Summary
Precise regulation of neurogenesis is achieved in specific regions of the vertebrate nervous system by formation of distinct neurogenic and non-neurogenic zones. We have investigated how neurogenesis becomes confined to zones adjacent to rhombomere boundaries in the zebrafish hindbrain. The non-neurogenic zone at segment centres comprises a distinct progenitor population that expresses fgfr2, erm, sox9b and the retinoic acid degrading enzyme cyp26b1. FGF receptor activation upregulates expression of these genes and inhibits neurogenesis in segment centres. Cyp26 activity is a key effector inhibiting neuronal differentiation, suggesting antagonistic interactions with retinoid signaling. We identify the critical FGF ligand, fgf20a, which is expressed by specific neurons located in the mantle region at the centre of segments, adjacent to the non-neurogenic zone. Fgf20a mutants have ectopic neurogenesis and lack the segment centre progenitor population. Our findings reveal how signaling from neurons induces formation of a non-neurogenic zone of neural progenitors.
Keywords: Neurogenesis, neural progenitors, neurogenic zone, FGF, feedback inhibition, retinoic acid
INTRODUCTION
During development of the vertebrate central nervous system, progenitor cells that comprise the neural epithelium differentiate to form a wide variety of neuronal and glial cell types at appropriate locations and in correct numbers. The specific cell types formed are regulated spatially by signals that underlie subdivision of the neural epithelium into domains with distinct identity or that specify distinct subtypes within these regions. Different neuronal and glial cell types are born at different times, with switches in the differentiation of a progenitor population to generate, for example, initially neurons and subsequently glial cells (Guillemot, 2007). This generation of differentiated cells over a prolonged period necessitates that sufficient neural progenitor cells are maintained throughout development. The differentiation of the correct number of neurons and maintenance of progenitors are regulated by extrinsic and intrinsic factors that promote or inhibit neurogenesis, and that regulate the proliferation of neural epithelial cells (Bertrand et al., 2002; Diez del Corral and Storey, 2001).
Neurogenesis is initiated by the upregulation of proneural transcription factors that trigger a downstream cascade of genes that control further steps of neuronal differentiation (Bertrand et al., 2002). The amount of neurogenesis is limited by a number of factors that inhibit the upregulation or the activity of proneural proteins. Some inhibitory factors are widely expressed in the neural epithelium, for example members of the SoxB1 (Sox1, Sox2, Sox3) family of proteins that have a major role in maintenence of neural progenitor cells (Bylund et al., 2003; Graham et al., 2003). Another crucial regulation of neurogenesis occurs through local cell-cell signaling in the process of lateral inhibition. Proneural genes upregulate the expression of Notch ligands, which by activating the Notch receptor in adjacent cells upregulate Hes/Her transcriptional repressors that inhibit neurogenesis (Fisher and Caudy, 1998). This Notch-mediated lateral inhibition ensures that progenitor cells are maintained within regions in which neurogenesis is occurring.
A further mechanism to regulate neurogenesis occurs in some regions of the neural epithelium in which there is a large scale spatial organisation of neurogenic and non-neurogenic zones. Such patterning has been found to be generated by spatially-restricted inhibitory mechanisms that confine neurogenesis to specific regions (Bally-Cuif and Hammerschmidt, 2003; Diez del Corral and Storey, 2001). For example, neurogenesis does not occur at the midbrain-hindbrain boundary, or in interproneuronal domains along the dorsoventral axis of the spinal cord, due to Notch-independent expression of specific Hes/Her genes (Bae et al., 2005; Geling et al., 2003; Geling et al., 2004) or to the expression of members of the Zic gene family (Brewster et al., 1998).
An important question is the identity of extrinsic signaling factors that regulate the promotion or inhibition of neurogenesis. A number of signals have been implicated in the control of neurogenesis, including retinoic acid (RA), Wnt, BMP, Shh and FGF family members (Dono, 2003; Ford-Perriss et al., 2001; Maden, 2007; Michaelidis and Lie, 2008; Pozniak and Pleasure, 2006; Sharpe and Goldstone, 2000; Sharpe and Goldstone, 1997; Shi et al., 2008; Wu et al., 2003). In some cases, these signals are expressed in specific centres within the neural epithelium or adjacent tissues and are involved in a coordinated regulation of neurogenesis and patterning along the anterior-posterior or dorsal-ventral axis. These factors can act in a cooperative or antagonistic manner, for example the promotion of neurogenesis by RA being opposed by FGF signaling in the caudal spinal cord (Diez del Corral et al., 2003; Diez del Corral and Storey, 2004). However, for many of these signals, their relationship with neurogenesis is complex and context-dependent; for example, FGF signaling has been implicated in the self renewal of neural stem cells (Gage et al., 1995; Gritti et al., 1996), and in the promotion or inhibition of neurogenesis (Borello et al., 2008; Ford-Perriss et al., 2001; Hardcastle et al., 2000; Topp et al., 2008).
Previous studies have shown that in the zebrafish hindbrain, neurogenesis becomes confined to zones that flank segment boundaries, and does not occur in the central region of each segment (Amoyel et al., 2005; Cheng et al., 2004). We set out to investigate how this stereotyped pattern of neurogenic and non-neurogenic progenitor zones is established. We report that FGFR activation occurs in the centre of segments, where it upregulates multiple genes including fgfr2, erm and sox9b, and is essential for the inhibition of neurogenesis. FGFR activation inhibits neurogenesis in part by upregulating the expression of an RA-degrading enzyme, Cyp26b1. We identify fgf20a as the critical activator of FGFR essential for the inhibition of neurogenesis and find that it is expressed by a subset of neurons located at segment centres. These findings reveal a mechanism for the establishment of non-neurogenic zones, in which FGF signaling from neurons inhibits neurogenesis in the adjacent neural epithelium.
MATERIAL AND METHODS
Zebrafish strains, husbandry and genotyping
Zebrafish embryos were staged according to hours post-fertilization (hpf) and morphological criteria (Kimmel et al., 1995). Fgf20a (Devoid of blastema, dob) mutant embryos (Whitehead et al., 2005) were obtained from homozygous fgf20a incrosses and raised at 25°C from 3 hpf until fixation. Transgenic embryos Tg(hsp70l:dnfgfr1-EGFP) and Tg(hsp70:ca-fgfr1) used for this work are heterozygotes from outcrosses. Tg(hsp70l:dnfgfr1-EGFP) and Tg(hsp70:ca-fgfr1) adult carriers were identified as described (Lee et al., 2005; Marques et al., 2008). Tg(hsp70l:dnfgfr1-EGFP) embryos were identified after heat shock by the fluorescence of the fgfr1-EGFP fusion protein. To confirm that phenotypes observed in embryos derived from Tg(hsp70:ca-fgfr1) zebrafish correlated with presence of the transgene, individual embryos were genotyped after in situ hybridisation. Briefly, after photographing embryos mounted in glycerol, they were washed overnight in PBT at 4°C, incubated in 50 µl TE for 10 min at 98°C, and then overnight at 55°C in TE containing 200 µg/ml proteinase K. After inactivation for 10 min at 98°C, genomic DNA was used for PCR amplification to detect Dsred transgene with the primers CATCCTGTCCCCCCAGTTCC and CCCAGCCCATAGTCTTCTTCTGC.
Heat shock and pharmacological treatments
To inducibly block FGFR activation, Tg(hsp70l:dnfgfr1-EGFP) embryos and wild type littermates were heat shocked for 30 min at 38.5°C. Treatments were started at the 22-somite stage and embryos fixed 24 h later. To induce constitutively active FGFR using Tg(hsp70:ca-fgfr1), 24 hpf embryos were heat shocked for 30 min at 38.5°C, transfered for 2 h at 28.5°C and then fixed. To pharmacologically block FGFR, 22-somite embryos were incubated for 24 h in 100 µM SU5402 (Calbiochem) or an equivalent dilution of DMSO carrier as control. To block Cyp26 activity, 24–26 hpf zebrafish embryos were treated with 50 µM R115866 (Janssen Pharmaceutica) for 24 h, or with equivalent dilutions of DMSO as control. To block RA signaling, 50 µM DEAB was added to 1–3 somite embryos. To block Cyp26 activity in DEAB treated embryos, R115866 was added at 50 µM at 8-somites and embryos fixed at 36 h. Embryos were in their chorion for all drug treatments.
