate region of the catalytic site (similar to many known kinase inhibitors) and the backbone carbonyl group of Leu74. The kaempferol heterocyclic ring system lies in the hydrophobic region of the catalytic site in what would be the position occupied by the ATP adenine. Kaempferol seems capable of forming numerous favorable contacts with the upper limit of the hydrophobic area shaped mainly by the aliphatic side chain of Val82 and the lower area defined by Leu150 and Leu200. Similar to kaempferol, the planar double ring of SL0101 occupies the ATP hydrophobic pocket, although rotated by about 90° and may form several hydrogen bonds with the catalytic pocket as well (Fig. 5B). Notably, a mutation of Val82 to phenylalanine in the RSK2 ATP site is associated with Coffin-Lowry syndrome, suggesting that Val82 plays an important role in RSK2 activity (26). Our modeling results indicated that kaempferol might form key interactions with Val82 and, hence, very likely affects RSK2 activity. To further examine this hypothesis and validate our docking outcomes, we constructed point mutants Val82Phe, Lys100Phe, or Leu147Phe in the pcDNA4-RSK2 FL (Fig. 5C) and transfected these individual constructs into 293 cells, and then an immunoprecipitation/kinase assay was conducted with the Xpress antibody and GST-NFAT3-261-365 fragment. The results showed that the phosphorylation of NFAT3-261-365 was significantly reduced when RSK2 Val82 or Lys100 was mutated but only slightly reduced in the Leu147Phe mutant. Indeed, in our homology model, Leu147 is located on the back of the ATP-binding pocket and does not form crucial interactions with kaempferol (Fig. 5B), which may explain why the phosphorylation capacity is only partially affected. Furthermore, we found that introduction of RSK2 FL induced cell proliferation, whereas NTDD RSK2 or CTDD RSK2 transfection resulted in suppressed cell proliferation (Fig. 5D). These results strongly suggested that kaempferol binding to Val82 and Lys100 is critical for its inhibition of RSK2 NH2-terminal kinase activity and cell proliferation.
Figure 5.
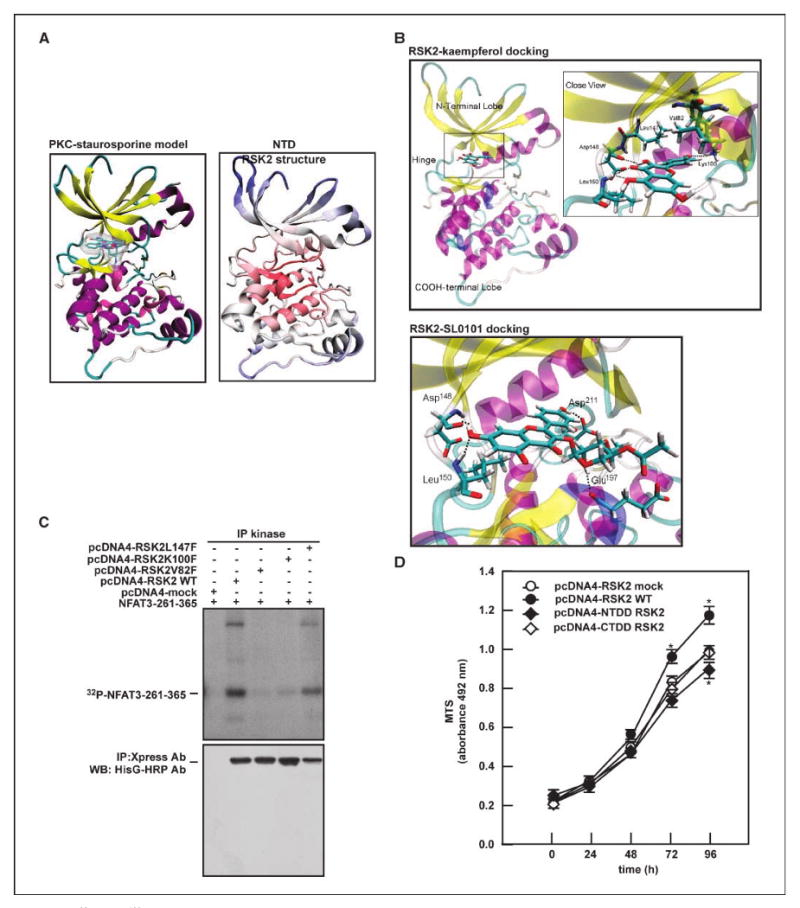
Val82 and Lys100 are critical amino acids required for RSK2 activity. A, a homology model of the RSK2 NTD (right) is compared with the crystal structure of PKC-staurosporine complex (left); both proteins are shown as cartoon, and staurosporine as atom-type colored licorice plus transparent molecular surface. B, docking of kaempferol or SL0101 to the RSK2 NTD. The protein is in transparent cartoon representation and the ligand is depicted as an atom-type colored stick model. In the close views of kaempferol and SL0101 binding, the amino acids that form interactions with the ligand are represented in atom-type sticks and the hydrogen bonds as dotted lines. C, Val 82 and Lys100 of RSK2 play a key role in RSK2 activity. The indicated mutant expression vectors of RSK2 were introduced into 293 cells. RSK2 kinase activity was determined with GST-NFAT3-261-365 and [γ-32P]ATP by immunoprecipitation/kinase assay as described in Supplementary Materials and Methods. D, kinase domain deletion of RSK2 suppresses RSK2 wild-type (WT)–induced cell proliferation. The indicated RSK2 deletion constructs were introduced into JB6 Cl41 cells and proliferation was measured at the indicated time points by MTS assay as described in Supplementary Materials and Methods. Points, mean of values and biological significance obtained from triplicate experiments; bars, SD. *, P < 0.05.
