Abstract
Apical constriction facilitates epithelial sheet bending and invagination during morphogenesis1, 2. Apical constriction is conventionally thought to be driven by the continuous purse-string-like contraction of a circumferential actin and Non-Muscle Myosin-II (myosin) belt underlying adherens junctions3–7. However, it is unclear whether other force-generating mechanisms can drive this process. Here, we use real-time imaging and quantitative image analysis of Drosophila gastrulation to show that apical constriction of ventral furrow cells is pulsed. Repeated constrictions, which are asynchronous between neighboring cells, are interrupted by pauses in which the constricted state of the cell apex is maintained. In contrast to the purse-string model, constriction pulses are powered by actin-myosin network contractions that occur at the medial apical cortex and pull discrete adherens junction sites inward. The transcription factors Twist and Snail differentially regulate pulsed constriction. Expression of snail initiates actin-myosin network contractions while expression of twist stabilizes the constricted state of the cell apex. Our results suggest a new model for apical constriction in which a cortical actin-myosin cytoskeleton functions as a developmentally controlled sub-cellular ratchet to incrementally reduce apical area.
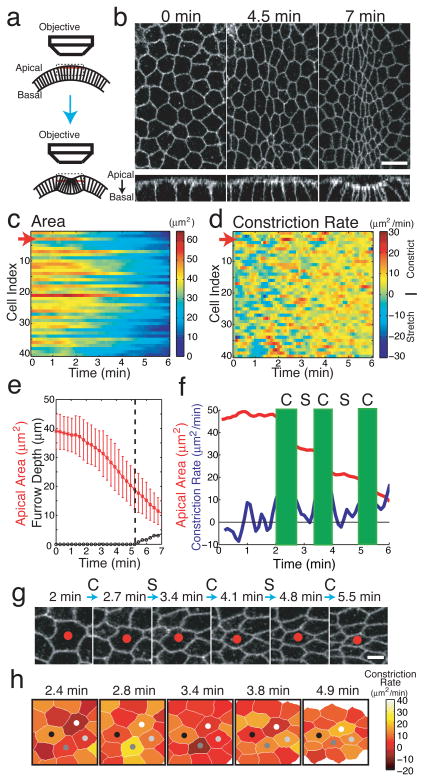
During Drosophila gastrulation, apical constriction of ventral cells facilitates the formation of a ventral furrow and the subsequent internalization of the presumptive mesoderm. Although myosin is known to localize to the apical cortex of constricting ventral furrow cells8–11, how myosin produces force to drive constriction is not known. Understanding this mechanism requires a quantitative analysis of cell and cytoskeletal dynamics. Therefore, we developed methods to visualize and quantify apical cell shape using Spider-GFP, a GFP-tagged transmembrane protein that outlines individual cells (Fig. 1a, 1b, Supplemental Figure 1, Video 1)12. Ventral cells constricted to ~50 % of their initial apical area before the onset of invagination and continued to constrict during invagination (Fig. 1c, 1e). Although the average apical area steadily decreased at a rate of ~5 μm2/min, individual cells exhibited transient pulses of rapid constriction that exceeded 10–15 μm2/min (Fig. 1d, 1f, 1g, and Video 2). During the initial 2 minutes of constriction, weak constriction pulses were often interrupted by periods of cell stretching. However, at 2 minutes constriction pulses increased in magnitude and cell shape appeared to be stabilized between pulses, leading to net constriction (Fig. 1d). These two phases likely correspond to the ‘slow/apical flattening’ and ‘fast/stochastic’ phases that have been previously described13, 14. Overall, cells underwent an average of 3.2 ± 1.2 constriction pulses over 6 minutes with an average interval of 82.8 ± 48 s between pulses (mean ± s.d., n = 40 cells, 126 pulses). Constriction pulses were mostly asynchronous between adjacent cells (Fig. 1h, Video 3). As a consequence, cell apices between constrictions appeared to be pulled by their constricting neighbors. Thus, apical constriction occurs via pulses of rapid constriction interrupted by pauses in which cells must stabilize their constricted state before re-initiating constriction.
Figure 1. Apical constriction of ventral furrow cells is pulsed.
a, Schematic of the imaging approach used to visualize ventral furrow cell apical constriction. We selected tangential z-slices 2 μm below the apical surface (red slices) to visualize cell outlines. b, Z-slices (top) and YZ cross-sections (bottom) of cell membranes visualized with Spider-GFP. Scale bar = 10 μm. Apical areas (c) and constriction rates (d) for individual cells of a representative embryo. Each row represents data (see colorbars) for an individual cell. e, Mean apical area (red) and furrow depth (black). Dotted line indicates when tissue invagination initiates. Error bars, s.d. (n = 41 cells). Quantification (f) and time-lapse images (g) of the constriction of an individual cell. The red arrows (c, d) and red dots (g) mark the cell that is quantified in f. C, contraction. S, stabilization. Scale bar = 4 μm. (h) Pulsed constriction is asynchronous in neighboring cells. Constriction rate is colorcoded (see colorbar) and mapped onto the corresponding cells in images at different timepoints.
