Abstract
The Epstein-Barr virus (EBV) growth-transforms B-lymphocytes. The virus-encoded nuclear antigen 2 (EBNA2) is essential for transformation and activates gene expression by association with DNA-bound transcription factors such as RBPJκ (CSL/CBF1). We have previously shown that EBNA2 contains symmetrically dimethylated Arginine (sDMA) residues. Deletion of the RG-repeat results in a reduced ability of the virus to immortalise B-cells. We now show that the RG repeat also contains asymmetrically dimethylated Arginines (aDMA) but neither non-methylated (NMA) Arginines nor citrulline residues. We demonstrate that only aDMA-containing EBNA2 is found in a complex with DNA-bound RBPJκ in vitro and preferentially associates with the EBNA2-responsive EBV C, LMP1 andLMP2A and promoters in vivo. Inhibition of methylation in EBV-infected cells results in reduced expression of the EBNA2-regulated viral gene LMP1, providing additional evidence that methylation is a prerequisite for DNA-binding by EBNA2 via association with the transcription factor RBPJκ.
Keywords: Epstein-Barr virus, EBV, nuclear antigen 2, EBNA2, methylation, Arginine, sDMA, aDMA, RBPJκ, DNA-binding
Introduction
The Epstein-Barr virus (EBV) is associated with various human malignancies and readily growth-transforms primary human B-lymphocytes generating lymphoblastoid cell-lines, the in vitro correlate of the tumour cells of EBV-associated post-transplant lymphoproliferative disease (PTLD) (for review, see (Rickinson and Kieff, 2007)). In EBV-transformed lymphocytes, 11 so-called latent genes are expressed. Of these, only the nuclear antigens EBNA-1, -2, -3a, -3c and the latent membrane protein LMP1 are necessary for transformation (reviewed in (Bornkamm and Hammerschmidt, 2001)).
EBNA2 is a multifunctional transcriptional activator (for a recent review, see (Palermo et al., 2008)). Although it self-associates (Harada, Yalamanchili, and Kieff, 2001), a property often observed for DNA-bound transcription factors, it does not bind directly to DNA but is tethered to promoter elements by interacting with DNA-bound cellular transcription factors. For example, it associates through its Trp-Trp-Pro (“WWP325”) motif at position 323-325 with the DNA-bound repressor RBPJκ (Henkel et al., 1994; Ling and Hayward, 1995; Zimber Strobl et al., 1993) thereby converting RBPJκ to the transcriptionally active form in an analogous fashion to the cellular transmembrane receptor, Notch (reviewed in (Zimber Strobl and Strobl, 2001)). A virus encoding an EBNA2 protein with a mutation in the WWP-motif is unable to immortalize B-lymphocytes and does not activate the viral oncogene LMP1 (Cohen, Wang, and Kieff, 1991). EBNA2 binds to a variety of basal transcription factors (Bornkamm and Hammerschmidt, 2001) and also forms complexes with proteins involved in RNA metabolism like the DEAD-box protein DDX20 (DP103/Gemin3) (Grundhoff et al., 1999) or the survival of motor neurons (SMN) protein (Barth et al., 2003; Voss et al., 2001). The binding of EBNA2 to a variety of other host proteins is reflected by its presence in high molecular weight complexes of different composition (Grässer et al., 1991; Tsui and Schubach, 1994; Wu, Krumm, and Schubach, 2000). In mitotic cells, the transcriptional activity of EBNA2 is inhibited through phosphorylation at Serine 243 (Yue, Gershburg, and Pagano, 2005; Yue, Shackelford, and Pagano, 2006). Figure 1 shows a schematic representation of EBNA2.
Fig. 1.
Schematic representation of the Epstein-Barr virus nuclear antigen 2 (EBNA2). EBNA2 of the standard B95.8 strain (accession number: AJ507799) of EBV consists of 487 amino acids (aa) present in a A-type viruses. The N-terminal dimerisation domain (“Dim”) is located next to a poly-proline stretch (“Pro”). The variable region (“variable”) differs between the A-type viruses and B-type viruses. B-type viruses have a reduced in vitro transformation potential. The binding site for RBPJκ (“RBPJκ”) is located around a Trp-Trp-Pro motif at aa 323-325. The adjacent Arginine-Glycine repeat (“ArgGly”) between aa 339-354 confers binding to the survival of motor neurons (SMN) protein and represents the second nuclear localization signal (“NLS”) in addition to the canonical NLS found at the extreme C-terminus between aa 468-487. The C-terminal acidic transactivation domain (“TAD”) between aa 424-468 interacts with various basal transcription factors.
EBNA2 features an Arginine-Glycine (RG-) repeat element at position 339-354 which contains symmetrically dimethylated Arginine (sDMA) residues that confer binding to the “Tudor” domain of the survival motor neuron protein (SMN) (Barth et al., 2003). EBNA2 might therefore represent the viral counterpart of the cellular SmD3 protein, which also associates with the Tudor domain of SMN via a symmetrically dimethylated RG repeat (Friesen and Dreyfuss, 2000). The deletion of the RG-repeat of EBNA2 results in a protein with a five-fold higher ability to stimulate expression of the viral oncogene LMP1 in reporter assays, but a recombinant virus featuring this deletion in EBNA2 has reduced transforming activity and needs an extended time span to induce transformed cell clones (Tong et al., 1994).
Methylation is a posttranslational modification that affects protein-protein interactions (Gary and Clarke, 1998) and plays a role in signal transduction, cellular proliferation, transcriptional processing and splicing of mRNA (Azzouz et al., 2005; Kim et al., 1997; Lee et al., 2005; Stallcup et al., 2003). In addition to Lysine residues, methylation on proteins also takes place at Arginines (Paik and Kim, 1967) which leads to three known forms in higher eukaryotes: ω-NG -MonoMethyl-Arginine (MMA), ω-NG,NG-asymmetric DiMethyl-Arginine (aDMA) and ω -NG,N′G-symmetric DiMethyl-Arginine (sDMA); the methylation of the internal guanidino nitrogen atom to form δ-NG-MonoMethylArginine has only been detected so far in yeast (for a recent review, see (Bedford and Clarke, 2009)). The methylation reactions are catalysed by Protein-Arginine-Methyl-Transferases (PRMTs), which can be classified as type I enzymes (PRMT-1, -2, -3, -4, -6) which generate aDMA and type II enzymes (PRMT5,-7) which generate sDMA (for review, see (Bedford and Richard, 2005)). So far, JmjD6 is the only Arginine-demethylating enzyme with a demonstrated activity towards histone H3R2 and histone H4R3 (Chang et al., 2007). In addition, MMA- and aDMA-modified Arginines may be deiminated by the enzyme PADI4 to form citrulline residues (Cuthbert et al., 2004; Wang et al., 2004). Here we show using newly developed monoclonal antibodies (mAbs) that sDMA- and aDMA- but not NMA- nor citrulline-containing EBNA2 is present in EBV-infected cells. Most importantly, we demonstrate that asymmetrically Arginine dimethylated EBNA2 preferentially associates with DNA-bound RBPJκ in vitro and with EBNA2-responsive promoters in vivo.