RSK2 protein level is higher in cancer tissues compared with normal tissues
Our earlier study showed that RSK2 overexpression in JB6 Cl41 cells enhanced proliferation and transformation induced by EGF or TPA and knockdown of RSK2 by small interfering RNA suppressed RasG12V-mediated Foci formation in NIH3T3 cells (22). These results suggested that the RSK2 protein level and activity might be higher in malignant cancer cells or tissues compared with normal cells or tissues. To examine this idea, we cultured several different malignant human cancer cell lines and nonmalignant human and mouse cell lines and then analyzed RSK2 protein level by Western blotting. The results indicated that many cancer cell lines, including H460, MCF-7, HCT-116, HCT-29, PC-2, Du-145, SoaS-2, A431, SK-MEL-5, SK-MEL-28, and RPMI 1640 (Fig. 6A), expressed higher RSK2 protein levels compared with nonmalignant human and mouse cells, such as HaCaT, JB6 Cl41, and NIH3T3 cells (Fig. 6A). Based on these results, we further hypothesized that kaempferol might suppress cancer cell proliferation. To examine this idea, we analyzed the effect of kaempferol on proliferation of various human malignant cancer cell lines, including A431 (skin epidermoid carcinoma), SK-MEL-5 and SK-MEL-28 (melanoma), and HCT-116 (colon cancer) by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay. The results indicated that kaempferol suppressed proliferation in a dose-dependent manner (Fig. 6B). In a human skin tissue array that included matched normal and various skin cancer tissues, we found by immunofluorescence assay that normal human skin, including epidermal and basal layers, exhibited RSK2 expression (Supplementary Fig. S1, H1–H10). Notably, we found that the RSK2 protein level was elevated by an average of ∼2-fold in almost all of the skin cancer tissues (Supplementary Fig. S1; Supplementary Table S1) compared with normal skin tissues (Fig. 6C). Importantly, a statistical comparison (by “two-sample unequal variance” Student's t test) of normal (9 samples) and cancer tissues (70 samples) indicated biological significance (P = 2.48E−16; Fig. 6C). Furthermore, in a comparison of RSK2 protein level in a tissue array of matched human normal skin (H1–H6 in Supplementary Fig. S1) and cancer tissues (A1–A6 in Supplementary Fig. S1), we found that the RSK2 protein level was substantially enhanced in skin cancer tissues (Fig. 6D, top). In a densitometric analysis of each matched sample, the RSK2 protein level was increased by ∼300% compared with normal control tissue sample (Fig. 6D, bottom left). Moreover, statistical analysis (homoscedastic Student's t test) to compare RSK2 protein level in all matched human normal and cancer tissues indicated that the difference in RSK2 expression was significant (P < 0.001; Fig. 6D, bottom right). These results indicated that RSK2 is highly abundant in cancer tissues compared with normal tissues and inhibition of RSK2 by dietary compounds such as kaempferol can suppress cancer cell proliferation.