To determine how myosin might generate force during pulsed constrictions, we simultaneously imaged myosin and cell dynamics using myosin regulatory light chain (MRLC, spaghetti squash, squ) fused to mCherry (Myosin-mCherry) and Spider-GFP. Discrete myosin spots and fibers present on the apical cortex formed a network that extended across the tissue (Fig. 2a, Supplemental Fig. 2a). These myosin structures were dynamic, with apical myosin spots repeatedly increasing in intensity and moving together (~40 nm/s) to form larger and more intense myosin structures at the medial apical cortex (Fig. 2c, Supplemental Fig. 2b, 2c, and Video 4). This process, which we refer to as myosin coalescence, resulted in bursts of myosin accumulation that were correlated with constriction pulses (Fig. 2b, 2c, 2d, Video 5). The peak rate of myosin coalescence preceded the peak constriction rate by 5–10 s, suggesting that myosin coalescence causes apical constriction (Supplemental Fig. 2d). Between myosin coalescence events, myosin structures, including fibers, remained present on the cortex, possibly maintaining cortical tension between constriction pulses (Fig. 2c). Contrary to the purse-string model, we did not observe significant myosin accumulation at cell-cell junctions. To confirm that constriction involved medial myosin coalescence and not contraction of a circumferential purse-string, we correlated constriction rate with myosin intensity at either the medial or junctional regions of the cell. Apical constriction was correlated more significantly with medial myosin (Fig. 2e), suggesting that in contrast to the purse-string model, constriction is driven by contractions at the medial apical cortex.
Figure 2. Constriction pulses are correlated with myosin coalescence.
a, Merged images of Myosin-mCherry (Z-projection, 5 μm depth, green) and Spider-GFP (individual z-slice 2 μm below the apical cortex, red). YZ cross-sections at lower magnification to illustrate furrow progression are shown at bottom. b, Mean apical area and myosin intensity (left) and myosin intensity for individual cells (right) for a representative embryo. Error bars, s.d. (n = 37 cells). c, Single channel and merged timelapse images of Myosin-mCherry (green) and Spider-GFP (red). Red arrows indicate spots that will coalesce. Blue arrow indicates myosin fiber that appears between contractions. d, Apical area and myosin intensity vs. time (top) and constriction rate and rate in change of myosin intensity vs. time (bottom) for an individual cell. e, Constriction rate is most highly correlated with medial myosin rather than junctional myosin. The diagram (top) illustrates the purse-string model for contraction in which we expect actin and myosin to become concentrated in the junctional region upon constriction. Data points represent correlation coefficients (r-values) for individual cells and the black bar indicates the mean (n = 37 cells). *, difference between the means is statistically significant (p<0.0001). Scale bars = 4 μm.
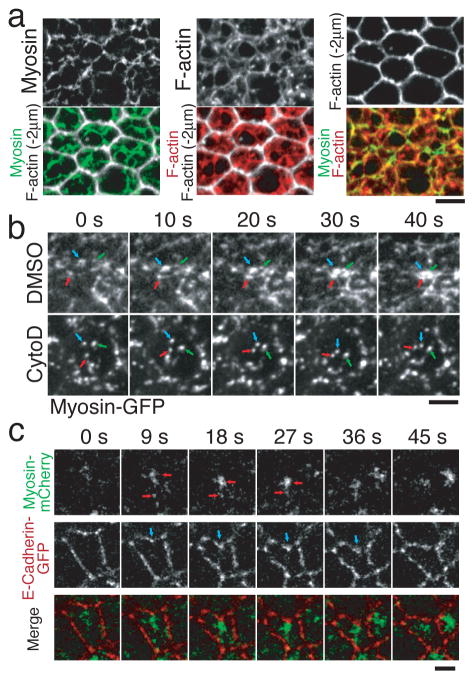
Myosin coalescence resembled contraction of a cortical actin-myosin network15, 16. Therefore, to determine whether apical constriction is driven by pulsed actin-myosin network contractions, we examined the organization of the cortical actin cytoskeleton. In fibroblasts and keratocytes, actin network contraction bundles actin filaments into fiber-like structures16, 17. Consistent with this expectation, we identified an actin filament meshwork underlying the apical cortex in which prominent actin-myosin fibers spanning the apical cortex appeared specifically in constricting cells (Fig. 3a, Supplemental Fig. 3a). An actin-myosin network contraction model would predict that myosin coalescence results from myosin spots exerting traction on each other through the cortical actin network. To test whether myosin coalescence requires an intact actin network, we disrupted the actin network with Cytochalasin D (CytoD). Actin network disruption with CytoD resulted in apical myosin spots that colocalized with actin structures and appeared specifically in ventral cells (Supplemental Fig. 3b, 3c). Myosin spots in CytoD injected embryos exhibited more rapid movement than in control-injected embryos, suggesting that apical myosin spots in untreated embryos are constrained by the cortical actin network (Supplemental Fig. 3d). Although myosin movement was uninhibited in CytoD treated embryos, myosin spots failed to coalesce and cells failed to constrict (Fig. 3b, Supplemental Fig. 3e). Because myosin coalescence requires an intact actin network, we propose that pulses of myosin coalescence represent actin-myosin network contractions.