In situ hybridization
Whole mount in situ hybridization was carried out as described (Xu and Wilkinson, 1998) with the following modifications. Embryos were prehybridised in hybridisation mix (50 % formamide, 5 × SSC, pH 4.5, 50 µg/ml yeast RNA, 100 µg/ml heparin, 0.2 % Tween20, 5 mM EDTA) for 2 h at 68°C. Hybridization was carried out at 68°C overnight, then the following washes carried out at 68°C: 5 min in 66 % formamide / 33% 2 × SSC; 5 min in 33% formamide / 66% 2 × SSC, 5 min in 2× SSC, 0.1% Tween20; 15 min in 0.2 × SSC, 0.1% Tween20; twice for 15 min in 0.1 × SSC, 0.1% Tween20. Final washes at room temperature were for 5 min each in 66% 0.1 × SSC, 33% PBT, then 33% 0.1 × SSC, 66% PBT and then PBT. For digoxigenin detection, embryos were blocked in 5% sheep serum and then incubated with anti-digoxigenin-AP antibody (Roche) (1:1500 dilution) at 4°C overnight. Finally, embryos were washed all day and overnight in PBT at room temperature and color developed with either NBT/BCIP (Roche) or Fast Red (Roche). Double fluorescent in situ hybridisation was performed as described (Julich et al., 2005), followed by detection using tyramide signal amplification (tyramide labelled with Alexa Fluor 488 or 594) following the manufacturer’s instructions (Molecular Probes). The following probes were used: erm (cb805), sox9b (MGC:76805), fgfr2 (gift of Ivor Mason), neurog1, neurod4, dla, and dld (Amoyel et al., 2005; Cheng et al., 2004), cyp26b1 (Hernandez et al., 2007), fgf20a (Whitehead et al., 2005), and sox3 (IMAGE clone id: 3726393). Photographs were taken using confocal microscopy (Leica TCS SP2).
Whole mount immunofluorescence
For whole mount antibody staining, embryos were fixed for 2 h at room temperature in 4% paraformaldehyde, rinsed in PBT, dechorionated and blocked for 1 h in 5% goat serum in PBT. Embryos were then incubated overnight at 4°C in the required antibody at the following dilutions: rabbit anti-Sox9 1:500 ((Morais da Silva et al., 1996); gift from Silvana Guioli), rabbit anti-EphA4 (1:450, (Irving et al., 1996)), mouse anti-HuC/D (1:100, Molecular Probes), mouse anti-neurofilament (1:25, Zymed), in 2.5% goat serum. Secondary goat antibodies used were Alexa Fluor conjugates (Invitrogen).
RESULTS
Expression of erm and fgfr2 is complementary to neurogenic zones
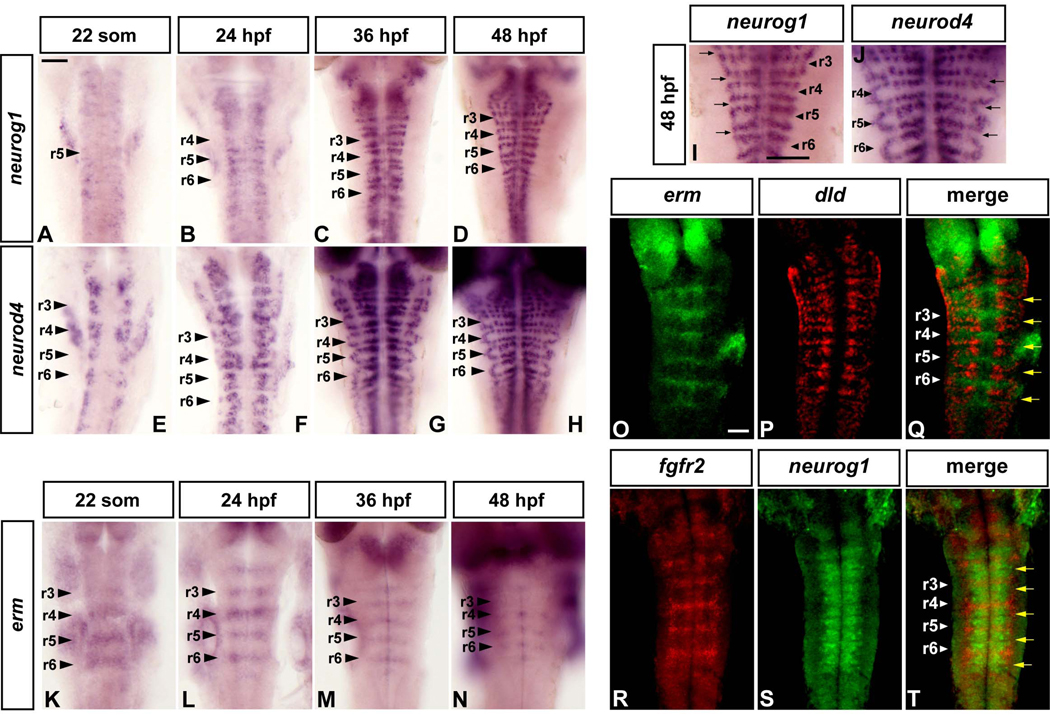
Previous studies have revealed that neurogenesis occurs in a dynamic pattern in the zebrafish hindbrain, initially at segment centres, then broadly throughout segments and later is restricted to zones flanking segment boundaries (Amoyel et al., 2005; Cheng et al., 2004). We further analysed the restriction of neurogenesis by detection of neurog1, dla and dld gene expression which mark the initiation of neurogenesis (Fig.1A–D, I, and data not shown), and neurod4 which is expressed downstream of proneural genes (Roztocil et al., 1997; Wang et al., 2003) (Fig.1E–H, J). We found that the widespread pattern of neurogenesis observed at the 22-somite stage (Fig.1A, E) becomes progressively restricted to zones adjacent to hindbrain boundaries, such that at 36 hpf and 48 hpf there is an absence of neuronal differentiation in the centre of rhombomeres (Fig.1C, D, G–J).
Figure 1. FGF signaling is restricted to non-neurogenic regions in the zebrafish hindbrain.
Dorsal views of flat mounted embryos, anterior to the top, at the indicated stages. Following in situ hybridisation, embryos of the same batch were developed for the same amount of time. Arrowheads indicate segment centres, and arrows point at hindbrain boundaries. All scale bars, 50 µm. (A–H): time course of neurog1 (A–D) and neurod4 (E–H) expression from 22 somites to 48 h. (I–J): Higher power views showing the spatial restriction of neurogenesis marked by neurog1 (same embryo as 1D) and neurod4 (1J, same embryo as 1H). (K–N): erm expression. (O–T): Double fluorescent in situ hybridization using probes for erm and dld (O–Q), and fgfr2 and neurog1 (R–T). Images shown are a merge of confocal stacks through the hindbrain at 36 hpf.
These observations raised the question of how neurogenesis becomes restricted during hindbrain development. We obtained clues in searches of publications and the ZFIN database (Thisse et al., 2004) for genes that have restricted expression within hindbrain segments. Among these genes, expression of two transcriptional targets of FGF signaling, erm and etv5, occurs within hindbrain segments at the 20–25-somite and later stages (Munchberg et al., 1999; Roussigne and Blader, 2006). We further analysed erm expression, and found that this occurs in segment centres from the 22-somite stage to 48 hpf, with the level of expression decreasing and becoming a narrower stripe at late stages (Fig.1K–N). To analyse the relationship with neurogenic zones, we carried out double fluorescent in situ hybridisations. We found that at 36 hpf erm is expressed in a complementary domain to dld, and thus marks the non-neurogenic zone in segment centres (Fig.1O–Q). Previous studies of fgfr gene expression in zebrafish have shown that in the hindbrain, fgfr1 is widely expressed, fgfr3 and fgfr4 are restricted to r1, and fgfr2 becomes upregulated in all segment centres (Tonou-Fujimori et al., 2002). We found that fgfr2 is progressively upregulated and restricted to segment centres from 24–48 hpf (Fig.S2A–D), and is complementary to neurog1 in neurogenic zones (Fig.1R–T). Taken together, these findings suggest that expression of fgfr2 and activation of the FGF pathway occurs in segment centres during the period that neurogenesis becomes restricted to zones adjacent to hindbrain boundaries. We therefore tested the possibility that FGF signaling is involved in the repression of neurogenesis in the hindbrain.
FGFR signaling inhibits neurogenesis in segment centres
Since fgfs have been implicated in early stages of hindbrain patterning (Maves et al., 2002; Walshe et al., 2002), analysis of a potential later role requires use a strategy that allows temporal control of FGFR inhibition or activation. To block FGFR activation, we used a transgenic heat shock inducible dominant-negative approach (Lee et al., 2005). A fluorescent signal due to expression of dn-FGFR1:EGFP fusion protein can be detected as early as 1 h after heat shock of Tg(hsp70l:dnfgfr1-EGFP) embryos and persists for at least 24 h (not shown). We found that following heat shock induction of dn-FGFR1 at the 22-somite stage, there is a rapid and persistent decrease in erm expression (not shown). It was important to ascertain that inhibition of FGFR at late stages does not phenocopy the loss of Fgf3/Fgf8, which are transiently expressed in rhombomere 4 and required for r5/r6 segmentation and correct formation of early-born reticulospinal neurons (Maves et al., 2002; Walshe et al., 2002). We found that induction of dn-FGFR1 at the 22-somite stage led to no change in segmental expression of EphA4 in r3/r5 or in formation of reticulospinal neurons (Fig.S1A–D).