Figure 6.
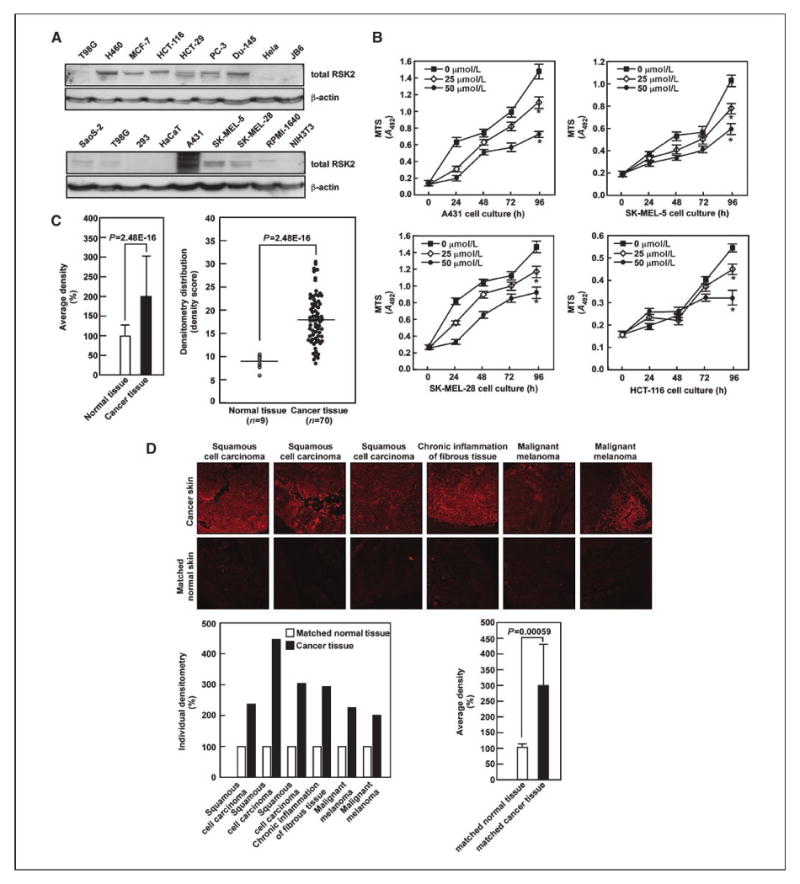
Kaempferol inhibits RSK2-mediated cancer cell proliferation. A, proteins extracted from different cells were examined for RSK2 protein level by Western blotting with an RSK2 antibody. B, kaempferol suppresses cancer cell proliferation. A431 (top left), SK-MEL-28 (bottom left), SK-MEL-5 (top right), and HCT-116 (bottom right) were seeded and treated with different doses of kaempferol and proliferation was measured by MTS assay at the indicated time points to assess time- and dose-dependent effects. Points, mean and biological significance of values obtained from triplicate experiments; bars, SD. *, P < 0.001. C, RSK2 protein level is higher in human skin cancer tissue compared with matched normal skin tissue. An analysis of a human skin tissue array, including matched normal and cancer tissues, was carried out as described in Materials and Methods. The light intensity was estimated using the ImageJ computer program. The overall average density of all samples is shown (left) along with the density of each individual sample (right). The results of the entire tissue array are shown in Supplementary Fig. S1 and the values obtained for each individual sample are shown in Supplementary Table S1. D, RSK2 protein level was compared in matched human normal and cancer tissues, including squamous cell carcinoma, chronic inflammation of fibrous tissue, and malignant melanoma, as described for C and Supplementary Fig. S1. Representative samples are provided (top), and the density of individual samples is shown (bottom left) along with the overall average density of the entire tissue array (bottom right). Significant differences were evaluated using the Student's t test as for Supplementary Table S1 (P = 0.00059).