Figure 3. Pulsed myosin coalescence and adherens junction bending require an actin-myosin network.
a, Cortical myosin (green), cortical F-actin (red), and F-actin 2μm below the apical cortex (white, to illustrate cell shape) were visualized in fixed embryos. b, Timelapse images of Myosin-GFP in control injected (DMSO) and cytochalasin D (CytoD) injected embryos. Arrows indicate individual myosin spots. Note that myosin spots move, but do not coalesce in CytoD treated embryos. c, Single channel and merged timelapse images of Myosin-mCherry (green) and E-Cadherin-GFP (red). Red arrows indicate myosin coalescence. Blue arrows indicate the site where adherens junctions bend inward beneath a myosin spot. Scale bars = 4 μm.
Because actin-myosin contractions occurred at the medial apical cortex, it was unclear how the actin-myosin network was coupled to adherens junctions. Therefore, we imaged E-Cadherin-GFP and Myosin-mCherry to examine the relationship between myosin and adherens junctions. Before apical constriction, adherens junctions are present ~4 μm below the apical cortex18. As apical constriction initiated, these subapical adherens junctions gradually disappeared while adherens junctions simultaneously appeared apically at the same level as myosin8, 19. This apical redistribution of adherens junctions occurred at specific sites along cell edges (midway between vertices, Supplemental Fig. 3f). As apical constriction initiated, these sites bent inwards. This bending depended on the presence of an intact actin network, consistent with actin-myosin network contraction generating force to pull junctions (Supplemental Fig. 3f). Indeed, myosin spots undergoing coalescence were observed to lead adherens junctions as they transiently bent inwards (Fig. 3c). Thus, pulsed actin-myosin network contraction at the medial cortex appears to pull the cell surface inward at discrete adherens junction sites, resulting in apical constriction.
The transcription factors Twist and Snail regulate apical constriction of ventral furrow cells20–23. Snail is a transcriptional repressor whose target(s) are currently unknown, while Twist enhances snail expression and activates the expression of fog and t48, which are thought to activate the Rho1 GTPase and promote myosin contractility8, 10, 19, 21, 24. To further examine the mechanism of pulsed apical constriction, we tested how Twist and Snail regulate myosin dynamics. In contrast to wild-type ventral cells, where myosin concentrated on the apical cortex (Fig. 2a), twist and snail mutants accumulated myosin predominantly at cell junctions, similar to lateral cells (Fig. 4a). These ventral cells failed to constrict productively, supporting our cortical actin-myosin network contraction model, rather than the purse-string model, for apical constriction. Importantly, twist and snail mutants differentially affected the coalescence of the minimal myosin that did localize to the apical cortex. While myosin coalescence was inhibited in snail mutants, it still occurred in twist mutants, as did pulsed constrictions (Fig. 4a, Video 6). This difference was also observed when Snail or Twist activity was knocked-down using RNAi (referred to as snailRNAi or twistRNAi) (Supplemental Fig. 4a, Video 7). The magnitude of constriction pulses in twistRNAi embryos was greater than that of twist mutant embryos, suggesting that the low level of Twist activity present in twistRNAi embryos enhances contraction efficiency by activating the expression of snail or other transcriptional targets. Myosin coalescence was inhibited in snail twist double mutants, demonstrating that the pulsed constrictions in twist mutants required snail expression (Fig. 4a, Video 6). Thus, the expression of snail, not twist, initiates the actin-myosin network contractions that power constriction pulses.
Figure 4. Snail and Twist function at distinct phases of pulsed constriction.
a, Timelapse images of Myosin-GFP Z-projections. Blue arrows indicate myosin spots that do not efficiently coalesce in snail mutants. Red arrows indicate myosin coalescence in twist mutants that appears to pull cell junctions. At least one coalescence event that pulled cell junctions occurred over a 6 minute period for 53 % of cells in the twist mutant compared to 4 % of cells in snail and snail twist mutants (n = 60 cells, 3 embryos per mutant). Scale bar = 4 μm. b, Timelapse images of Spider-GFP in snailRNAi or twistRNAi embryos. P[sna] indicates twist independent snail expression. Scale bar = 4 μm. c, Quantification of apical area (red) and constriction rate (blue) for individual cells in snailRNAi or twistRNAi embryos. d, Ratchet model of apical constriction. Myosin (green) contracts an apical actin network (red) that is coupled to adherens junctions (blue) driving constriction. Contractions are pulsed, interrupted by a phase in which the constricted state of the cell is stabilized.