Results
EBNA2 contains either symmetrically or asymmetrically dimethylated residues within its Arginine-Glycine (RG-) repeat
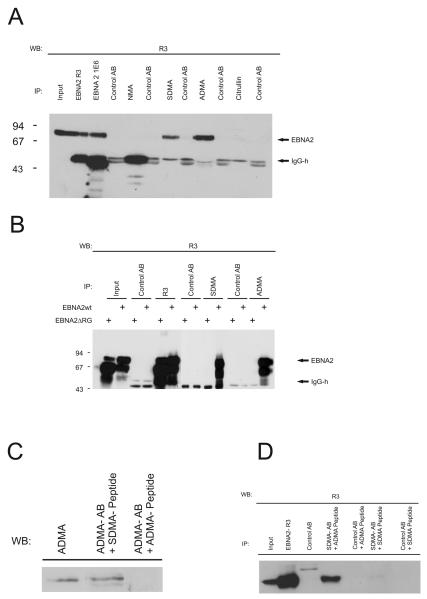
We had previously shown that the RG-repeat of EBNA2 contains sDMA-modified residues (Barth et al., 2003). To test whether EBNA2 also contains aDMA-modified, non-methylated Arginines or citrulline residues instead of Arginines, we generated mouse or rat monoclonal antibodies (mAbs) against the various possible variations in its RG-repeat. KLH-coupled peptides corresponding to the non-methylated, symmetrically or asymmetrically dimethylated RG-repeat of EBNA2 or a peptide containing citrulline instead of Arginine residues were used for the immunisation, while the same peptides bound to OVA were then used in an ELISA screening assay (see Table S1, supplementary data). Only those antibodies that reacted exclusively with their cognate peptide were established as stable clones. The specificity of the antibodies was confirmed in a dot-blot assay (Figure S1, supplementary data). The antibodies were then tested by immunoprecipitation using extracts of EBV-positive B95.8 cells. The precipitated EBNA2 was detected using the previously described antibody R3 that binds to a C-terminal epitope outside the RG-repeat (Kremmer et al., 1995). As shown in Figure 2A, only the sDMA and aDMA-specific but not the NMA- or citrulline-specific antibodies yielded a signal for EBNA2 indicating that both methylated forms are present in EBV-infected cells. EBNA2 was then immunoprecipitated from extracts of 293-T cells transiently expressing either EBNA2-wt or the mutant EBNA2-ΔRG with a deletion of the RG repeat (Tong et al., 1994). As shown in Figure 2B, the sDMA and aDMA-specific antibodies reacted with EBNA2-wt but not the deletion mutant demonstrating that the correct epitope on EBNA2 was recognised. Moreover, the signal for EBNA2 could be inhibited by preincubation of the 6F12 antibody with the aDMA-containing peptide but not the sDMA-peptide demonstrating again that the antibody is specific for aDMA-EBNA2 (Figure 2C). Vice versa, we were able to inhibit the immunoprecipitation of EBNA2 by the sDMA-specific antibody 13B10 using the sDMA peptide but not the aDMA-peptide (Figure 2D).
Fig. 2.
Detection of methylated EBNA2 species. (A) Monoclonal antibodies (mAbs) directed against the non-methylated (NMA), sDMA-, aDMA-, or citrulline containing Arginine-Glycine (RG)-repeat of EBNA2 were tested by precipitation using extracts of EBV-positive B95.8 cells. For each antibody, an appropriate isotype control was tested in parallel to exclude unspecific binding to the protein G sepharose. Precipitated EBNA2 protein was visualized using the EBNA2-specific mAb R3 which binds outside the RG-domain of EBNA2. (B) Immunoprecipitation of EBNA2 from transiently transfected cells. HEK 293-T cells expressing either EBNA2-wt or the EBNA2-RG mutant were precipitated with R3 and the various methylation specific monoclonal antibodies as indicated using appropriate isotype control antibodies. The position of EBNA2 is indicated by an arrow. (C) Specific inhibition of aDMA-antibody by aDMA peptide. The aDMA-specific antibody 6F12 was used in a Western blot either untreated or pre-incubated with aDMA or sDMA-peptide as indicated. (D) Specific inhibition of sDMA-antibody by sDMA peptide. Extract of B95.8 cells was used for immunoprecipitation using either untreated or antibody 7D9 preincubated with aDMA or sDMA peptide. Irrelevant control antibody was used as an internal control. Precipitated EBNA2 was detected by Western blot using the R3 antibody.
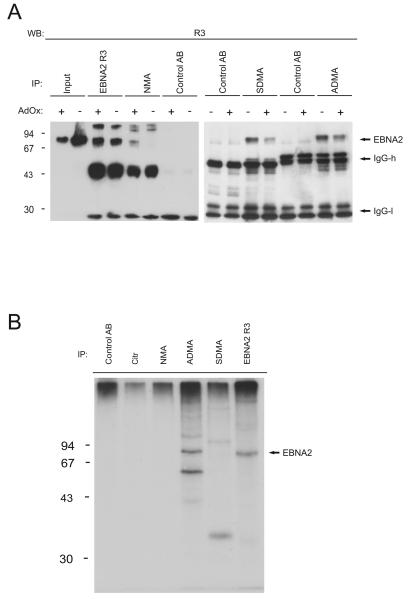
EBV-positive B95.8 cells were then treated with the methylation inhibitor AdOx prior to immunoprecipitation of EBNA2. As can be seen in Figure 3A, the overall amount of EBNA2 remained essentially unchanged while the signal obtained with the sDMA- or aDMA-specific antibodies decreased. In addition, we now detect a signal for EBNA2 with the NMA-specific antibody. We observed additional immunoreactive bands above the EBNA2 signal which might be derived from IgG molecules that contained incompletely reduced disulfide bonds. The appearance of unmethylated EBNA2 in the AdOx-treated cell extract confirms the specificity of the NMA-antibody. Our data demonstrate that EBNA2 does not exist in a nonmethylated state or in a form that has the Arginines in the RG-repeat converted to citrullines. Most importantly, the antibody 6F12 (see below) developed against a peptide containing aDMA reacted with EBNA2 indicating that a substantial fraction contains aDMA in addition to the sDMA-modified EBNA2 establishing aDMA as a novel modification of EBNA2. We obtained comparable signals for sDMA- vs. aDMA-containing EBNA2 but since the antibodies used may display different affinities for their antigens, this does not provide absolute quantification of the relative abundance of the two methylated forms.
Fig. 3.
(A) Inhibition of methylation by AdOx treatment. EBNA2 was precipitated with the indicated antibodies from B95.8 cell extract treated with the methylation inhibitor AdOx. Precipitated EBNA2 was visualised using the R3 antibody. The IgG heavy (“IgG-h”) and light (“IgG-l”) chains of the antibodies released from the beads were also detected by the secondary peroxidase coupled anti-rat antibody. Co-electrophoresed molecular mass marker proteins (× 10−3 kDa) were, in descending order: phosphorylase B, bovine serum albumin (BSA), ovalbumin (OVA), and carboanhydrase. (B) Only aDMA-EBNA2 is phosphorylated. The different antibodies as indicated were used to precipitate EBNA2 from 32P-labelled B95.8 cell extracts. The bound EBNA2 was analysed by SDS-PAGE and autoradiography. The position of EBNA2 is indicated. Co-electrophoresed 14C-labelled molecular mass marker proteins (× 10−3 Da) were, in descending order: phosphorylase B, bovine serum albumin (BSA), ovalbumin (OVA), and carboanhydrase.