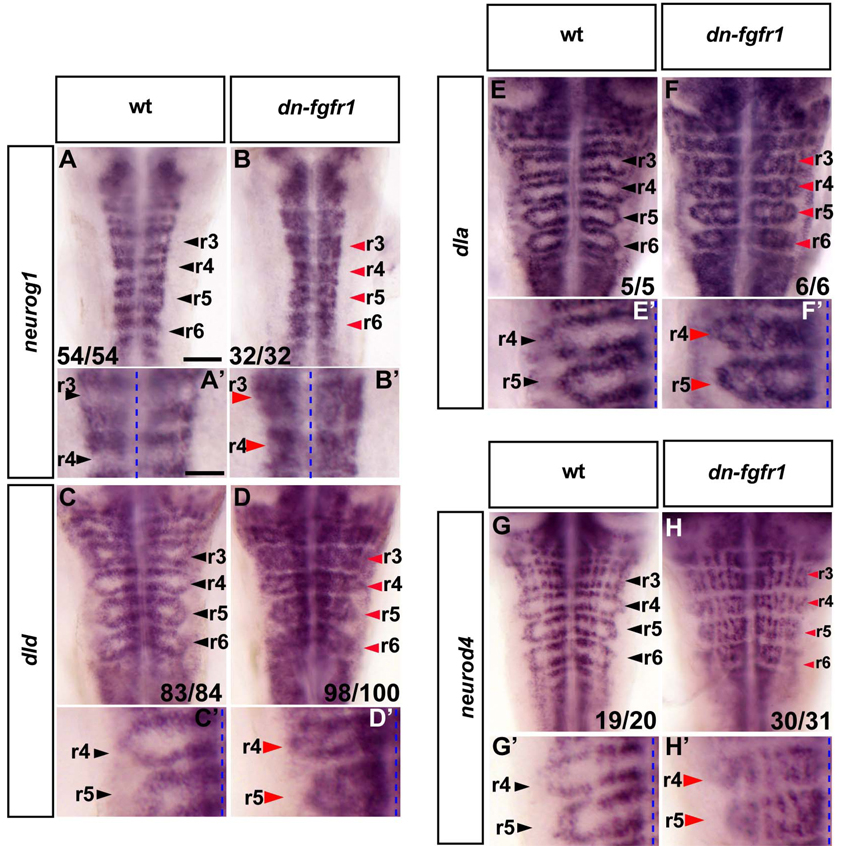
To test whether blocking FGFR activation affects neurogenesis, we analysed markers of the initiation and later steps of neuronal differentiation. To inhibit FGFR activation, we induced dn-FGFR1 expression by heat shock of 22-somite embryos for 30 min and then incubated for 24 h before fixation. We found that this led to ectopic expression of neurog1, dld, dla and neurod4 in the centre of segments (Fig.2A–H, A’–H’). In contrast, there was no ectopic expression in hindbrain boundaries, consistent with previous studies that have implicated Notch activation in the inhibition of neurogenesis (Cheng et al., 2004). Interestingly, not all cells in segment centres upregulate proneural genes following FGFR inhibition, but rather neurogenesis ocuurs in a punctate manner in longitudinal columns co-extensive with the normal sites of differentiation in neurogenic zones (Fig.2G’–H’). These observations suggest that the ectopic differentiation in segment centres is subject to dorsoventral patterning and lateral inhibition of neurogenesis as occurs in the normal neurogenic zones. Similar phenotypes were observed following treatment of embryos with the FGFR inhibitor SU5402 (Fig.S1E–H, G’, H’). Since FGFR2 is upregulated in segment centres, we carried out morpholino oligonucleotide mediated knockdowns to analyse whether it is required for the inhibition of neurogenesis. We did not detect any change in neurogenesis (data not shown), suggesting that other FGF receptors mediate sufficient levels of FGF signaling.
Figure 2. Blocking FGFR activation results in proneural gene expression and differentiating neurons in segment centres.
In situ hybridisation of 40 hpf zebrafish to detect proneural gene expression (neurog1, dld, dla; A–F and A’–F’) or differentiating neurons (neurod4; G–H and G’–H’) in either wild type (wt) or dominant negative fgfr1 embryos (Tg(hsp70l:dnfgfr1-EGFP)). Heat shock was started at the 22 somite stage. Black arrowheads indicate segment centres and red arrowheads indicate ectopic neurogenesis. Scale bar for A–H, 50 µm. (A’–H’): higher power view of images in A–H. Scale bar, 25 µm. Punctate line: midline. See also Figure S1.
Since FGF signaling can promote the proliferation of neural progenitors, we analysed whether blocking with dn-FGFR1 or inducible expression of constitutively active FGFR1 (Marques et al., 2008) affects cell proliferation in the hindbrain. We found that there was no significant change in the number of pH3-expressing cells after blocking endogenous FGFR activation, or increased FGFR1 activation (data not shown). The inhibition of neurogenesis by FGFR activation in segment centres is thus not associated with a role in the promotion of cell proliferation.
FGFR signaling regulates segment centre restricted gene expression
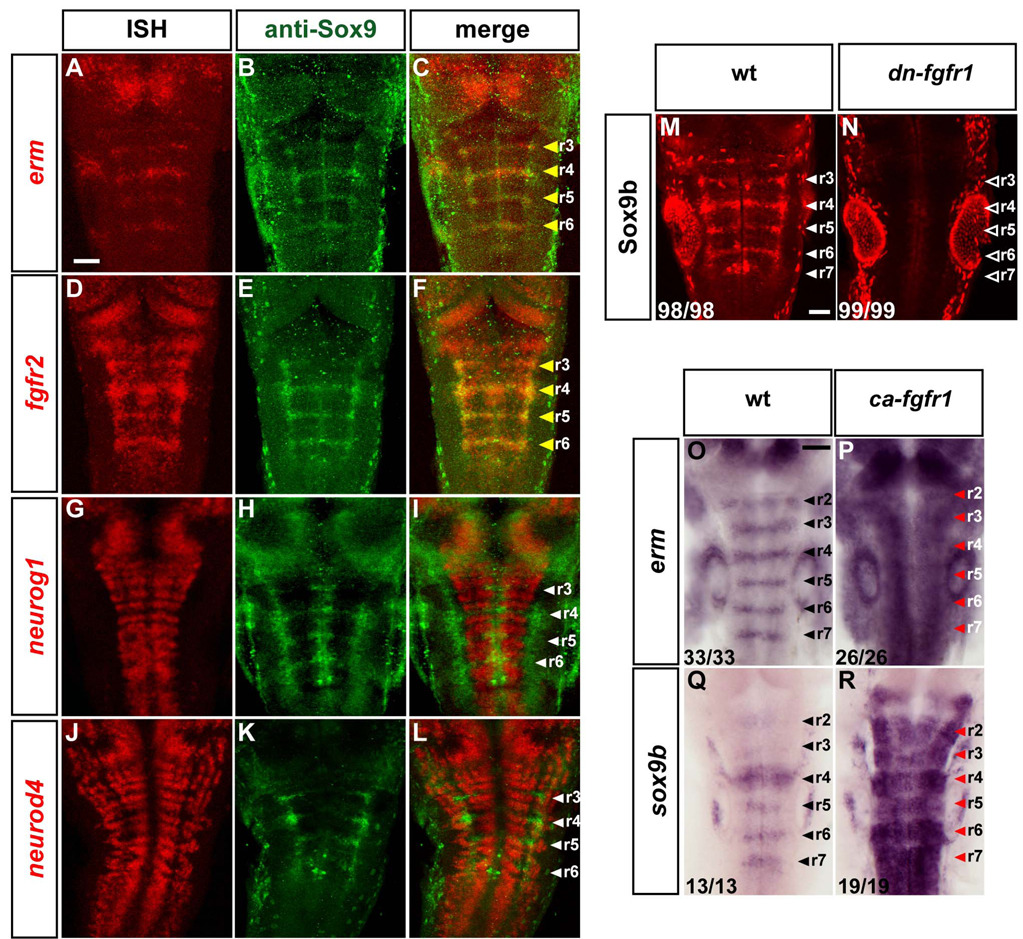
Our results raise the question of whether the inhibition of neurogenesis by FGFR signaling involves formation of progenitors in segment centres that have a distinct specification from those within neurogenic zones. Previous studies have shown that in other tissues Sox9 is upregulated by FGF signaling (Govindarajan and Overbeek, 2006; Murakami et al., 2000; Schmahl et al., 2004), and in the nervous system Sox9 has been implicated in glial cell differentiation (Stolt et al., 2003). It was therefore intriguing that sox9b is expressed in a stripe within hindbrain segments (Yan et al., 2005). We found that expression of Sox9b protein co-localises with erm and fgfr2 mRNA in segment centres and is excluded from neurogenic zones (Fig.3A–L). These data raise the possibility that Sox9b is expressed downstream of FGFR signaling in the hindbrain. To test this, we first analysed the effect of transgenic expression of dn-FGFR1 and found that Sox9b expression is absent when FGFR signaling is blocked (Fig.3M, N). In order to check that the decrease in Sox9b expression is not due to a general loss of neural progenitors, we analysed sox3 expression which marks progenitors throughout the nervous system (Pevny and Placzek, 2005; Uwanogho et al., 1995). We found that inhibition of FGFR signaling at these stages has no detectable effect on sox3 expression (Fig.S2I, J).