Net apical constriction was inhibited in both snailRNAi and twistRNAi embryos (Supplemental Fig. 4b). Therefore, we wondered why the pulsed contractions that we observed in twistRNAi embryos failed to constrict cells. Using Spider-GFP to visualize cell outlines, we found that while constriction pulses were inhibited in snailRNAi embryos, constriction pulses still occurred in twistRNAi embryos (Fig. 4b, 4c, Supplemental Fig. 4c, Video 8). However, the constricted state of cells in twistRNAi embryos was not stabilized between pulses, resulting in fluctuations in apical area with little net constriction (Fig. 4b, 4c). This stabilization defect was not due to lower snail activity because these fluctuations continued when snail expression was driven independent of twist using the P[sna] transgene (Fig. 4b)20. While the frequency and magnitude of constriction pulses in such embryos were similar to control embryos, stretching events were significantly higher in twistRNAi; P[sna] embryos, suggesting a defect in maintaining cortical tension (Supplemental Fig. 4d). This defect might result from a failure to establish a dense actin meshwork, because both twist mutants and twistRNAi embryos had a more loosely arranged apical meshwork of actin spots and fibers than constricting wild-type cells (Supplemental Fig. 4e). Therefore, twist expression stabilizes the constricted state of cells between pulsed contractions.
In conclusion, we propose a ‘ratchet’ model for apical constriction in which phases of actin-myosin network contraction and stabilization are repeated to incrementally constrict the cell apex (Fig. 4d). In contrast to the purse-string model, we find that apical constriction is correlated with pulses of actin-myosin network contraction that occur on the apical cortex. Pulsed cortical contractions could allow dynamic rearrangements of the actin network to optimize force-generation as cells change shape. Because contractions are asynchronous, cells must resist pulling forces from adjacent cells between contractions. A cortical actin-myosin meshwork appears to provide the cortical tension necessary to stabilize apical cell shape and promote net constriction. The transcription factors Snail and Twist are most important for the contraction and stabilization phases of constriction, respectively. Thus Snail and Twist activities are temporally coordinated to drive productive apical constriction. Remarkably, despite the dynamic nature of the contractions in individual cells, the behavior of the system at the tissue-level is continuous, similar to convergent extension in Xenopus25. Therefore, pulsed contraction may represent a conserved cellular mechanism that drives precise tissue-level behavior.
METHODS SUMMARY
Image acquisition and analysis
Two-color imaging was performed at room temperature using a Leica SP5 confocal microscope, 63×/1.3 NA glycerin objective, an Argon ion laser, and a 561 nm Diode laser. Spider-GFP images represent confocal slices 2 μm below the apical cortex whereas myosin images represent maximum intensity Z-projections of an apical section 5 μm in depth. Using MATLAB (MathWorks, Natick, MA), we developed software to track cells and measure apical area and myosin intensity. Data points were smoothed using a Gaussian smoothing filter with sigma = 15–18 s (3 time points). Myosin intensity was measured from maximum intensity Z-projections as the sum intensity of all pixels in a cell. Mean myosin intensity was calculated for junctional and medial pools of myosin by creating masks that selected regions < 0.3 μm or > 0.3 μm from the cell boundary, respectively.
Embryo fixation and staining
Heat fixation and staining with anti-myosin heavy chain (MHC) antibody did not preserve the normal organization of apical myosin observed in live squ-GFP26, squ-mCherry (Myosin-mCherry), and GFP-zipper (GFP-MHC)27 embryos. Therefore, endogenous GFP fluorescence was used to visualize myosin. squ-GFP embryos were fixed with 10% paraformaldehyde/heptane for 20 minutes, manually devitellinized, stained with Alexa-568 Phalloidin (Invitrogen) to visualize actin, and mounted in AquaPolymount (Poysciences, Inc.).
Drug/RNAi injection
CytoD was injected laterally at mid-late cellularization with 0.5 mg/ml CytoD in 10 % DMSO (Calbiochem). Double stranded RNA against snail and twist (2 mg/ml) were injected laterally into freshly laid eggs that were incubated 2.5–3 hours before imaging gastrulation.
Methods
Fly stocks and genetics
Fluorescent fusion protein stocks Spider-GFP (III) (95-1)12, Myosin-GFP (II or III) (squAX3; squ-GFP, gift of Roger Karess)26, E-Cadherin-GFP (II) (ubi-DE-cad-GFP)13, and Moesin-GFP (III)28 are described in the indicated references. Myosin-mCherry (III) (squ-mCherryA11) was recombined with Spider-GFP or Moesin-GFP to generate Myosin-mCherry Spider-GFP/TM3 and Myosin-mCherry Moesin-GFP. Homozygous Myosin-mCherry Spider-GFP flies could not be maintained as a stock. Therefore, Myosin-mCherry Spider-GFP/Myosin-mCherry flies were used for two-color imaging. The dynamics of Myosin-mCherry and Spider-GFP observed in the two-color strain are indistinguishable from the dynamics of Myosin-GFP and Spider-GFP alone. This suggests that the behavior of myosin we observe with 2-color imaging reflects normal myosin dynamics. E-Cadherin-GFP Myosin-mCherry flies used for imaging were ubi-DE-cad-GFP shgR69/CyO; squ-mCherryM1.