Discussion
Flavonoids such as myricetin, quercetin, kaempferol, luteolin, and apigenin are well-known compounds found in edible plants. The highest total flavonoid content is observed in onion leaves (e.g., quercetin at 1,497.5 mg/kg, luteolin at 391.9 mg/kg, and kaempferol at 832.0 mg/kg; ref. 27). The recently isolated compound SL0101 [kaempferol 3-O-(3″,4″-di-O-acetyl-α-l-rhamnopyranoside)] was shown to inhibit RSK2 activity (IC50 = 89 nmol/L in vitro) and suppress proliferation of MCF-7 breast and LNCaP prostate cancer cells (19, 21). We found that kaempferol also suppressed RSK2 activity with an IC50 of ∼7 μmol/L in vitro (22). However, accumulating evidence indicates that the side chain of some food compounds is cleaved in the intestinal flora, which increases the absorption of that particular compound (28, 29). Furthermore, kaempferol can be detected in human plasma (800 nmol/L) and urine after oral administration, suggesting that kaempferol can be absorbed in the intestine (30). This evidence indicates that kaempferol is a major dietary flavonoid, which can be absorbed from intestine. Recently, we found that kaempferol selectively inhibits RSK2 activity in vitro and in vivo and also suppresses EGF-induced anchorage-independent cell transformation through the inhibition of histone H3 phosphorylation at Ser10 (22). Other research groups have shown that kaempferol induces apoptosis (31) in partially differentiated colon cancer (32), suggesting that kaempferol might function as an anticancer agent. Furthermore, combination treatment with kaempferol and quercetin in gut (HuTu-80 and Caco-2) and breast (PMC42) cancer cells suppresses proliferation that is associated with a decreased expression of nuclear proliferation antigen Ki67 (33). In this study, we found that the RSK2 protein level was increased in many human skin tumors, including melanoma and squamous cell carcinoma, and chronic inflammatory fibrous tissues, and many human cancer cell lines. By comparing the efficacy of kaempferol in cancer cells (Fig. 6D), we found that cancer cells are more sensitive to kaempferol compared with nonmalignant cells. Proliferation was inhibited in JB6 cells by ∼15% at 15 μmol/L and 39% at 60 μmol/L kaempferol (22). However, proliferation of cancer cells, such as A431, SK-MEL-5, SK-MEL-28, and HCT-116, was inhibited by kaempferol by about 24% to 27% at 25 μmol/L and 45% to 60% at 50 μmol/L. These results clearly showed that cancer cells, which contain a greater abundance of RSK2 protein, are more sensitive to the effect of kaempferol on proliferation. These findings support the importance of developing therapeutic agents targeting RSK2 and kaempferol binding may be a good starting point for selective drug design. However, although kaempferol is one of the most common and abundant dietary phytochemicals with potent chemopreventive activity against RSK2 activity, its effect in in vivo animal models has not yet been studied.
A published solved structure for the NTD RSK2 is not yet available. However, a homology model based on a complex formed between protein kinase A (PKA) and ANP has been reported (24). We chose PKC instead of PKA as the template for building an NTD RSK2 homology model because in the PKC X-ray structure, the ATP-binding site is occupied by the large and flat staurosporine (a well-known PKC inhibitor), which forces the catalytic pocket to adopt an open conformation that is optimal for accommodating small molecules, thus facilitating docking experiments. Very recently, the crystal structure of the NTD RSK1 in complex with staurosporine was solved (24). The NTD RSK1 shares a 95% amino acid identity with the NTD RSK2. We compared the structures of the two proteins and the root mean square deviation between our NTD RSK2 homology model and the solved RSK1-NTD structure was only 1.9 Å, which implies that they likely exhibit very similar conformations. Furthermore, the staurosporine molecule presented in both ATP-binding sites superimposes almost entirely, adopting an identical interaction profile within the pocket, which further validates our NTD RSK2 homology model. Based on docking results of kaempferol and SL0101, point and deletion truncation mutants of RSK2 clearly showed that kaempferol binding to Val82 and Lys100 is required for the inhibitory effect of kaempferol against RSK2 activity (Figs. 