Figure 3. FGF signaling maintains a Sox9b expressing population in segment centres.
(A–L): In situ hybridisation for erm, fgfr2, neurog1 or neurod4 (red) followed by immunostaining with anti-Sox9 antibody (green). Images shown are a projection of confocal stacks. Scale bar, 50 µm. White arrowheads indicate segment centres, and yellow arrowheads indicate colocalization of Sox9b with fgfr2 and erm. The staining in segment centres is due to Sox9b, as it is lost in Sox9b morphant embryos (not shown). (M–N): Whole mount immunostaining of Sox9b in 36 hpf wild type (wt; M) or transgenic dominant-negative fgfr1 embryos (Tg(hsp70l:dnfgfr1-EGFP); N). White arrowheads indicate segment centres in wt embryos, and open arrowheads in transgenic embryos point at centres where Sox9b expression is absent. Images shown are merged confocal stacks. Scale bar, 20 µm. (O–R): In situ hybridizations of 26 hpf wild type embryos (left) or embryos expressing constitutively-active FGFR1 (Tg(hsp70:ca-fgfr1)), using erm (O–P) or sox9b (Q–R) probes. Embryos were heat shocked at 24 hpf and fixed 2 h later. Arrowheads indicate segment centres. Red arrowheads indicate upregulation of erm or sox9b expression. Scale bar, 50 µm. See also Figure S2.
Since another possible explanation of decreased Sox9b expression is that it is secondary to loss of the expressing cell population, we tested the effect of expressing constitutively activated FGFR1. We analysed expression of known targets of FGFR activation, and found that there is a high level of erm expression throughout the embryo 2 h after induction of activated FGFR1 (Fig.3O, P). Transgenic embryos with overactivation of FGFR1 have a major increase in sox9b levels, mostly in areas within the nervous system where it is normally expressed (Fig.3Q, R).
The observation that fgfr2 expression occurs in segment centres (Fig.1R–T) (Tonou-Fujimori et al., 2002) raised the possibility that this gene is upregulated by the FGF pathway. In agreement with this, we found that fgfr2 expression was abolished following induction of dn-fgfr1 expression (Fig.S2E–F) and that transgenic expression of activated fgfr1 leads to widespread expression of fgfr2 (Fig.S2G, H). Taken together, these results have identified a distinct population of neural progenitors in the centre of hindbrain segments, in which erm, sox9b and fgfr2 are expressed downstream of FGFR activation.
Cyp26b1 is a target of FGFR signaling in the hindbrain
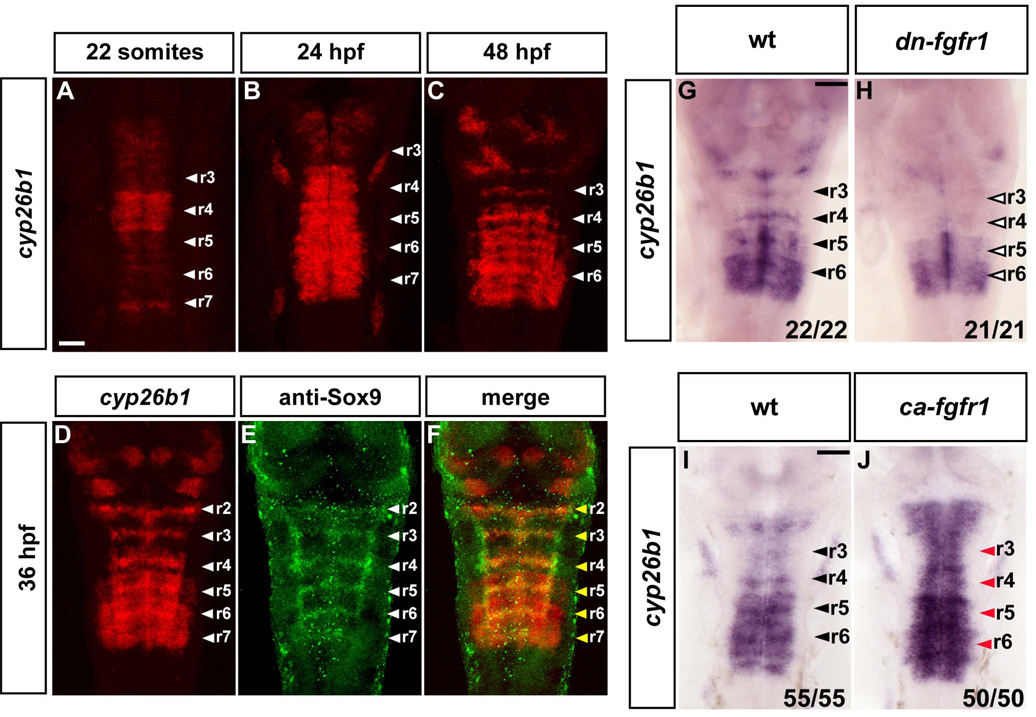
A clue to a further signaling pathway that may regulate neurogenesis in the hindbrain came from the observation that cyp26b1 is expressed in segment centres (Gu et al., 2005; Reijntjes et al., 2007; Zhao et al., 2005). Cyp26b1 belongs to the Cyp26 family of proteins which catabolize retinoic acid (RA) (Fujii et al., 1997; White et al., 1996; White and Schilling, 2008). Since RA induces neuronal differentiation in different contexts (Maden, 2007), an enzyme that degrades RA constitutes a good candidate to be required for the inhibition of neurogenesis. We therefore analysed cyp26b1 expression in more detail and found that it occurs in a complex and dynamic pattern in the hindbrain between the 22 somite stage to 48 hpf (Fig.4A–F). Cyp26b1 expression occurs throughout this period in segment centres, overlapping with Sox9b expression (Fig.4D–F), as well as at lower levels in some hindbrain boundaries. In addition, cyp26b1 is expressed more broadly throughout segments in a dynamic pattern in the posterior hindbrain (Fig.4B–D), which at 36 hpf occurs in a graded manner in r5 and r6.
Figure 4. Cyp26b1 is expressed in segment centres and regulated by FGF signaling.
(A–D): In situ hybridisations to detect the time course of cyp26b1 expression. Images are a merge of confocal stacks of Fast Red staining. Scale bar, 50 µm. White arrowheads indicate segment centres. (D–F): In situ hybridisation of cyp26b1 followed by immunostaining with anti-Sox9 antibody. Yellow arrowheads show colocalization of cyp26b1 with Sox9b in segment centres. (G–H): In situ hybridization of 40 hpf embryos to detect cyp26b1 expression in wild type (wt; G) or dominant negative FGFR1 embryos (Tg(hsp70l:dnfgfr1-EGFP)) (H). Heat shocks were started at the 22 somite stage. Black arrowheads indicate segment centres, and open arrowheads indicate the disappearance of cyp26b1 expression from centres. Scale bar, 50 µm. (I–J): In situ hybridization of 26 hpf embryos to detect cyp26b1 in either wild type (wt; I) or constitutively active fgfr1 embryos, Tg(hsp70:ca-fgfr1) (J). Heat shocks were started at 24 hpf and embryos fixed 2 h later. Black arrowheads indicate segment centres, and red arrowheads centres in embryos with cyp26b1 upregulation. Scale bar, 50 µm.
The expression of cyp26b1 in segment centres prompted us to analyse whether this was dependent upon FGFR activation. We found that expression of dn-FGFR1 led to loss of cyp26b1 expression in segment centres, whereas segmental expression in r5 and r6 was not affected (Fig.4G, H). Conversely, expression of constitutively active FGFR1 led to upregulation of cyp26b1 in the hindbrain 2 h after induction of the transgene (Fig.4I, J). cyp26b1 is therefore upregulated downstream of FGFR signaling in segment centres.