Strategies were used to visualize both homozygous and hemizygous twist and snail mutants. First, the squ-GFP transgene was jumped onto the CyO balancer, generating CyO-Myosin-GFP. Chromosomes containing snaIIG05, twiey53, and snaIIG05 twiey53 were marked with Df(2L)dpp[s7-dp35] 21F1–3;22F1–2 (halo) to allow homozygous embryos to be distinguished from their heterozygous siblings. We then rebalanced twist and snail mutants to obtain halo twiey53/CyO-Myosin-GFP, halo snaIIG05/CyO-Myosin-GFP, and halo snaIIG05 twiey53/CyO-Myosin-GFP. Homozygous mutants were selected by identifying the halo mutant phenotype. Myosin accumulation in ventral cells was delayed in twist and snail mutants, occurring after cephalic furrow initiation, in contrast to wild-type embryos, where myosin appears before the cephalic furrow is observed. The results presented are representative from 9 halo snaIIG05 movies, 6 halo twiey53 movies, and 3 halo snaIIG05 twiey53 movies.
Alternatively, a compound 2nd chromosome stock homozygous for Myosin-GFP (III) (C(2)v; Myosin-GFP) was used. In the C(2)v stock, the right arms (containing twist) and the left arms (containing halo and snail) of chromosome 2 assort independently. For snail mutants C(2)v; Myosin-GFP was crossed to halo snaIIG05/CyO and halo embryos were identified. For twist mutants, C(2)v; Myosin-GFP was crossed to halo twiey53/CyO and 1/3 of the non-halo progeny were twist mutants. Both strategies identified the distinct phenotypes for snail and twist mutants described in Fig. 4a.
To provide twist-independent snail expression we used the P[snac] (referred to as P[sna]) transgene, which contains the snail cDNA downstream of the proximal element (2×) of the twist promoter20. One zygotic copy of P[snac] was sufficient to induce invagination in a snail mutant background, indicating that it provides enough activity to rescue snail-dependent contraction.
Construction of Myosin-mCherry
Myosin regulatory light chain (MRLC, spaghetti squash, or squ) including its native promoter was tagged at the C-terminus with mCherry. A 2 kb genomic fragment containing the squ open reading frame and promoter was inserted into the KpnI and SalI sites of pBluescript. The squ 3′ UTR (800 bp) was then cloned into the downstream BamHI and XbaI sites. The sequence for mCherry, including a short linker region, was cloned into the ClaI and BamHI sites in between the squ ORF and 3′ UTR. The 3.5 kb KpnI/XbaI fragment containing myosin-mCherry was then cloned into pCasPer4 and sent to BestGene Inc. to make transgenic flies. Myosin-mCherry complemented the null squAX3 allele demonstrating that it is functional.
Timelapse image acquisition
Egg collections were performed in plastic cups covered with apple juice plates. Flies were allowed to lay eggs for 2–4 hrs at 25°C before removing the plate and collecting embryos undergoing cellularization. Embryos were dechorionated with 50 % bleach, washed with water, and were then mounted on a slide with embryo glue (Scotch tape resuspended in heptane) with the ventral side facing up. A chamber was made using two No. 1.5 coverslips as spacers and was filled with Halocarbon 27 oil for imaging. Embryos were not compressed. Mesoderm invagination occurred with a timeframe similar to that deduced from fixed embryos, and embryos imaged under these conditions could subsequenty hatch, demonstrating that our imaging conditions had minimal impact on development.
Single-color images of Spider-GFP and two-color images Myosin-mCherry Spider-GFP were obtained using a Leica SP5 confocal microscope, 63×/1.3 NA glycerin objective, an Argon ion laser, and a 561nm Diode laser. Images were aquired using a pinhole setting of two Airy units. For simultaneous two-color images, we set the excitation bandpass to 495–550 nm to detect GFP and 578–650 nm to detect mCherry. There was minimal bleadthrough between the two channels. Images were acquired using at a resolution of 141 nm/pixel. Myosin-GFP images were obtained using a PerkinElmer Ultraview spinning disk confocal, 60×/1.2 NA water objective, Argon/Krypton laser, and Orca ER 4 (Hamamatsu) CCD camera.
Image processing and analysis
The images presented were processed using ImageJ (http://rsb.info.nih.gov/ij/) and Adobe Photoshop CS (Adobe Systems, Inc., San Jose, CA). Unless otherwise noted, Spider-GFP images represent a single confocal slice 2 μm below the apical cortex whereas Myosin-GFP, Myosin-mCherry, and Moesin-GFP represent maximum intensity z-projections 5 μm in depth. A gaussian smoothing filter with a radius of 1 pixel was used to reduce noise in published images.
To visualize apical cell shape, we manually selected Z-slices at a depth that was ~2 μm below the apical surface and the apical myosin. Cell outlines at 2 μm depth were very similar in dimensions to more apical cell outlines, however they were easier to visualize and lacked membrane irregularities (i.e. in Fig. 3c) that complicated image analysis. Shifts in Z-position did not result in discontinuities in the data (Fig. 1e) and the pulsed behavior that we describe was also observed when a single Z-slice was used.