Only ADMA-modified EBNA2 is phosphorylated
EBNA2 is a phosphoprotein (Grässer et al., 1992; Petti, Sample, and Kieff, 1990). Hyperphosphorylation was shown to inhibit transcriptional activation of EBNA2 during mitosis (Yue et al., 2004). To test whether the aDMA- or sDMA-modified EBNA2 are phosphorylated, EBV-positive B95.8 B-cells were metabolically labelled with H332PO4 and the cell extract was immunoprecipitated with the different antibodies and subsequently analysed by SDS-PAGE and autoradiography (Grässer et al., 1991). As can be seen in Figure 3B, the precipitation with the aDMA-specific antibody as well as the R3-antibody but not the sDMA-, -NMA or citrulline-specific antibodies yielded signals for EBNA2. This data indicates that only the aDMA-modified EBNA2 is detectably phosphorylated. We noticed additional bands co-precipitated by the aDMA- and sDMA-specific antibodies. As EBNA2 forms high-molecular weight complexes, these phosphoproteins might represent factors associated with either aDMA or sDMA-modified EBNA2. The bands that were precipitated with the aDMA-specific antibody migrated to different positions in the gel than those precipitated by the sDMA-specific antibody indicating that the sDMA and -aDMA forms of EBNA2 associate with different cellular proteins. We also tested whether the induction of lytic EBV-replication by TPA influences the methylation status of EBNA2 as it was shown that phosphorylation of Serine 243 by the viral kinase BGLF4 was induced during lytic cycle replication (Yue, Gershburg, and Pagano, 2005). The induction of the lytic cycle was confirmed using recently developed antibodies against the viral DNA polymerase (Barth et al., 2008) or against the BZLF1 protein (Young et al., 1991). We found no change in the methylation of the protein upon lytic cycle induction by TPA-treatment (see supplemental Figure S2A), while the AdOx-treatment reduced the methylation of EBNA2 but did not induce the lytic cycle (see supplemental Figure S2B).
aDMA and sDMA-containing EBNA2 is detectable in high-molecular weight complexes
EBNA2 forms high molecular weight complexes consistent with its association with multiple proteins (Grässer et al., 1991; Wu et al., 1996). Nuclear extracts were separated by 5-40% sucrose gradient centrifugation (Grässer et al., 1991) and tested in a Western blot using the antibody R3 or the aDMA-specific clone 6F12. As shown in Figure 4, the majority of EBNA2 detectable by R3 migrated in fractions 3-6 representing dimeric EBNA2 in complexes of about 300-400 kDa. In contrast, the aDMA-specific clone 6F12 yielded the strongest signal in fractions 10-13 corresponding to a molecular mass of about 500-700 kDa and a distinct signal in the bottom fraction 17 corresponding to a very high molecular mass. Extreme care was taken to avoid resuspending particulate material that might have sedimented at the bottom of the centrifuge tubes. Schubach and co-workers have described an EBNA2 species of 3-4 MDa detectable in gel filtration experiments (Tsui and Schubach, 1994)which is consistent with the EBNA2 species present in fraction 17. Using 6F12 (aDMA antibody), we also observed an immunoreactive band of approx. 200kDa in fractions 3-6 that might represent dimeric EBNA2 as the samples were not boiled prior to the gel electrophoresis. Because the sDMA-specific antibodies did not work in Western blot analysis, we subjected the fractions of the gradients to an immunoprecipitation employing EBNA2-R3, the sDMA- or aDMA-specific antibodies and an irrelevant control antibody. The precipitated EBNA2 was visualised using R3. As can be seen in Figure 4, the control antibody gave no signal, while the precipitation with R3 and the other antibodies yielded signals with a peak in fractions 3-6. R3 and the aDMA- and sDMA-specific mAbs also precipitated higher molecular weight species (fraction 17). The major part of aDMA-containing EBNA2 sediments in the fractions 10-13 corresponding to the higher molecular weight complexes while the precipitation mainly yielded a signal in the fractions 3-6 for the lower molecular mass complexes. This result indicates that the former complexes contain proteins bound to the modified RG-repeat which render this epitope inaccessible to the antibody by immunoprecipitation. Because we clearly detect some EBNA2 precipitated from fraction 17 presenting very high molecular weight complexes, we assume that sDMA-containing EBNA2 is also present in the fractions 10-13 but cannot be precipitated because of RG-bound proteins.
Fig. 4.
Methylated EBNA2 is present in high molecular weight complexes. Native cell extracts were subjected to 5-40% sucrose gradient centrifugation. The fractions obtained were either concentrated by ethanol precipitation and then analysed by Western blot analysis with the indicated antibodies (panels labelled “WB”) or subjected to precipitation followed by Western blot analysis (panel labelled “IP/WB”) with the indicated antibodies. The GST-specific mAb 6G9 (Kremmer, unpublished) was used as a control for the precipitation. The bands at approx. 50 kDa represent the heavy chains of the antibodies used for precipitation that are detected by the secondary peroxidase-coupled anti-rat antibody. Molecular mass marker proteins were described in Figure 2.