Loss of Cyp26 activity results in ectopic initiation of neurogenesis
The expression pattern of cyp26b1 and its regulation by the FGFR pathway raised the possibility that catabolism of RA is required for the inhibition of neurogenesis in segment centres. Since Cyp26 family members have essential early roles in anteroposterior patterning of the hindbrain (Hernandez et al., 2007; Uehara et al., 2007; White et al., 2007), we took a pharmacological approach, using R115866 which is a specific inhibitor of Cyp26 enzymes that results in increased RA signaling in vivo (Stoppie et al., 2000). We found that treatment of embryos with R115866 from 24–40 hpf led to ectopic expression of neurog1 and dla in segment centres (Fig.5A–D). However, expression of neurod4 was not upregulated in segment centres following inhibition of Cyp26 activity during this period (Fig.5E, F).
Figure 5. Blocking Cyp26 activity results in premature neurogenesis.
(A–F): In situ hybridization of 40 hpf embryos to detect expression of neurog1 (A–B), dla (C–D) or neurod4 (E–F) in DMSO or R115866 treated embryos. Treatments were started at 24–26 h. Black arrowhead points at r5. Scale bar, 50 µm. (A’–F’): higher power views of r4 and r5 shown in A–F (black arrowheads). Red arrowheads indicate ectopic proneural expression. Scale bar, 25 µm. (G–J). Blocking RA signaling with DEAB partially rescues loss of Cyp26. In situ hybridization of 36 hpf embryos to detect expression of neurog1 in DMSO (G), R115866 (H), DEAB (I) or R115866 + DEAB (J) treated embryos.
These results suggest that inhibition of Cyp26 activity leads to the ectopic upregulation of proneural gene expression that initiates neurogenesis, but is not sufficient for the progression of neuronal differentiation marked by neurod4. This is in contrast to the effect of FGFR inhibition, which leads to ectopic expression of both proneural markers and neurod4 in segment centres (Fig.2). We therefore analysed whether blocking of Cyp26 activity affects expression of segment centre markers. We found that in most embryos treated with R115866, fgfr2 (54% of embryos, n=68), cyp26b1 (72%, n=25) and Sox9b (84%, n=44) are still expressed in segment centres. These findings suggest that expression of Cyp26 contributes to inhibition of the initiation of neurogenesis, but FGFR signaling also induces other genes that regulate different aspects of maintaining an undifferentiated population.
The finding that excess RA due to cyp26 inhibition leads to ectopic neurogenesis raises the question of whether retinoid signaling is required for neurogenesis within the normal neurogenic zones. We found that treatment of embryos with DEAB to block the RA synthesis enzyme, RALDH2, has no effect on the expression of neurogenic markers (Fig.5G, I). To determine whether DEAB does affect RA signaling in the context of hindbrain neurogenesis, we tested whether co-treatment with DEAB rescues the effect of excess RA due to blocking of Cyp26 enzymes. We found that the ectopic neurogenesis that occurs following Cyp26 inhibition is suppressed by co-treatment with DEAB (Fig.5H, J). These results reveal that the level of RA affects neurogenesis in segment centres, whereas RA is not essential for neurogenesis adjacent to hindbrain boundaries.
Fgf20a is expressed by neurons at segment centres
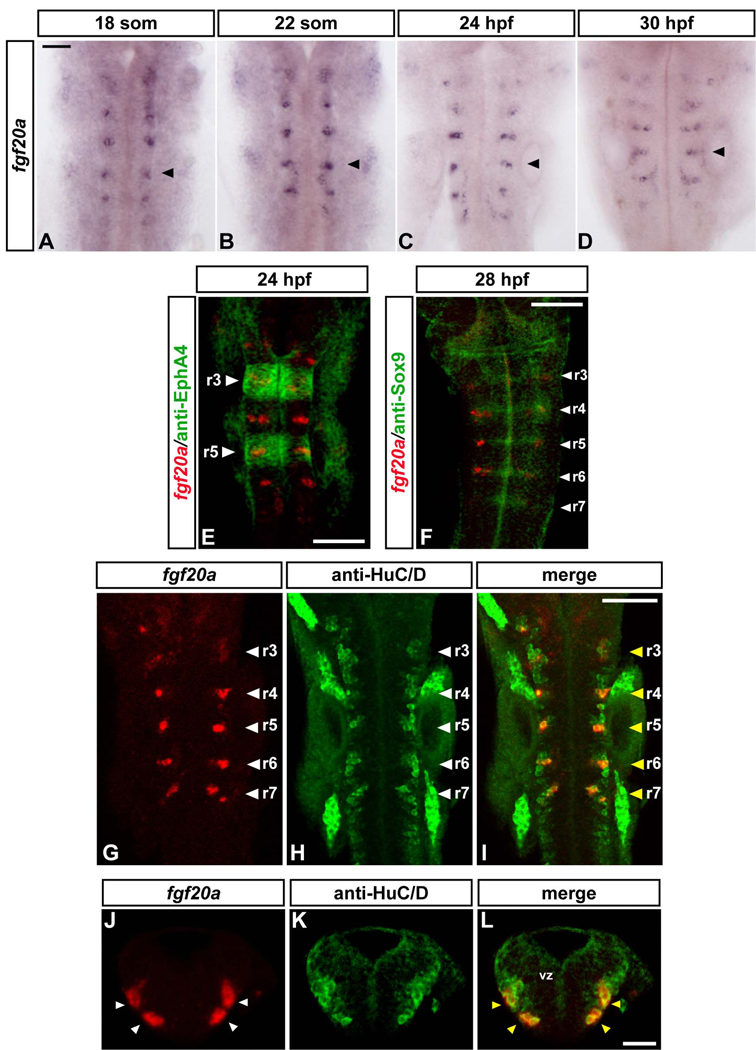
Our findings raise the important questions of the identity and site of expression of the FGF(s) that activate FGF receptors in segment centres. We therefore carried out in situ hybridisation analyses of zebrafish FGF genes to determine their expression pattern during the relevant period of hindbrain development. This identified fgf20a as a potential candidate. Fgf20a starts to be expressed in the hindbrain at the 14-somite stage in a few isolated cells (data not shown) and by the 18-somite stage is detected in a cluster of cells in each of rhombomeres 2–7 (Fig.6A). Similar fgf20a expression in discrete cell populations is observed at least until 36 hpf (Fig.6B–D, Fig.S3J–L) and is undetectable by 48 hpf (data not shown). The fgf20a expressing cells are located at segment centres (Fig.6E), at the same anteroposterior location as Sox9b expression (Fig.6F). To determine whether fgf20a expression occurs in progenitors or neurons, we carried out double staining with the pan-neuronal marker HuC/D and analysed transverse sections. We found that fgf20a expressing cells are located in the mantle zone and correspond to a subset of neurons (Fig.6G–L; Fig.S3A–I). The location of these neurons at segment centres suggests that fgf20a is a good candidate for restricting neurogenesis in the hindbrain.
Figure 6. Fgf20a is expressed in neurons at segment centres.
(A–D): Time course of fgf20a expression at 18 somites (A), 22 somites (B), 24 hpf (C) and 30 hpf (D). Embryos belong to the same batch and were developed for the same amount of time. Black arrowheads point at the centre of r5. Scale bar, 50 µm. (E–I): Merge of confocal stacks of double-stained embryos. Scale bar, 100 µm. (E) In situ hybridisation of fgf20a (red) and anti-EphA4 staining (green) to reveal r3 and r5 (white arrowheads) at 24 h. (F) In situ hybridisation of fgf20a (red) and antibody staining for Sox9b (green) in 28 hpf embryo. White arrowheads indicate segment centres. (G–I): In situ hybridisation of fgf20a (red) and antibody staining for the pan-neuronal marker HuC/D in 24 hpf embryos. White arrowheads indicate segment centres, and yellow arrowheads show colocalization of fgf20a with specific HuC/D-expressing neurons. (J–L): Double labelling of fgf20a (red) and HuC/D (green). Images show a merge of confocal stacks through r4 at 24 hpf in transverse sections. Dorsal is to the top. In r4, fgf20a expressing cells form clusters (white arrowheads) in the mantle zone and colocalize with specific HuC/D expressing neurons (yellow arrowheads). vz: ventricular zone. Scale bar, 50 µm. See also Figure S3.