4D and 5B and C). The MAPK signal transduction network comprises a very complicated and highly interactive series of phosphorylation cascades (16, 34). The most thoroughly characterized MAPK family members include the ERKs, c-Jun NH2-terminal kinases, and p38 proteins, which are regulated and activated by MEK kinases or MAPK kinase kinases via MEK, MKK3/7, or MKK3/6. RSK2 is a member of the p90RSK family that includes RSK1, RSK2, RSK3, RSK4, MSK1, and MSK2 (3, 4). Our previous work indicated that RSK2 plays an important role in cell proliferation, differentiation, and transformation and inhibits apoptosis by interacting with multiple transcription factors or other proteins, including p53 (13), NFAT3 (14), and histone H3 (13, 22). Therefore, finding a natural dietary compound that inhibits RSK2 activity would be valuable and useful for chemopreventive application. In our previous study, we found that kaempferol suppressed RSK2 activity in vitro and ex vivo (22) and inhibited proliferation and cell transformation induced by tumor promoters such as EGF or TPA (22). However, the molecular mechanism(s) of the inhibitory effects of kaempferol is not yet fully understood. In this study, we provide evidence that kaempferol directly binds to the NTD RSK2 to prevent its activation. Furthermore, we found that the RSK2 protein level was elevated in cancer cell lines and tissues compared with normal cells and tissues (Fig. 6). Kaempferol treatment suppressed proliferation in human skin cancer cells, including A431 skin epidermoid carcinoma, SK-MEL-5 and SK-MEL-28 melanomas, and HCT-116 colon cancer cells, in a dose-dependent manner (Fig. 6B). The overall results indicated that inhibition of RSK2 activity with dietary natural compounds such as kaempferol might provide a safe and effective chemopreventive application to suppress redevelopment of cancer after primary treatment or intervention.
Acknowledgments
Grant support: The Hormel Foundation and NIH grants CA077646, CA111536, CA120388, ES016548, and R37 CA081064.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Sebolt-Leopold JS. Development of anticancer drugs targeting the MAP kinase pathway. Oncogene. 2000;19:6594–9. doi: 10.1038/sj.onc.1204083. [DOI] [PubMed] [Google Scholar]
- 2.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev. 2004;68:320–44. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones SW, Erikson E, Blenis J, Maller JL, Erikson RL. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc Natl Acad Sci U S A. 1988;85:3377–81. doi: 10.1073/pnas.85.10.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frodin M, Jensen CJ, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–34. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frodin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151:65–77. doi: 10.1016/s0303-7207(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 6.Nebreda AR, Gavin AC. Perspectives: signal transduction. Cell survival demands some RSK. Science. 1999;286:1309–10. doi: 10.1126/science.286.5443.1309. [DOI] [PubMed] [Google Scholar]
- 7.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–63. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 8.Sassone-Corsi P, Mizzen CA, Cheung P, et al. Requirement of RSK-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–91. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 9.Blenis J. Signal transduction via the MAP kinases: proceed at your own RSK. Proc Natl Acad Sci U S A. 1993;90:5889–92. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis RJ. Transcriptional regulation by MAP kinases. Mol Reprod Dev. 1995;42:459–67. doi: 10.1002/mrd.1080420414. [DOI] [PubMed] [Google Scholar]
- 11.Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–77. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Matsuda K, Bialek P, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry syndrome. Cell. 2004;117:387–98. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 13.Cho YY, He Z, Zhang Y, et al. The p53 protein is a novel substrate of ribosomal S6 kinase 2 and a critical intermediary for ribosomal S6 kinase 2 and histone H3 interaction. Cancer Res. 2005;65:3596–603. doi: 10.1158/0008-5472.CAN-04-3935. [DOI] [PubMed] [Google Scholar]