Only aDMA-containing EBNA2 is detectable in DNA-binding complexes in vitro
We next tested for the presence of methylated EBNA2 in DNA-associated complexes. For this purpose, we analysed the ability of the methylation-specific antibodies to induce a supershift in an electrophoretic mobility shift assay (EMSA). We had originally shown that EBNA2 is tethered via RBPJκ to its cognate promoter sequences (Zimber Strobl et al., 1993). It is important to point out that although considerable evidence, including data from chromatin immunoprecipitation experiments (Bark-Jones, Webb, and West, 2006), supports the fact that EBNA2 stably associates with DNA via RBPJκ, we and others have found that EBNA2 can destabilise the single RBPJκ-DNA complex detected in gel retardation assays carried out using C promoter probes containing one RBPJκ binding site ((Waltzer et al., 1994; Waltzer et al., 1996) and our unpublished data). It is therefore important to ensure that the in vitro assays used accurately reflect the behaviour of RBPJκ and EBNA2 in vivo. Using an LMP 2A/TP1 promoter probe containing two RBPJκ sites allows the detection of the RBPJκ and/or EBNA2-containing complexes: complex I represents one molecule of RBPJκ bound to DNA, complex III corresponds to two molecules of RBPJκ, and complex IV contains two molecules of RBPJκ and one molecule of EBNA2, while cell proteins non-specifically bound to the promoter are present in complex II. Complex IV can be “supershifted” by R3 (Meitinger et al., 1994). Conversely, the interaction of EBNA2 with the DNA-bound RBPJκ can be destroyed by the antibody 6C8 which recognizes the “WWP”-motif of EBNA2 necessary for binding to RBPJκ (Sauder et al., 1996). Prior experiments had shown that EBNA2 generated in a reticulocyte-based system was able to induce an EBNA2-specific shift that could be supershifted with R3 antibody and was also destroyed by the 6C8 antibody as was observed for EBNA2 from native cell extracts (Meitinger et al., 1994; Sauder et al., 1994; Zimber Strobl et al., 1993). In addition, cell extract as a source for RBPJκ can also be substituted by IVT-RBPJκ from the reticulocyte system (Maier et al., 2005). The in vitro generated EBNA2 reacted with the antibody 6F12 (aDMA) in a Western blot and was also precipitated using the sDMA-specific antibody 7D9 indicating that both sDMA and aDMA modified EBNA2 were generated (Figure S3, supplementary data) in line with our previous report that the reticulocyte-based EBNA2 contains sDMA residues (Barth et al., 2003). As can be seen in Figure 5A, EBNA2 alone did not bind to the probe (lane 2) while the IVT-RBPJκ yielded a band (lane 3) that could be shifted with IVT-EBNA2 (lane 4) and could furthermore be supershifted by R3 (lane 5). The WWP mAb 6C8 destroyed this complex (lane 6). Significantly, only the aDMA-specific clone generated a supershift (lane 11), while neither the NMA-, the sDMA- nor the control antibodies bound to the RBP-Jκ-EBNA2 DNA bound complex (lanes 7-10). The same result was also obtained when gel shift experiments were carried out using extracts from EBV-infected cells (Figure 5B). Here, the R3 antibody supershifted complex IV (Figure 5B, lane 4) which was destroyed by 6C8 (lane 5), while only the aDMA-specific antibody induced a supershift (lane 10). Moreover, when reticulocyte-derived EBNA2, was used in conjunction with cell extract derived from the EBNA2-deficient, EBV-positive P3HR1 cell line, we again obtained an EBNA2-specific band that was only supershifted by R3 and the aDMA-specific clone 6F12, but not by the sDMA- or NMA-specific antibodies (data not shown). These data therefore support the notion that the aDMA-modification of EBNA2 is necessary to promote the association of EBNA2 with DNA-bound RBPJκ. To exclude the possibility that the gel shift buffer conditions prevented binding of the sDMA-specific antibody to its epitope, immunoprecipitations were carried out using the same nuclear extracts and conditions. We could clearly precipitate EBNA2 with the sDMA-specific antibody (supplementary Figure S4A). Furthermore, we found that the antibodies still bound to their OVA-conjugated peptides in a dot-blot assay when the blot stripes were incubated with up to 2M NaCl excluding grossly different binding affinities as an explanation for the observed results (data not shown).
Fig. 5.
aDMA-modified EBNA2 is present in DNA-binding complexes. (A) EBNA2 and RBPJκ synthesized in a coupled in vitro transcription-translation system derived from rabbit reticulocytes (Promega) were used in a gel shift (EMSA) (Meitinger et al., 1994) after preincubation of the proteins with the indicated antibodies. The complexes III and IV which are formed by RBPJk and RBPJκ plus EBNA2, respectively, as well as complex IV supershifted with mAb R3 (lane 4) or aDMA-specific mAb 6F12 (lane 11) are indicated. (B) EBNA2-containing Raji cell extract was incubated with the indicated antibodies and then assayed in a gel shift assay. R3 recognizes EBNA2 regardless of its methylation status and induces a “supershift” indicated by the upper arrow (Zimber Strobl et al., 1993), the mAb 6C8 directed against the “WWP”-repeat of EBNA2 destroys the EBNA2/RBPJκ-complex IV (Sauder et al., 1994). Antibodies used are indicated above each lane. Control antibodies corresponded to the respective IgG-subtype of each antibody. To efficiently separate the high molecular weight complexes, the electrophoresis was carried out for an extended time. Therefore, uncomplexed 32P-labelled probe ran out of the gel. The position of the RBPJκ-containing complexes I-IV as described in the text are indicated; the arrow points at the EBNA2-containing complex IV that is supershifted by R3 and aDMA-6F12 but destroyed by WWP-6C8.
Methylation is a prerequisite for interaction of EBNA2 with DNA-bound RBPJκ
Next, we analysed nuclear extracts of cells treated or untreated with the methylation inhibitor AdOx in a gel shift assay. We observed a decrease in methylation of EBNA2 in these extracts while the level of the protein was unaffected (see above). As can be seen in Figure 6A, the EBNA2-derived shift observed with untreated cell extract was stronger than the one obtained with the AdOx-treated extract demonstrating again that methylation of EBNA2 influences its interaction with DNA-bound RBPJκ. The determination of the signal strength showed a decrease in signal intensity of 30-67% for the AdOx-treated samples (compare, i.e., lanes 2 and 3). Again, we only observed a supershift with R3 (lanes 6 and 7) and the aDMA-specific antibody (lanes 18 and 19). Here, untreated extract showed a supershift with the aDMA antibody (lane 18) while essentially no supershift with this antibody was detectable with the AdOx-treated sample (lane 19). For better clarity, the upper part of the gel depicted in Figure 6A as well as a quantification of the signal from the supershift of the treated and the corresponding untreated samples is shown in Figure 6B.
Fig. 6.
Methylation of EBNA2 is a prerequisite for DNA-binding. (A) Extracts of Raji cells either treated (lanes designated “+”) or untreated (lanes designated “−”) with the methylation inhibitor AdOx were used in the gel shift experiments. The antibodies used are given above each lane. The upper arrow indicates the supershifted EBNA2-containing complex IV. (B) The Figure shows the quantification upper section of the supershift shown in Figure 5A together with the quantification of the supershifted EBNA2-containing complex obtained using the PhosphoImager® (GE Healthcare, Freiburg, Germany). (C) Remethylation generates DNA-binding EBNA2. The DNA probe (lane 1) was either incubated with in vitro generated RBPJκ alone (“IVT RBPJκ”, lane 2), with IVT-RBPJκ plus unmethylated E.coli-EBNA2 alone (“E.coli E2 + jκ”, lane 3) or remethylated E.coli-EBNA2 (“Rem. E2”) together with in vitro synthesized RBPJκ (lane 4). The indicated combinations of E.coli-EBNA2, remethylated EBNA2 or Reticulocyte-based EBNA2 (”Retic E2”) were tested with IVT-RBPJκ and control- antibody (“Co AB”, lanes 6 and 7), EBNA2-specific R3 antibody (“R3”, lanes 8 and 9), sDMA-specific antibody (“sDMA”, lanes 10 and 11) or the aDMA-specific antibody (“aDMA”, lanes 12 and 13). The shifted and supershifted EBNA2-containing complexes are indicated. (D) aDMA modification of EBNA2 by in vitro methylation. EBNA2 was synthesized in vitro in the E. coli-based RTS 500 system (Roche, Penzberg, Germany). This unmethylated E. coli-EBNA2 was then pre-incubated with unprogrammed reticulocyte lysate. The left lane contains reticulocyte-derived EBNA2 (“IVT-EBNA2 Retic. Lysate”) and the adjacent lane features E. coli-synthesized EBNA2 (“IVT-EBNA2 E.coli lysate”) as detected by the EBNA2-specific R3 antibody. The next lane contains unmethylated E.coli-EBNA2 (“IVT-EBNA2 E.coli lysate”), and the rightmost lane contains E.coli-EBNA2 remethylated with the reticulocyte lysate (“IVT-EBNA2 E.coli.-remethylated”). In these two lanes, EBNA2 was stained using the aDMA-specific antibody 6F12.