Fgf20a maintains segment centre markers and restricts neurogenesis
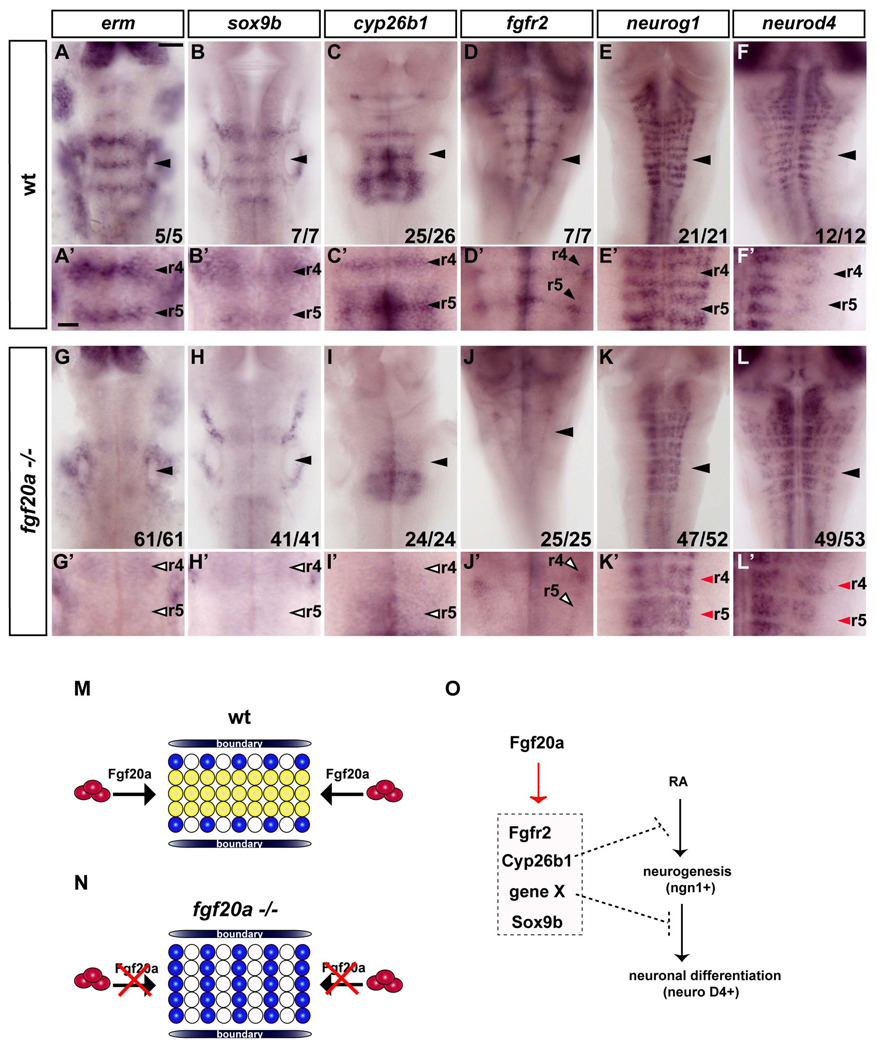
To analyse the role of fgf20a, we analysed mutants which harbor a temperature sensitive null allele of the fgf20a gene (Whitehead et al., 2005). We found that fgf20a mutants have normal segmental expression of ephrinB3 (data not shown) and thus appears not to be required for the early role of Fgf signaling in r5/r6 segmentation. In contrast, there is a major decrease in erm expression in segment centres in 24 hpf fgf20a mutants, whereas expression in other tissues such the midbrain-hindbrain boundary appears to be at normal levels (Fig.7A, A’, G, G’). Similarly, there is decreased expression of sox9b, cyp26b1 and fgfr2 (Fig.7B–D, B’–D’, H–J, H’ –J’) in segment centres in fgf20a mutants.
Figure 7. Fgf20a is required for inhibition of neurogenesis in segment centres.
In situ hybridisations of wild type (A–F) or fgf20a homozygous embryos (G–L) raised at 25°C. (A’–L’) show higher power images of A–L. Scale bar, 50 µm for A–L; 20 µm for A’–L’. erm expression in segment centres is significantly reduced in fgf20a mutants (open arrowheads in G’). Markers of segment centres, sox9b, cyp26b1 and fgfr2 are greatly decreased in fgf20a −/− embryos (open arrowheads in H’–J’). (K–L): fgf20a mutant embryos have ectopic neurogenesis in segment centres, detected by neurog1 (K, E) and neurod4 expression (L, F). Red arrowheads indicate ectopic neurogenesis in segment centres (K’, L’). (M–N) Model of the patterning of neurogenesis by fgf20a in hindbrain segments. In wild type embryos (M), fgf20a secreted from neurons in the adjacent mantle region (red ovals) prevents neuronal differentiation (blue circles) in segment centres by maintaining a population of progenitors (yellow circles). (N) In fgf20a mutants there is ectopic neurogenesis and low level expression of segment centre markers. (O) Summary of the regulation of genes in the non-neurogenic zone of progenitors in segment centres. fgf20a upregulates a set of genes that control different aspects of maintaining an undifferentiated population.
These findings reveal that fgf20a is required for the FGFR-dependent maintenance of segment centre markers. We therefore analysed whether loss of fgf20a function affects neurogenesis in the hindbrain. We found that in 40 hpf fgf20a mutants there is ectopic expression of neurog1 and neurod4 in segment centres (Fig.7E, E’, F, F’, K, K’, L, L’), as occurs following the blocking of FGFR activation by expression of dn-fgfr1 or by SU5402 treatment. Taken together, our data show that fgf20a, which is expressed by early born neurons located at segment centres, has a crucial role in maintaining segment centre markers and in spatial restriction of neurogenesis in the hindbrain.
DISCUSSION
Generation of the appropriate number and type of neural cell types requires precise regulation of cell differentiation and maintenance of progenitors. In some regions of the nervous system, this involves formation of spatially segregated neurogenic and non-neurogenic regions that are induced downstream of axial patterning mechanisms or by localised inhibitory signals within the neural epithelium (Bae et al., 2005; Bally-Cuif and Hammerschmidt, 2003; Bertrand et al., 2002; Brewster et al., 1998; Kawauchi et al., 2005; Saarimaki-Vire et al., 2007). In the zebrafish hindbrain, neurogenesis becomes restricted to zones adjacent to segment boundaries and is absent in segment centres and boundaries. Previous studies suggest that Notch activation underlies the inhibition of neurogenesis at segment boundaries (Cheng et al., 2004). Here, we show that the non-neurogenic zone in segment centres is marked by the expression of a number of genes, including fgfr2, erm, sox9b and cyp26b1. Overactivation of FGFR leads to upregulation of segment centre markers within 2 h suggesting that they are early targets of FGF signaling, whereas blocking FGFR activation leads to downregulation of these markers. Furthermore, inhibition of FGFR leads to ectopic neurogenesis in segment centres in longitudinal columns co-extensive with the normal pattern of neuronal differentiation adjacent to boundaries. We identify fgf20a as the critical activator of FGFR in segment centres. Fgf20a is expressed by a subset of neurons located in the mantle layer at the centre of segments, and in the fgf20a mutant, there is a spreading of neurogenesis and downregulation of segment centre marker expression. These studies have uncovered a mechanism in which signaling from specific early-born neurons underlies formation of a non-neurogenic zone (Fig.7M, N).
Roles of FGF in the inhibition of neurogenesis
Previous studies have found diverse roles of FGF signaling in the regulation of neurogenesis. In some contexts, specific FGFs promote neuronal differentiation, for example FGF15 in the mouse cerebral cortex (Borello et al., 2008) and FGF8 in the early Xenopus forebrain (Hardcastle et al., 2000). However, a more common role of FGF signaling is to promote the proliferation of neural epithelial cells and maintain progenitors required for subsequent differentiation to post-mitotic neurons. For example, a null mutation in FGF2 leads to a reduced number of neurons in the mouse neocortex (Ortega et al., 1998) and cortex (Vaccarino et al., 1999), consistent with a role in maintaining the neural stem cell pool (Zheng et al., 2004). Our findings suggest that in hindbrain segment centres, FGF signaling maintains a distinct progenitor zone by inhibiting neuronal differentiation, but is not required for the promotion of cell proliferation. Similarly, the sites of FGF pathway activation in the adult zebrafish brain correspond to radial glial cells, some of which may act as neural progenitors, but do not correlate with cell proliferation (Topp et al., 2008).
Roles of FGF signaling sources in local inhibition of neurogenesis occur in other regions of the developing nervous system. For example, FGFs expressed at the mid-hindbrain boundary act through multiple FGF receptor family members in mouse to maintain a zone of progenitors (Jukkola et al., 2006; Saarimaki-Vire et al., 2007). Similarly, in the olfactory epithelium, localised expression of FGF8 in the nasal pit maintains an adjacent zone of proliferating progenitor cells, with neuronal differentiation occuring distal from the FGF source (Kawauchi et al., 2005). Our findings are suggestive of an analogous role of fgf20a in the local inhibition of neurogenesis in segment centres in the hindbrain. Whereas in these other examples, an FGF source within the neural epithelium underlies the local inhibition of neurogenesis, in the hindbrain it is due to an FGF expressed by early-born neurons.