We developed software to measure apical area, constriction rate, and myosin intensity using MATLAB (MathWorks, Natick, MA). Raw images were bandpass filtered with effective cutoff wavelengths of 1.4 μm (low pass) and 17.9 μm (high pass). After thresholding, a series of morphological operations was applied to extract cell outlines (Supplemental Figure 1). Indexed cells were automatically tracked based on distances between cell centroids at subsequent time points. Cell properties were measured at each time point. We manually removed cells with errors in the segmentation to ensure that all cells in the dataset were correctly identified. Data for apical area and myosin were smoothed using a gaussian smoothing filter (sigma = 15–18 s, 3 time points) and constriction rates and myosin rates were calculated from the smoothed data. Unless otherwise stated, a constriction pulse was defined as an event where the constriction rate exceeded one standard deviation above the mean (>10.8 μm2/min). To measure myosin intensity, we first clipped intensity values below two standard deviations above the mean to separate myosin structures from unselective background labeling. Myosin intensity was then measured from maximum intensity Z-projections (two highest values averaged) as the sum intensity of all pixels in a given cell. The correlation between constriction rate and the myosin intensity rate of change was determined by calculating the correlation coefficients between these two data sets for individual cells. To examine the time-dependence of this correlation, the data for constriction rate and myosin rate of change were shifted in time relative to one another.
Furrow depth (Fig. 1e) was calculated by measuring the distance between vitelline membrane and apical cortex in YZ cross-sections.
Myosin-GFP spot velocity was measured using the Manual Tracking plugin for ImageJ. We tracked individual spots or other distinct myosin structures for the duration of their lifetime and averaged the 3 highest velocities to calculate maximum velocity. For wild-type movies, we calculated the velocity of myosin structures coalescing. We chose coalescence events that occurred early or occurred near the midline to minimize the effects of tissue movement during invagination.
Drug injection
Embryos were dechorionated in 50% bleach for 2 min, washed with water, mounted on the edge of a glass slide (ventral side up) using embryo glue, and dessicated for 4–8 minutes. A 3:1 mixture of halocarbon 700:halocarbon 27 was used for injection. Embryos were injected laterally at mid-late cellularization (furrow canals had passed the base of the nuclei) with ~1% egg volume of control or drug solution. We injected 0.5 mg/ml CytochalasinD (Calbiochem) in 10% DMSO.
RNAi
Primers for dsRNA were designed using E-RNAi (http://www.dkfz.de/signaling2/e-rnai/)29. Primers included the sequence of the T7 promoter (TAATACGACTCACTATAGGG) followed by the following recognition sequences:
Twi01-F: GCCAAGCAAGATCACCAAAT
Twi01-R: GACCTCGTTGCTGGGTATGT
Twi02-F: GGAGCTGCAGAACAATGTGA
Twi02-R: TGCTGTTGCTGGTGGATTAG
Sna01-F: CGGAACCGAAACGTGACTAT
Sna01-R: GCGGTAGTTTTTGGCATGAT
Sna02-F: ATCATGCCAAAAACTACCGC
Sna02-R: AGCGACATCCTGGAGAAAGA
Control-F: GAATGCTATGGGAGGCGATA
Control-R: TCAGCTTAGGCTCTGGGTGT
Primer pairs were used to amplify a PCR product from genomic DNA. PCR products were directly used in a transcription reaction with T7 polymerase using the MEGAscript transcription kit (Ambion). Annealing was performed by adding 10mM EDTA, 0.1% SDS, and 0.1M NaCl to the reaction and incubating this mixture in a water bath heated to above 90°C, which was allowed to cool for several hours. The dsRNA was purified by phenol/chloroform extraction and resuspended in injection buffer (5mM KCl, 0.1mM NaPhosphate, pH 7.0). We injected a 2 mg/ml solution of snail or twist dsRNA into the embryo. Identical results were obtained with either dsRNA fragment used to knock-down twist or snail. Sna01 and Twi01 were used for all the experiments in the manuscript, with the exception of Supplemental Fig. 4e, in which Twi02 was used. Control primers amplified a portion of an unknown ORF CG3651, which has no function during ventral furrow formation. Injections were performed as described for the drug injections, except egg collections were performed after 30 minutes to inject embryos at the earliest possible stage. To provide twist-independent snail expression we crossed Spider-GFP virgin females to P[snac]20 males and collected embryos for twist dsRNA injection.
Statistics
Statistical significance between means was determined using an unpaired t-test. P-values were calculated using Prism 5 (Graphpad Software, Inc.).
Supplementary Material
Video 1. Ventral furrow apical constriction visualized with Spider-GFP. The focal plane was adjusted to stay 2 μm below the apical surface along the ventral midline. Time interval between acquired frames was 6 s and the acquisition length was 8 minutes. Video is 36× faster than real-time.
Video 2. Pulsed constriction of ventral furrow cells. Cell outlines are labeled with Spider-GFP. Time interval between acquired frames was 6 s and the acquisition length was 5 minutes. Video is 72× faster than real-time.