Methylation converts non-binding EBNA2 into a DNA-associated form
We then assayed the DNA-binding properties of non-methylated EBNA2 produced in an E.coli-based transcription-translation system (“E.coli E2”). By precipitation with the NMA-specific antibody we could show that the E. coli-based EBNA2 was indeed not methylated and initial experiments indicated that the E.coli-EBNA2 did not bind to DNA (data not shown). To determine whether methylation of EBNA2 by a methyl transferase activity in the reticulocyte system converted EBNA2 into the RBPJκ-associated, DNA-binding form, we incubated the E.coli-EBNA2 with the un-programmed reticulocyte-based transcription-translation extract supplemented with the methyl donor SAM and carried out the EMSA using this remethylatedEBNA2 (“Rem. E2”) in conjunction with IVT RBPJκ. Gel shift assays were carried out with the same amount of E.coli-EBNA2 as the reticulocyte-EBNA2 (“Retic E2”) and IVT-RBPJκ. The E. coli-EBNA2, the remethylated-EBNA2 or the reticulocyte-EBNA2 did not produce a supershift alone (Figure 6C, lanes 3, 4 and 5, respectively). The remethylated and the reticulocyte EBNA2 were then either assayed with control-antibody (lanes 6 and 7, respectively), R3 (lanes 8 and 9), the sDMA-specific antibody (lanes 10 and 11) or aDMA-specific antibody (lanes 12 and 13). As can be seen, only R3 and the aDMA- but not the sDMA-antibody induced a supershift. The NMA-specific antibody also did not generate a supershift (Figure S5, supplementary information). Western blot analysis (Figure 6D) confirmed that the C-terminal antibody R3 recognized EBNA2 from both the reticulocyte-and the E.coli-based IVT system while the aDMA-specific antibody 6F12 only reacted with the remethylated-EBNA2 but not the E.coli-EBNA2 showing that aDMA-EBNA2 can be generated in vitro.
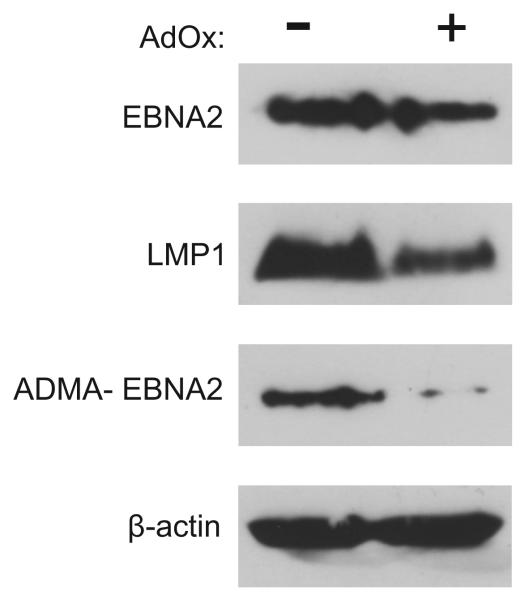
To determine whether demethylation of EBNA2 in vivo results in a reduction in EBNA2-activated transcription of its target promoters, we investigated the expression of the EBNA2-responsive LMP1 gene. EBV-infected B95.8 monkey cells or the human IB4 lymphoblastoid cell line immortalized with B95.8 virus were treated with AdOx. Although the overall amount of EBNA2 was only reduced by 10-20%, we observed a 70% reduction in the amount of aDMA-EBNA2 detected by Western blot and a corresponding decrease in the expression of LMP1 by 40-50% while the β–actin loading control was unaffected (Figure 7). Comparable results were obtained using Raji cells (data not shown). These results support the notion that methylation is a prerequisite for binding of EBNA2 to DNA.
Fig. 7.
Inhibition of EBNA2 methylation reduces LMP1 expression. B95.8 cells were untreated (“−“) or treated (“+”) with the methylation inhibitor AdOx and analysed by Western blot for the expression of EBNA2, LMP1, aDMA-EBNA2. Levels of EBNA2 (“EBNA2”, top panel), LMP1, aDMA-modified EBNA2 and β-actin as loading control were determined using the antibodies R3, S12, 6F12 and AC15 (Sigma), respectively.
aDMA modified EBNA2 preferentially binds to promoters in vivo
To investigate the effect of EBNA2 methylation on the binding of EBNA2 to promoters in vivo, we carried out chromatin immunoprecipitation experiments using a matched pair of EBNA2 negative and positive Burkitt's lymphoma cell-lines. The group I Burkitt's lymphoma (BL) cell-line Mutu I was derived from a tumour biopsy and maintains the characteristic restricted form of latency (latency I) found in BL, associated with the expression of only EBNA 1. The Mutu III cell-line was derived from Mutu I cells that had drifted in culture to express the full panel of EBV latent genes including EBNA2 (latency III). These cell-lines therefore allow an examination of the transcriptional effects of EBNA2 in the same cell background. The presence or absence of EBNA2 in the two cell lines was confirmed by Western blot analysis (supplemental Figure S6 and data not shown). Primers were designed to amplify three EBNA2 target EBV promoters; the C promoter (Cp), the LMP1 promoter (LMP1p) and the LMP2A promoters (LMP2Ap) (Figure 8A, 8B and 8C, respectively). Using the R3 antibody, we observed the expected profile of EBNA2 binding to Cp, peaking in the region encompassing the RBPJκ site (−368 to −374, (Waltzer et al., 1994)) detected by the −430 to −355 amplicon (−430) (Figure 8D). These results were in line with our previous observations using the anti-EBNA2 PE2 mouse mAb (Bark-Jones, Webb, and West, 2006). The aDMA specific antibody detected high levels of EBNA2 binding consistent with the presence of aDMA-EBNA2 in promoter-bound complexes (Figure 8E). In contrast, the sDMA specific antibody detected significantly less binding of sDMA-modified EBNA2 to the promoter. Similar results demonstrating comparatively less association of sDMA-modified EBNA2 with promoter sequences were obtained using LMP1 and LMP2A promoter primers, with the peak of EBNA2 binding detected by the primer sets encompassing one or more of the RBPJκ sites (Figure 8F, G, H, I). Control experiments confirmed that both aDMA and sDMA-specific antibodies were able to precipitate equivalent amounts of EBNA2 protein from cross-linked chromatin under ChIP conditions (supplemental Figure S4B), indicating that the reduced signal obtained in the real-time PCR analysis reflects a decreased association of sDMA-EBNA2 with promoter DNA. These data therefore indicate that aDMA-modified EBNA2 is likely to represent the predominant form of EBNA2 found at target promoters in vivo.
Fig.8.