Antagonism between FGF and RA signaling in neurogenesis
Previous work has shown that retinoic acid has an important role in the promotion of neurogenesis in a number of regions of the nervous system (Maden, 2007). Studies of the caudal spinal cord in chick embryos have revealed an antagonistic relationship between RA and FGF signaling in the control of neuronal differentiation (Diez del Corral et al., 2003). FGF8 expression in caudal regions represses expression of RALDH2 required for synthesis of RA, whereas RA attenuates FGF8 expression. Consequently, there are counter-gradients of FGF and RA activity such that neurogenesis is inhibited in caudal regions where there is high FGF and low RA signaling, and initiated more rostrally where there is low FGF and high RA. By analogy, it was possible that the expression of the RA catabolising enzyme cyp26b1 in hindbrain segment centres is required for the inhibition of neurogenesis downstream of FGFR activation. Consistent with this, we found that cyp26b1 expression in segment centres requires FGFR activation, and that inhibiton of Cyp26 proteins leads to ectopic expression of proneural and Delta genes that mark the initiation of neurogenesis. However, in contrast to the effect of blocking FGFR activation, Cyp26 inhibition did not lead to expression of neurod4, which marks a later step of neuronal differentiation. Furthermore, expression of fgfr2 and Sox9b was maintained following Cyp26 inhibition, and thus the expression of these genes is regulated by FGFR independently of inhibition of RA signaling. Based upon these findings, we propose that FGFR activation in segment centres upregulates multiple genes that regulate different aspects of neurogenesis (Fig. 7O): decreased RA signaling due to expression of Cyp26b1 contributes to inhibition of the onset of neurogenesis in segment centres, but unidentified targets inhibit subsequent steps of neuronal differentiation.
Intriguingly, inhibition of RA synthesis by DEAB does not affect neurogenesis in the normal neurogenic zones adjacent to boundaries, yet DEAB suppresses ectopic neurogenesis in segment centres that occurs following inhibition of Cyp enzyme activity. RA is therefore not essential for neurogenesis in the hindbrain, but increased RA is sufficient to drive initiation of neurogenesis, and Cyp26 enzymes are required to prevent this in segment centres. The simplest explanation of these findings is that another neurogenic factor(s) acts in parallel with RA and is present at a sufficient level adjacent to hindbrain boundaries.
Significance of signaling from neurons
Our findings suggest that the formation of non-neurogenic zones is due to feedback inhibition in which the generation of fgf20a-expressing neurons limits subsequent neurogenesis. Feedback inhibition mediated by other signals has been found to limit the amount of neurogenesis. In the case of lateral inhibition by Notch ligands expressed by nascent neurons, this acts at short-range within neurogenic zones and occurs transiently since it is relieved once the differentiating neuron has migrated into the mantle layer. A more sustained feedback inhibition occurs in the olfactory epithelium. The BMP family member GDF11 is expressed by differentiating progenitors and olfactory receptor neurons downstream of the proneural gene Mash1, and acts to limit the amount of further neurogenesis and maintain progenitors (Wu et al., 2003). GDF11-expressing nascent neurons are distributed widely in the neural epithelium and adjacent mantle layer and do not mediate a spatial patterning of neurogenesis.
Our studies raise the question of whether formation of a non-neurogenic zone in segment centres has roles other than in limiting the amount of neurogenesis. One model is that some cells in segment centres migrate into the neurogenic zones and thus provide a supply of progenitors for subsequent differentiation distal from the FGF source, as may occur in the nasal epithelium (Kawauchi et al., 2005). Similarly, cells in the non-neurogenic zone at the mid-hindbrain boundary in zebrafish later contribute to neurogenesis throughout the adjacent region (Tallafuss and Bally-Cuif, 2003). Another non-mutually exclusive possibility is that the upregulation of Sox9 expression in hindbrain segment centres underlies a switch from neuronal to glial cell differentiation as found in the mouse spinal cord (Stolt et al., 2003). One role of the non-neurogenic zone may therefore be to enable generation of glial cell types.
FGF20 and homeostasis
The generation and maintenance of the correct number of progenitors and differentiated derivatives is crucial both during development and tissue homeostasis in the adult. It is intriguing that FGF20, which is expressed in the substantia nigra, has been implicated in the survival of dopaminergic neurons (Murase and McKay, 2006; Ohmachi et al., 2000), a role that may underlie the reported association between FGF20 haplotypes and Parkinsons disease (van der Walt et al., 2004). It will be interesting to determine whether FGF20 also contributes to maintenance of progenitors in the adult nervous system, analogous to the developmental role that we have uncovered. Roles of FGF20 in tissue homeostasis have been revealed in studies of the zebrafish fin. The normal regeneration of the fin following injury does not occur in the fgf20a mutant, due to a requirement for fgf20a in formation of the blastema that generates the missing differentiated tissues (Whitehead et al., 2005). Furthermore, fgf20a is required for the homeostatic maintenance of tissue during cell turnover in uninjured fins (Wills et al., 2008). Taken together with our findings, these studies raise the interesting possibility that FGF20 acts in diverse tissues to maintain progenitor cells by participating in feedback loops that underlie homeostasis.
Supplementary Material
Acknowledgements
We thank Sebastian Gerety, Francois Guillemot, James Briscoe, Qiling Xu and Javier Terriente for advice and comments on the manuscript; Silvana Gioli for anti-Sox9 antibody; Cecilia Moens for cyp26b1 probe, Ivor Mason for fgfr2 probe, David Raible and Alex Nechiporuk for fgf genes, and Janssen Pharmaceutica for Cyp26 inhibitor. This work was supported by the MRC, a Marie Curie Intra-European Fellowship (R.G-Q), and the NIH and Pew Charitable Trusts (KDP).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rosa Gonzalez-Quevedo, Division of Developmental Neurobiology, MRC National Institute for Medical Research, Mill Hill, London, NW7 1AA, UK..
Yoonsung Lee, Department of Cell Biology, Duke University Medical Center, Durham, NC 27710, USA..
Kenneth D. Poss, Department of Cell Biology, Duke University Medical Center, Durham, NC 27710, USA.
David G. Wilkinson, Division of Developmental Neurobiology, MRC National Institute for Medical Research, Mill Hill, London, NW7 1AA, UK..
References
- Amoyel M, Cheng YC, Jiang YJ, Wilkinson DG. Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development. 2005;132:775–785. doi: 10.1242/dev.01616. [DOI] [PubMed] [Google Scholar]
- Bae YK, Shimizu T, Hibi M. Patterning of proneuronal and interproneuronal domains by hairy- and enhancer of split-related genes in zebrafish neuroectoderm. Development. 2005;132:1375–1385. doi: 10.1242/dev.01710. [DOI] [PubMed] [Google Scholar]
- Bally-Cuif L, Hammerschmidt M. Induction and patterning of neuronal development, and its connection to cell cycle control. Curr Opin Neurobiol. 2003;13:16–25. doi: 10.1016/s0959-4388(03)00015-1. [DOI] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Borello U, Cobos I, Long JE, Murre C, Rubenstein JL. FGF15 promotes neurogenesis and opposes FGF8 function during neocortical development. Neural Develop. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster R, Lee J, Ruiz i Altaba A. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393:579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cheng YC, Amoyel M, Qiu X, Jiang YJ, Xu Q, Wilkinson DG. Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev Cell. 2004;6:539–550. doi: 10.1016/s1534-5807(04)00097-8. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Storey KG. Markers in vertebrate neurogenesis. Nat Rev Neurosci. 2001;2:835–839. doi: 10.1038/35097587. [DOI] [PubMed] [Google Scholar]
- Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bioessays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- Dono R. Fibroblast growth factors as regulators of central nervous system development and function. Am J Physiol Regul Integr Comp Physiol. 2003;284:R867–R881. doi: 10.1152/ajpregu.00533.2002. [DOI] [PubMed] [Google Scholar]
- Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Ford-Perriss M, Abud H, Murphy M. Fibroblast growth factors in the developing central nervous system. Clin Exp Pharmacol Physiol. 2001;28:493–503. doi: 10.1046/j.1440-1681.2001.03477.x. [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc Natl Acad Sci U S A. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A, Itoh M, Tallafuss A, Chapouton P, Tannhauser B, Kuwada JY, Chitnis AB, Bally-Cuif L. bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development. 2003;130:1591–1604. doi: 10.1242/dev.00375. [DOI] [PubMed] [Google Scholar]