Video 3. Pulsed constriction is asynchronous. The color of the cell indicates the constriction rate (as illustrated by the colorbar in Fig. 1h) for a given time point. Time interval between acquired frames was 6s and the acquisition length was 6 minutes. Video is 18× faster than real-time.
Video 4. Apical myosin dynamics during ventral furrow cell apical constriction. Z-projections of Myosin-mCherry (green, 5 μm depth) are merged with a single Spider-GFP z-slice located 2 μm below the apical cortex (red). Time interval between acquired frames was 5 s and the acquisition length was 7.5 minutes. Video is 25× faster than real-time.
Video 5. Myosin coalescence accompanies apical constriction. Z-projections of Myosin-mCherry (green, 5 μm depth) are merged with a single Spider-GFP z-slice located 2 μm below the apical cortex (red and bottom). Time interval between acquired frames was 6 s and the acquisition length was 2.6 minutes. Video is 50× faster than real-time.
Video 6. Twist and Snail differentially regulate myosin dynamics. Z-projections of Myosin-GFP (5 μm depth) for the indicated genetic backgrounds. Note that myosin coalescence is more prominent in twist mutant embryos than snail mutant embryos. This results in more movement of myosin structures since they are being pulled. Time interval between acquired frames was 10 s and the acquisition length was 16.7 minutes. Video is 100× faster than real-time.
Video 7. twistRNAi and snailRNAi mimic the mutant phenotypes. Z-projections of Myosin-GFP for the indicated knock-downs. Time interval between acquired frames was 10 s and the acquisition length was 16.7 minutes. Video is 100× faster than real-time. Note that the phenotypes are not as severe as the null mutants due to remaining Twist and Snail activity.
Video 8. Constriction pulses in twistRNAi embryos are not stabilized. Cell outlines 2 μm below the apical cortex are labeled using SpiderGFP. Cell shape is relatively constant in snailRNAi embryos, while fluctuations in cell size and shape occur in twistRNAi embryos. Time interval between acquired frames was 6 s and the acquisition length was 7 minutes. Video is 36× faster than real-time.
Acknowledgments
E.F.W. is an investigator of the Howard Hughes Medical Institute. We thank Dan Kiehart and Roger Karess for providing flies; Joe Goodhouse for assisting with microscopy; and Stefano De Renzis, Xuemin Lu, Trudi Schupbach, Anna Sokac, Florian Ulrich, and Yu-Chiun Wang for helpful comments on the manuscript. This work is supported by grant #PF-06-143-01-DDC from the American Cancer Society to A.C.M., NIH/NIGMS P50 GM071508 to M.K., and by National Institute of Child Health and Human Development grant 5R37HD15587 to E.F.W.
Footnotes
Author Contributions
Biological reagents and fly stocks were made by A.C.M. and E.F.W. and experiments were performed by A.C.M. Image analysis methods were developed by M.K. and the live-imaging data were analyzed by A.C.M. and M.K. The first draft of the manuscript was written by A.C.M. All authors participated in discussion of the data and in producing the final version of the manuscript.
References
- 1.Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol. 2007;8:633–44. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 2.Leptin M. Gastrulation movements: the logic and the nuts and bolts. Dev Cell. 2005;8:305–20. doi: 10.1016/j.devcel.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Alberts B, et al. Molecular Biology of the Cell. Garland Science; New York, NY: 2002. [Google Scholar]
- 4.Baker PC, Schroeder TE. Cytoplasmic filaments and morphogenetic movement in the amphibian neural tube. Dev Biol. 1967;15:432–50. doi: 10.1016/0012-1606(67)90036-x. [DOI] [PubMed] [Google Scholar]
- 5.Burnside B. Microtubules and microfilaments in newt neuralation. Dev Biol. 1971;26:416–41. doi: 10.1016/0012-1606(71)90073-x. [DOI] [PubMed] [Google Scholar]
- 6.Hildebrand JD. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J Cell Sci. 2005;118:5191–203. doi: 10.1242/jcs.02626. [DOI] [PubMed] [Google Scholar]
- 7.Karfunkel P. The activity of microtubules and microfilaments in neurulation in the chick. J Exp Zool. 1972;181:289–301. doi: 10.1002/jez.1401810302. [DOI] [PubMed] [Google Scholar]
- 8.Dawes-Hoang RE, et al. folded gastrulation, cell shape change and the control of myosin localization. Development. 2005;132:4165–78. doi: 10.1242/dev.01938. [DOI] [PubMed] [Google Scholar]
- 9.Fox DT, Peifer M. Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development. 2007;134:567–78. doi: 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- 10.Nikolaidou KK, Barrett K. A Rho GTPase signaling pathway is used reiteratively in epithelial folding and potentially selects the outcome of Rho activation. Curr Biol. 2004;14:1822–6. doi: 10.1016/j.cub.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 11.Young PE, Pesacreta TC, Kiehart DP. Dynamic changes in the distribution of cytoplasmic myosin during Drosophila embryogenesis. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–5. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. J Cell Sci. 2001;114:493–501. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- 14.Sweeton D, Parks S, Costa M, Wieschaus E. Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development. 1991;112:775–89. doi: 10.1242/dev.112.3.775. [DOI] [PubMed] [Google Scholar]