Chromatin immunoprecipitation analysis. Location of the amplicons generated by the C promoter (A) or LMP1 promoter (B) or LMP2A promoter (C) real-time PCR primer sets. Numbers refer to the start of the amplicons generated by the indicated primer sets relative to the transcription start sites. The RBPJκ and PU.1 binding sites are indicated by grey and black boxes, respectively. The LMP1 promoter sequence has been inverted for simplicity and lies in the reverse orientation in the EBV genome. (D) Chromatin was immunoprecipitated from Mutu I (I) or Mutu III cells (III) using the R3 rat mAb and analysed using Cp-specific primers. To allow comparison between antibodies and experiments relative ChIP signals were calculated by expressing the percentage input signal relative to the Mutu III signal obtained with the furthest downstream primer set. Results show the mean +/− standard deviation for at least 3 independent experiments (n) carried out on at least 2 different batches of chromatin (c). (E) Chromatin immunoprecipitations carried out using the aDMA EBNA2 specific (6F12, open bars) and sDMA EBNA2-specific (7D9, black bars) mouse mAbs, analysed using Cp specific primers. (F) Chromatin immunoprecipitations carried out using the R3 rat mAb analysed using LMP1p-specific primers. (G) Chromatin immunoprecipitations carried out using the aDMA-EBNA2 and sDMA EBNA2-specific mouse mAbs analysed using LMP1p specific primers. (H) Chromatin immunoprecipitations carried out using the R3 rat mAb analysed using LMP2Ap-specific primers. (I) Chromatin immunoprecipitations carried out using the aDMA-EBNA2 and sDMA EBNA2-specific mouse mAbs analysed using LMP2Ap specific primers.
Discussion
Here, we provide the first demonstration of aDMA-methylation as a novel modification of EBNA2. In addition, essentially all EBNA2 molecules in the cell are converted into methylated forms and citrulline-containing EBNA2 was not detectable. Our data show that methylation influences the interaction of EBNA2 with RBPJκ and implicate methylation in the regulation of the activity of this oncoprotein. It has been demonstrated that histones undergo various secondary modifications including Arginine methylation (Jenuwein and Allis, 2001). In addition to methylation at Lysine residue 7, histone H3 may be methylated at Arginine 8 by either PRMT5 or PRMT1. The latter modification was shown to activate transcription, while methylation by PRMT5 inactivated transcription ((Pal et al., 2004) and references therein). We assume that PRMT1 generates aDMA-modified histone H3, while PRMT5 produces sDMA-histone H3. No clear picture has emerged for methylation of transcription factors. For example, aDMA-STAT1 generated by PRMT1 (Mowen et al., 2001) as well as sDMA-IL-2 generated by PRMT5 (Richard, Morel, and Cleroux, 2005) were both activating. We have not formally shown that EBNA2 is methylated by PRMT1 but it is likely that either PRMT1 or another type I methyltransferase is responsible for the generation of aDMA-containing EBNA2. In our assays, we find predominantly aDMA-modified EBNA2 at promoters in vivo, indicating that type I methyltransferases produce a positive effect on transcription in this context.
The data obtained from our ChIP analyses indicated that less sDMA-modified EBNA2 associated with three EBV target promoters in vivo, compared to the signals obtained using the R3 or aDMA-specific antibodies. These data indicate that the aDMA-modified form of EBNA2 preferentially associates with promoters in vivo. Nonetheless, these ChIP experiments do detect some sDMA association with promoters in contrast with the results of the gel shift experiments where the sDMA-specific antibody was not able to supershift an EBNA2-RBPJκ DNA complex at all. It is possible that the sDMA antibody epitope is occluded in the EBNA2-RBPJκ-DNA complex formed in vitro, possibly by other factors present in the complex. The association of EBNA2 and/or RBPJκ with the additional components of the transcriptional machinery present at promoters in vivo may result in the release of these factors or induce a conformational change in the complex that re-exposes the epitope. It is also possible that aDMA-EBNA2 binds to RBPJκ directly while sDMA-EBNA2 associates with other factors present at the promoters in vivo.
We have demonstrated previously that EBNA2 targets the survival of motor neurons (SMN) protein via its RG-repeat modified by the type II PRMT5 (Barth et al., 2003) and that co-expression of SMN activates transcription by EBNA2 (Voss et al., 2001). However, a ChIP analysis using antibodies against SMN did not yield a signal from the LMP1 promoter although SMN was clearly precipitated (data not shown). It is still unclear why the deletion of the RG-repeat reduces transformation by EBNA2 while the expression of the viral oncogene LMP1 is strongly up-regulated. One possibility is that the deletion of the RG-repeat without insertion of a spacer to replace the deleted amino acids results in an altered EBNA2 protein that is able to interact with RBPJκ while the presence of the non-methylated RG-repeat inhibits association with DNA-bound RBPJκ. This data is in line with our finding that inhibition of EBNA2 methylation results in a decrease of LMP1 protein synthesis.
Interestingly, although AdOx treatment reduced the level of methylated EBNA2, it did not completely ablate it. It is unclear whether EBNA2 undergoes demethylation (deimination) within its RG-repeat as we saw no citrulline-containing EBNA2 with our antibodies. Because the half-life of EBNA2 is very long (Grässer et al., 1991), it is unlikely that proteolytic degradation of EBNA2 removes “active” EBNA2 from the cell. However, DNA-bound aDMA-EBNA2 was phosphorylated, while the sDMA-EBNA2 was not. It is possible that during mitosis, as shown by others (Yue et al., 2004), the DNA-bound aDMA-EBNA2 is reversibly inactivated through phosphorylation at Ser243.
In summary, we show that EBNA2 contains both sDMA and aDMA-residues and that methylation is required for DNA-binding by EBNA2, with aDMA-modified EBNA2 preferentially associated with promoters in vitro and in vivo. In addition, only aDMA-containing EBNA2 is phosphorylated pointing at further functional differences between these subspecies of EBNA2. Further studies will be necessary to determine how differential methylation of EBNA2 regulates its association with the components of the transcriptional machinery and affects its biological activities
Materials and Methods
Generation of monoclonal antibodies (mAbs)
Lou/C rats or BALB/c mice were immunized with KLH-coupled peptides corresponding to the Arginine-Glycine repeat of EBNA2 that contained either non-methylated Arginines (NMA), symmetrically (sDMA) or asymmetrically (aDMA) dimethylated Arginine residues or a peptide containing citrulline instead of Arginine residues. The peptides were based on the sequence NH2-C-GQSRGRGRGRGRGRGK GKSRDK; the aminoterminal, non-EBNA2-derived Cysteine residue was added for covalent coupling to KLH or OVA. The screening was carried out by ELISA with the peptides coupled to OVA using an irrelevant peptide as a control (Barth et al., 2008). Positive clones were then tested against the other peptides and only those that reacted exclusively with their cognate peptide were established. KLH- and OVA-coupled peptides were purchased from PSL, Heidelberg, Germany. The rat mAb 8C12 (IgG2a) reacts with NMA-EBNA2, the mouse mAb 13B10 (IgG2a) recognizes sDMA-EBNA2, the mouse mAb 6F12 (IgG2b) binds to aDMAEBNA2 and the rat antibody 4A6 (IgG1) reacts with citrulline-containing EBNA2-derived peptide. The antibodies R3, 1E6 and 6C8 were described previously (Kremmer et al., 1995). R3 and 1E6 bind to a C-terminal epitope, 6C8 recognizes the WWP-repeat at aa 320 and destroys the EBNA2-RBPJκ-interaction (Sauder et al., 1994). S12 binds to LMP1 (Mann, Staunton, and Thorley Lawson, 1985), the monoclonal antibody 3F10 (Roche, Penzberg, Germany) binds to the HA-tag. Mouse AC-15 anti β–actin was from Sigma (München, Germany).