- Geling A, Plessy C, Rastegar S, Strahle U, Bally-Cuif L. Her5 acts as a prepattern factor that blocks neurogenin1 and coe2 expression upstream of Notch to inhibit neurogenesis at the midbrain-hindbrain boundary. Development. 2004;131:1993–2006. doi: 10.1242/dev.01093. [DOI] [PubMed] [Google Scholar]
- Govindarajan V, Overbeek PA. FGF9 can induce endochondral ossification in cranial mesenchyme. BMC Dev Biol. 2006;6:7. doi: 10.1186/1471-213X-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Gritti A, Parati EA, Cova L, Frolichsthal P, Galli R, Wanke E, Faravelli L, Morassutti DJ, Roisen F, Nickel DD, et al. Multipotential stem cells from the adult mouse brain proliferate and self-renew in response to basic fibroblast growth factor. J Neurosci. 1996;16:1091–1100. doi: 10.1523/JNEUROSCI.16-03-01091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Xu F, Wang X, Gao X, Zhao Q. Molecular cloning and expression of a novel CYP26 gene (cyp26d1) during zebrafish early development. Gene Expr Patterns. 2005;5:733–739. doi: 10.1016/j.modgep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Hardcastle Z, Chalmers AD, Papalopulu N. FGF-8 stimulates neuronal differentiation through FGFR-4a and interferes with mesoderm induction in Xenopus embryos. Curr Biol. 2000;10:1511–1514. doi: 10.1016/s0960-9822(00)00825-3. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving C, Nieto MA, DasGupta R, Charnay P, Wilkinson DG. Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev Biol. 1996;173:26–38. [Google Scholar]
- Jukkola T, Lahti L, Naserke T, Wurst W, Partanen J. FGF regulated gene-expression and neuronal differentiation in the developing midbrain-hindbrain region. Dev Biol. 2006;297:141–157. doi: 10.1016/j.ydbio.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Julich D, Hwee Lim C, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Lewis J, Jiang YJ, Holley SA. beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol. 2005;286:391–404. doi: 10.1016/j.ydbio.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I, Calof AL. Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development. 2005;132:5211–5223. doi: 10.1242/dev.02143. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132:5173–5183. doi: 10.1242/dev.02101. [DOI] [PubMed] [Google Scholar]
- Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8:755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- Michaelidis TM, Lie DC. Wnt signaling and neural stem cells: caught in the Wnt web. Cell Tissue Res. 2008;331:193–210. doi: 10.1007/s00441-007-0476-5. [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet. 1996;14:62–68. doi: 10.1038/ng0996-62. [DOI] [PubMed] [Google Scholar]
- Munchberg SR, Ober EA, Steinbeisser H. Expression of the Ets transcription factors erm and pea3 in early zebrafish development. Mech Dev. 1999;88:233–236. doi: 10.1016/s0925-4773(99)00179-3. [DOI] [PubMed] [Google Scholar]
- Murakami S, Kan M, McKeehan WL, de Crombrugghe B. Upregulation of the chondrogenic Sox9 gene by fibroblast growth factors is mediated by the mitogen-activated protein kinase pathway. Proc Natl Acad Sci U S A. 2000;97:1113–1118. doi: 10.1073/pnas.97.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase S, McKay RD. A specific survival response in dopamine neurons at most risk in Parkinson's disease. J Neurosci. 2006;26:9750–9760. doi: 10.1523/JNEUROSCI.2745-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmachi S, Watanabe Y, Mikami T, Kusu N, Ibi T, Akaike A, Itoh N. FGF-20, a novel neurotrophic factor, preferentially expressed in the substantia nigra pars compacta of rat brain. Biochem Biophys Res Commun. 2000;277:355–360. doi: 10.1006/bbrc.2000.3675. [DOI] [PubMed] [Google Scholar]
- Ortega S, Ittmann M, Tsang SH, Ehrlich M, Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc Natl Acad Sci U S A. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Pozniak CD, Pleasure SJ. A tale of two signals: Wnt and Hedgehog in dentate neurogenesis. Sci STKE. 2006;2006:pe5. doi: 10.1126/stke.3192006pe5. [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Rodaway A, Maden M. The retinoic acid metabolising gene, CYP26B1, patterns the cartilaginous cranial neural crest in zebrafish. Int J Dev Biol. 2007;51:351–360. doi: 10.1387/ijdb.062258sr. [DOI] [PubMed] [Google Scholar]
- Roussigne M, Blader P. Divergence in regulation of the PEA3 family of ETS transcription factors. Gene Expr Patterns. 2006;6:777–782. doi: 10.1016/j.modgep.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- Saarimaki-Vire J, Peltopuro P, Lahti L, Naserke T, Blak AA, Vogt Weisenhorn DM, Yu K, Ornitz DM, Wurst W, Partanen J. Fibroblast growth factor receptors cooperate to regulate neural progenitor properties in the developing midbrain and hindbrain. J Neurosci. 2007;27:8581–8592. doi: 10.1523/JNEUROSCI.0192-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J, Kim Y, Colvin JS, Ornitz DM, Capel B. Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development. 2004;131:3627–3636. doi: 10.1242/dev.01239. [DOI] [PubMed] [Google Scholar]
- Sharpe C, Goldstone K. Retinoid signalling acts during the gastrula stages to promote primary neurogenesis. Int J Dev Biol. 2000;44:463–470. [PubMed] [Google Scholar]
- Sharpe CR, Goldstone K. Retinoid receptors promote primary neurogenesis in Xenopus. Development. 1997;124:515–523. doi: 10.1242/dev.124.2.515. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppie P, Borgers M, Borghgraef P, Dillen L, Goossens J, Sanz G, Szel H, Van Hove C, Van Nyen G, Nobels G, et al. R115866 inhibits all-trans-retinoic acid metabolism and exerts retinoidal effects in rodents. J Pharmacol Exp Ther. 2000;293:304–312. [PubMed] [Google Scholar]
- Tallafuss A, Bally-Cuif L. Tracing of her5 progeny in zebrafish transgenics reveals the dynamics of midbrain-hindbrain neurogenesis and maintenance. Development. 2003;130:4307–4323. doi: 10.1242/dev.00662. [DOI] [PubMed] [Google Scholar]
- Thisse B, Heyer V, Lux A, Alunni V, Degrave A, Seiliez I, Kirchner J, Parkhill JP, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. [DOI] [PubMed] [Google Scholar]
- Tonou-Fujimori N, Takahashi M, Onodera H, Kikuta H, Koshida S, Takeda H, Yamasu K. Expression of the FGF receptor 2 gene (fgfr2) during embryogenesis in the zebrafish Danio rerio. Mech Dev. 2002;119 Suppl 1:S173–S178. doi: 10.1016/s0925-4773(03)00112-6. [DOI] [PubMed] [Google Scholar]
- Topp S, Stigloher C, Komisarczuk AZ, Adolf B, Becker TS, Bally-Cuif L. Fgf signaling in the zebrafish adult brain: association of Fgf activity with ventricular zones but not cell proliferation. J Comp Neurol. 2008;510:422–439. doi: 10.1002/cne.21802. [DOI] [PubMed] [Google Scholar]
- Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dolle P, Sakai Y. CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol. 2007;302:399–411. doi: 10.1016/j.ydbio.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, Hung YT. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat Neurosci. 1999;2:246–253. doi: 10.1038/6350. [DOI] [PubMed] [Google Scholar]
- van der Walt JM, Noureddine MA, Kittappa R, Hauser MA, Scott WK, McKay R, Zhang F, Stajich JM, Fujiwara K, Scott BL, et al. Fibroblast growth factor 20 polymorphisms and haplotypes strongly influence risk of Parkinson disease. Am J Hum Genet. 2004;74:1121–1127. doi: 10.1086/421052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I. Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol. 2002;12:1117–1123. doi: 10.1016/s0960-9822(02)00899-0. [DOI] [PubMed] [Google Scholar]
- Wang X, Emelyanov A, Korzh V, Gong Z. Zebrafish atonal homologue zath3 is expressed during neurogenesis in embryonic development. Dev Dyn. 2003;227:587–592. doi: 10.1002/dvdy.10331. [DOI] [PubMed] [Google Scholar]
- White JA, Guo YD, Baetz K, Beckett-Jones B, Bonasoro J, Hsu KE, Dilworth FJ, Jones G, Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J Biol Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn. 2008;237:2775–2790. doi: 10.1002/dvdy.21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- Wills AA, Kidd AR, 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135:3063–3070. doi: 10.1242/dev.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. Autoregulation of neurogenesis by GDF11. Neuron. 2003;37:197–207. doi: 10.1016/s0896-6273(02)01172-8. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wilkinson DG. In situ hybridisation of mRNA with hapten labelled probes. In: Wilkinson D, editor. In situ hybridisation: A practical approach. Oxford: Oxford University Press; 1998. pp. 87–106. [Google Scholar]
- Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT, Singer A, Kimmel C, Westerfield M, Postlethwait JH. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dobbs-McAuliffe B, Linney E. Expression of cyp26b1 during zebrafish early development. Gene Expr Patterns. 2005;5:363–369. doi: 10.1016/j.modgep.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Zheng W, Nowakowski RS, Vaccarino FM. Fibroblast growth factor 2 is required for maintaining the neural stem cell pool in the mouse brain subventricular zone. Dev Neurosci. 2004;26:181–196. doi: 10.1159/000082136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.