- 15.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 16.Verkhovsky AB, Svitkina TM, Borisy GG. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol. 1997;139:397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–63. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolsch V, Seher T, Fernandez-Ballester GJ, Serrano L, Leptin M. Control of Drosophila gastrulation by apical localization of adherens junctions and RhoGEF2. Science. 2007;315:384–6. doi: 10.1126/science.1134833. [DOI] [PubMed] [Google Scholar]
- 20.Ip YT, Maggert K, Levine M. Uncoupling gastrulation and mesoderm differentiation in the Drosophila embryo. Embo J. 1994;13:5826–34. doi: 10.1002/j.1460-2075.1994.tb06926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–76. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 22.Leptin M, Grunewald B. Cell shape changes during gastrulation in Drosophila. Development. 1990;110:73–84. doi: 10.1242/dev.110.1.73. [DOI] [PubMed] [Google Scholar]
- 23.Seher TC, Narasimha M, Vogelsang E, Leptin M. Analysis and reconstitution of the genetic cascade controlling early mesoderm morphogenesis in the Drosophila embryo. Mech Dev. 2007;124:167–79. doi: 10.1016/j.mod.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Costa M, Wilson ET, Wieschaus E. A putative cell signal encoded by the folded gastrulation gene coordinates cell shape changes during Drosophila gastrulation. Cell. 1994;76:1075–89. doi: 10.1016/0092-8674(94)90384-0. [DOI] [PubMed] [Google Scholar]
- 25.Keller R, Shook D, Skoglund P. The forces that shape embryos: physical aspects of convergent extension by cell intercalation. Phys Biol. 2008;5:15007. doi: 10.1088/1478-3975/5/1/015007. [DOI] [PubMed] [Google Scholar]
- 26.Royou A, Sullivan W, Karess R. Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. J Cell Biol. 2002;158:127–37. doi: 10.1083/jcb.200203148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franke JD, Montague RA, Kiehart DP. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15:2208–21. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 28.Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP. GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol. 1997;191:103–17. doi: 10.1006/dbio.1997.8707. [DOI] [PubMed] [Google Scholar]
- 29.Arziman Z, Horn T, Boutros M. E-RNAi: a web application to design optimized RNAi constructs. Nucleic Acids Res. 2005;33:W582–8. doi: 10.1093/nar/gki468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1. Ventral furrow apical constriction visualized with Spider-GFP. The focal plane was adjusted to stay 2 μm below the apical surface along the ventral midline. Time interval between acquired frames was 6 s and the acquisition length was 8 minutes. Video is 36× faster than real-time.
Video 2. Pulsed constriction of ventral furrow cells. Cell outlines are labeled with Spider-GFP. Time interval between acquired frames was 6 s and the acquisition length was 5 minutes. Video is 72× faster than real-time.
Video 3. Pulsed constriction is asynchronous. The color of the cell indicates the constriction rate (as illustrated by the colorbar in Fig. 1h) for a given time point. Time interval between acquired frames was 6s and the acquisition length was 6 minutes. Video is 18× faster than real-time.
Video 4. Apical myosin dynamics during ventral furrow cell apical constriction. Z-projections of Myosin-mCherry (green, 5 μm depth) are merged with a single Spider-GFP z-slice located 2 μm below the apical cortex (red). Time interval between acquired frames was 5 s and the acquisition length was 7.5 minutes. Video is 25× faster than real-time.
Video 5. Myosin coalescence accompanies apical constriction. Z-projections of Myosin-mCherry (green, 5 μm depth) are merged with a single Spider-GFP z-slice located 2 μm below the apical cortex (red and bottom). Time interval between acquired frames was 6 s and the acquisition length was 2.6 minutes. Video is 50× faster than real-time.
Video 6. Twist and Snail differentially regulate myosin dynamics. Z-projections of Myosin-GFP (5 μm depth) for the indicated genetic backgrounds. Note that myosin coalescence is more prominent in twist mutant embryos than snail mutant embryos. This results in more movement of myosin structures since they are being pulled. Time interval between acquired frames was 10 s and the acquisition length was 16.7 minutes. Video is 100× faster than real-time.
Video 7. twistRNAi and snailRNAi mimic the mutant phenotypes. Z-projections of Myosin-GFP for the indicated knock-downs. Time interval between acquired frames was 10 s and the acquisition length was 16.7 minutes. Video is 100× faster than real-time. Note that the phenotypes are not as severe as the null mutants due to remaining Twist and Snail activity.
Video 8. Constriction pulses in twistRNAi embryos are not stabilized. Cell outlines 2 μm below the apical cortex are labeled using SpiderGFP. Cell shape is relatively constant in snailRNAi embryos, while fluctuations in cell size and shape occur in twistRNAi embryos. Time interval between acquired frames was 6 s and the acquisition length was 7 minutes. Video is 36× faster than real-time.