Dot blot assay
Approx. 0.1 μg each of the OVA-coupled peptides (se e above) dissolved in PBS were spotted onto a nitrocellulose membrane (Whatman Protran®, Dassel, Germany). OVA with HA-peptide reactive with the HA-specific antibody 3F10 served as an internal control. The membrane strips containing the different peptide conjugates were blocked for 30 min at 25°C with 1% non-fat dried milk dissolved in PBS, and then incubated with the different antibodies diluted 1:10 in the milk/PBS solution overnight. The bound antibodies were visualized by the ECL®-method (GE Healthcare, München, Germany) was carried out as described (Barth et al., 2003) using goat-anti-rat or- anti mouse-antibody coupled to horseradish peroxidase as secondary antibody.
Cell lines and tissue culture
HEK 293-T cells were cultured in DMEM medium (GIBCO), supplemented with 10% FCS and antibiotics, non-adherent cell lines were grown in RPMI 1640 medium (GIBCO), supplemented with 10% FCS, Na-Pyruvate and antibiotics. The EBV-infected cell lines Raji, B95.8, IB4 and P3HR1 as well as 293-T cells were previously described (Barth et al., 2003; Grässer et al., 1993 ; Voss et al., 2001). Mutu I and Mutu III cell lines were a kind gift from Prof. Martin Rowe, Birmingham, U.K. (Sample et al., 1991). Mutu I does not express EBNA2 while Raji, Mutu III and B95.8 contain EBNA2. P3HR1 contains a non-transforming virus that has a deletion in the EBNA2 gene. The metabolic labelling of B95.8 cells with H 323PO4 (Hartmann, Braunscheig, Germany) and the analysis of precipitated EBNA2 by SDS-PAGE and autoradiography was carried out as described (Grässer et al., 1992).
Transfections
For transient expression of EBNA2 proteins, 5×106 293-T cells were transfected with 8μg/10cm dish of the pSG5-EBNA2-wt or pSG5-EBNA2-ΔRG expression vectors (Barth et al., 2003) using Nanofectin® (PAA, Cölbe, Germany). Western blotting by the ECL®-method (GE Healthcare, München, Germany) was carried out as described (Barth et al., 2003).
Preparation of native whole cell extract
Raji or B95.8 cells were treated for 72h with the methylation inhibitor AdOx (Sigma, München, Germany) at 20μM. Lytic viral replication was induced by treatment of the cells for 48h with 32pM of the phorbol ester TPA (Barth et al., 2008) and cells were lysed for 30 min on ice in buffer 1 (PBS supplemented with 0,5% IGEPAL (Sigma) and 0,15M NaCl) containing protease inhibitors (Pepstatin, 2μg/mL, PMSF 10μM, Leupeptin 1μg/mL, Aprotinin 1μg/mL). The lysa te was centrifuged in a tabletop centrifuge at 15.000 × g for 15 min, and the supernatant was used for further analysis.
Preparation of native nuclear extracts
Raji or B95.8 cells either treated with AdOx or TPA (see above) were cultivated for 72h or 48h, respectively. Nuclear cell extracts were prepared in a buffer containing 20mM HEPES pH 7,9, 420mM NaCl, 1,5mM MgCl2, and 2mM EDTA pH 8,5 (Dignam, Lebovitz, and Roeder, 1983). Gel shift analysis was carried out exactly as described (Zimber Strobl et al., 1993).
Immunoprecipitation
The rat monoclonal antibody (mAb) R3 (rat IgG2a) recognizes a C-terminal epitope of EBNA2 (Kremmer et al., 1995) and induces a “supershift” in electrophoretic mobility shift assays (Zimber Strobl et al., 1993) while the clone 6C8 (rat IgG2a) binds to the Trp-Trp-Pro motif of EBNA2 and interferes with binding to RBPJκ (Sauder et al., 1994). For immunoprecipitation and gel-shift analysis, appropriate mouse or rat IgG isotype controls were used. For precipitation, 400μL of mAb supernatant were coupled to 100μL of settled protein-G-sepharose (“PGS”, GE Healthcare, München, Germany) for 1 h at 4°C under agitation, sedimented at 5.000 rpm in a tabletop centrifuge and washed once with 1 mL of lysis buffer 1. For precipitation experiments either 400μg protein of native whole cell extract or 100μg protein of native nuclear extract was added and incubated for 2h at 4°C under agitation, washed three times with lysis buffer 2 (PBS with 0,5% IGEPAL and 0,5M NaCl) and once with lysis buffer 1. The pellet was resuspended in 2 × SDS sample-buffer and incubated for 10 minutes at RT or heated at 98°C. For Western blot analysis using the aDMA-specific antibody 6F12, cell extracts or precipitated material was resuspended for up to 3h in sample buffer w/o β-mercaptoethanol at RT without heating. Peptide inhibition of the aDMA -specific antibody in Western blot analysis was carried out by incubating 300μL of 6F12 tissue culture supernatant with 20 μg of peptide dissolved in 10 μL PBS for 1h prior to addition of the antibody solution to the Western blot. Inhibition of the sDMA-specific antibodies in immunoprecipitation experiments, 300 μL of 13B10 antibody solution was bound for 2h to protein G Sepharose (PGS), then 20μg peptide in 20 μL of PBS were added and incubated for 18h at 4°C. The PGS was then washed and used for immunoprecipitation.
Sucrose-gradient centrifugation
200μL of native nuclear extract were loaded on a 5-40% sucrose-gradient (10mM HEPES pH 7,9, 5mM Na3PO4, 5mM KCl, 0,5mM MgCl2, 1mM DTT) and centrifuged at 265.000 × g for 6h at 4°C. Typically, 17 fractions were collected (Grässer et al., 1991) and either concentrated by ethanol-precipitation prior to Western blotting or subjected directly to precipitation as outlined above.
Electrophoretic mobility shift assay
Preparation of native nuclear extracts and electrophoretic mobility shift assay (EMSA) was carried out as described above (Sauder et al., 1994; Zimber Strobl et al., 1993). The gels were documented using a PhosphoImager®(Amersham). In vitro transcription-translation of EBNA2 was performed using the TNT® Coupled Reticulocyte Lysate System (Promega, Mannheim, Germany) as described (Barth et al., 2003) following the instruction of the manufacturer. Typically, 50 μL of the transcription-translation mix were programmed with 1 μg of vector DNA using T7 RNA polymerase. For in vitro generation of RBPJκ in this system, we used the vector AJ247 (Maier et al., 2005) kindly supplied by B. Kempkes, Helmholtz Zentrum München. To generate non-methylated protein, EBNA2 was vitro transcribed-translated using the E. coli-based “Rapid Translation System” (RTS 500®, Roche, Penzberg, Germany). For this purpose, the coding region of EBNA2 was PCR-amplified using the primers EBNA2-5′Nde (5′-ccg gaa ttc cat atg cct aca ttc tat ctt gc) and EBNA2-3′Sal (5′-gcg aat tcg tcg act tac tgg atg gag ggg cga g). The PCR product was inserted as a NdeI-SalI fragment into the pIVEX2.3-MCS vector supplied with the kit. This vector was then used to program the “RTS 500” reaction. To methylate this E. coli-derived EBNA2, 40μL of the TRS 500 reaction mixture were incubated for 2h at 30°C with 25μL of the reticulocyte lysate (see above) supplemented with 2μL of the 10× reaction buffer and S-Adenosyl-Methionine (SAM) at 1.5 μM as a methyl group donor.