- 14.Cho YY, Yao K, Bode AM, et al. RSK2 mediates muscle cell differentiation through regulation of NFAT3. J Biol Chem. 2007;282:8380–92. doi: 10.1074/jbc.M611322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson G, English JM, White MA, Cobb MH. ERK5 and ERK2 cooperate to regulate NF-κB and cell transformation. J Biol Chem. 2001;276:7927–31. doi: 10.1074/jbc.M009764200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–44. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho YY, Bode AM, Mizuno H, et al. A novel role for mixed-lineage kinase-like mitogen-activated protein triple kinase α in neoplastic cell transformation and tumor development. Cancer Res. 2004;64:3855–64. doi: 10.1158/0008-5472.CAN-04-0201. [DOI] [PubMed] [Google Scholar]
- 18.Bonni A, Brunet A, West AE, et al. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–62. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 19.Clark DE, Errington TM, Smith JA, et al. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res. 2005;65:3108–16. doi: 10.1158/0008-5472.CAN-04-3151. [DOI] [PubMed] [Google Scholar]
- 20.David JP, Mehic D, Bakiri L, et al. Essential role of RSK2 in c-Fos-dependent osteosarcoma development. J Clin Invest. 2005;115:664–72. doi: 10.1172/JCI22877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JA, Poteet-Smith CE, Xu Y, et al. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 2005;65:1027–34. [PubMed] [Google Scholar]
- 22.Cho YY, Yao K, Kim HG, et al. Ribosomal S6 kinase 2 is a key regulator in tumor promoter induced cell transformation. Cancer Res. 2007;67:8104–12. doi: 10.1158/0008-5472.CAN-06-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TL, Gussio R, Smith JA, et al. Homology model of RSK2 N-terminal kinase domain, structure-based identification of novel RSK2 inhibitors, and preliminary common pharmacophore. Bioorg Med Chem. 2006;14:6097–105. doi: 10.1016/j.bmc.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Ikuta M, Kornienko M, Byrne N, et al. Crystal structures of the N-terminal kinase domain of human RSK1 bound to three different ligands: implications for the design of RSK1 specific inhibitors. Protein Sci. 2007;16:2626–35. doi: 10.1110/ps.073123707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong SP, Ma WY, Dong Z. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J Biol Chem. 2000;275:20980–4. doi: 10.1074/jbc.M909934199. [DOI] [PubMed] [Google Scholar]
- 26.Jacquot S, Merienne K, De Cesare D, et al. Mutation analysis of the RSK2 gene in Coffin-Lowry patients: extensive allelic heterogeneity and a high rate of de novo mutations. Am J Hum Genet. 1998;63:1631–40. doi: 10.1086/302153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miean KH, Mohamed S. Flavonoid (myricetin, quercetin, kaempferol, luteolin, and apigenin) content of edible tropical plants. J Agric Food Chem. 2001;49:3106–12. doi: 10.1021/jf000892m. [DOI] [PubMed] [Google Scholar]
- 28.Day AJ, DuPont MS, Ridley S, et al. Deglycosylation of flavonoid and isoflavonoid glycosides by human small intestine and liver α-glucosidase activity. FEBS Lett. 1998;436:71–5. doi: 10.1016/s0014-5793(98)01101-6. [DOI] [PubMed] [Google Scholar]
- 29.Walle T. Absorption and metabolism of flavonoids. Free Radic Biol Med. 2004;36:829–37. doi: 10.1016/j.freeradbiomed.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 30.DuPont MS, Day AJ, Bennett RN, Mellon FA, Kroon PA. Absorption of kaempferol from endive, a source of kaempferol-3-glucuronide, in humans. Eur J Clin Nutr. 2004;58:947–54. doi: 10.1038/sj.ejcn.1601916. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen TT, Tran E, Ong CK, et al. Kaempferol-induced growth inhibition and apoptosis in A549 lung cancer cells is mediated by activation of MEK-MAPK. J Cell Physiol. 2003;197:110–21. doi: 10.1002/jcp.10340. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura Y, Chang CC, Mori T, et al. Augmentation of differentiation and gap junction function by kaempferol in partially differentiated colon cancer cells. Carcinogenesis. 2005;26:665–71. doi: 10.1093/carcin/bgi003. [DOI] [PubMed] [Google Scholar]
- 33.Ackland ML, van de Waarsenburg S, Jones R. Synergistic antiproliferative action of the flavonols quercetin and kaempferol in cultured human cancer cell lines. In Vivo. 2005;19:69–76. [PubMed] [Google Scholar]
- 34.Bode AM, Dong Z. Mitogen-activated protein kinase activation in UV-induced signal transduction. Sci STKE. 2003;2003:RE2. doi: 10.1126/stke.2003.167.re2. [DOI] [PubMed] [Google Scholar]