Chromatin immunoprecipitation
Mutu cells were diluted to 5×105 cells/mL 24 hrs prior to chromatin preparation. Cells were then resuspended at 1×107 cells/mL in fresh media and chromatin prepared as described previously (Bark-Jones, Webb, and West, 2006). For immunoprecipitations carried out with the anti-sDMA EBNA2 (7D9) and anti-aDMA EBNA2 (6F12) mouse antibodies, 50μL of protein A/G sepharose beads (Sigma) were precoated with 13.5 μg rabbit anti-mouse imunoglobulins (Dako) overnight and then washed in immunoprecipitation dilution buffer (Bark-Jones, Webb, and West, 2006). Beads were then further incubated in 500 μL immunoprecipitation dilution buffer with 100μL of either control, 7D9 or 6F12 hybridoma supernatent for 3 hrs at 4°C with rotation. For the R3 rat monoclonal antibody, 50μL of protein A/G sepharose beads were directly precoated by incubation with 500 μL immunoprecipitation dilution buffer and 100μL hybridoma supernatent. Pre-coated beads were then washed and blocked by incubation with 350μg single-stranded sonicated salmon testes DNA (Sigma) for 1 hour at 4°C with rotation and then re-washed. Chromatin (105μL) was diluted ten-fold in immunoprecipitation dilution buffer and pre-cleared using 45μL of pre-blocked protein A Sepharose slurry at 4°C for one hour. Input control samples (40μL) were then removed from the supernatants and stored at −20°C and the diluted chromatin added to the antibody and salmon sperm coated beads and the immune complexes collected by rotation at 4°C overnight. Immune complexes were then washed, and precipitated chromatin eluted and proteinase K digested as described previously (Bark-Jones, Webb, and West, 2006). DNA was purified using the QIAquick Gel extraction Kit (Qiagen) and eluted in 110μL sterile millipore water.
Real-time PCR
Quantitative PCR (Q-PCR) was performed using an Applied Biosystems 7500 real time PCR machine. A series of dilutions of input control DNA (1/4, 1/16, 1/64 and 1/256) from Mutu 1 and Mutu III cells were used to create input standard curves for each primer set. Results were expressed as percentage input control following subtraction of the background signal obtained from control immunoprecipitations carried out using isotype matched control antibodies. Primers specific for the C promoter were described previously (Bark-Jones, Webb, and West, 2006). LMP1 promoter primers amplified the region from −320 to −235 relative to the LMP1 transcription start site (left primer 5′ GCA GAT TAC ACT GCC GCT TC 3′, right primer 5′ GGC CAA GTG CAA CAG GAA 3′), the region from +31 to +117 (left primer 5′ CCT GAG GAT GGA ACA CGA C 3′, right primer 5′ AGA GGA GGA GAA GGA GAG CAA 3′) and the region from +572 to +634 (left primer 5′ GGA GAT TCT CTG GCG ACT TG 3′, right primer 5′ TGA GCA GGA TGA GGT CTA GGA 3′). LMP2A promoter primers amplified the region from −272 to −209 relative to the LMP2A transcription start site (left primer 5′ GAT AGC CTC GCG ACT CGT GGG AA 3′, right primer 5′ AAT CTT CAC ACA CTG CTG CTG 3′), the region from +148 to +228 (left primer 5′ CCA ATA TCC ATC TGC TTC TGG 3′, right primer 5′ GGC TCT TCA TTA GAT TCA CGT TC 3′) and the region from +379 to +470 (left primer 5′ CTC ATC TCA ACA CAT ATA TGA AGA AGC 3′, right primer 5′ TTG ATG TGA CTT GTG ATG CAA T 3′).
Supplementary Material
Acknowledgements
We thank Ruth Nord for expert technical assistance, Richard Zimmermann, Dept. of Biochemistry, Universitätsklinikum Homburg, for the synthesis of E.coli-EBNA2 in the RTS 500 system, Bettina Kempkes, Helmholtz Zentrum München, for the RBPJk vector construct and Martin Rowe, CRUK Institute for Cancer Studies, Birmingham, UK for the Mutu cell lines and BZ.1 antibody. This work was supported by Deutsche Forschungsgemeinschaft (DFG) through grant KR2218/2-1 to E.K. and GR950/12-1 to F.G. and by the Wellcome Trust through grant 078291 to M.W. We thank Lothar Strobl, Helmholtz Zentrum München, for critical reading of the manuscript.
The abbrevations used are
- EBV
Epstein-Barr virus
- EBNA2
Epstein-Barr virus-encoded nuclear antigen 2
- EMSA
electrophoretic mobility shift assay
- NMA
non-methylated Arginine
- sDMA
symmetrically dimethylated Arginine
- aDMA
asymmetrically dimethylated Arginine
- AdOx
oxidized Adenosine
References
- Azzouz TN, Pillai RS, Dapp C, Chari A, Meister G, Kambach C, Fischer U, Schumperli D. Toward an assembly line for U7 snRNPs: interactions of U7-specific Lsm proteins with PRMT5 and SMN complexes. J Biol Chem. 2005;280(41):34435–40. doi: 10.1074/jbc.M505077200. Epub 2005 Aug 8. [DOI] [PubMed] [Google Scholar]
- Bark-Jones SJ, Webb HM, West MJ. EBV EBNA 2 stimulates CDK9-dependent transcription and RNA polymerase II phosphorylation on serine 5. Oncogene. 2006;25(12):1775–85. doi: 10.1038/sj.onc.1209205. [DOI] [PubMed] [Google Scholar]
- Barth S, Liss M, Voss MD, Dobner T, Fischer U, Meister G, Grässer FA. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine- glycine repeat to the survival motor neuron protein. J Virol. 2003;77(8):5008–13. doi: 10.1128/JVI.77.8.5008-5013.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jaker C, Hock J, Meister G, Grässer FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–75. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18(3):263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Bornkamm GW, Hammerschmidt W. Molecular virology of Epstein-Barr virus. Philos Trans R Soc Lond B Biol Sci. 2001;356(1408):437–59. doi: 10.1098/rstb.2000.0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318(5849):444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Cohen JI, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65(5):2545–54. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118(5):545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11(5):1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275(34):26370–5. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol. 1998;61:65–131. doi: 10.1016/s0079-6603(08)60825-9. [DOI] [PubMed] [Google Scholar]
- Grässer FA, Göttel S, Haiss P, Boldyreff B, Issinger OG, Mueller Lantzsch N. Phosphorylation of the Epstein-Barr virus nuclear antigen 2. Biochem Biophys Res Commun. 1992;186(3):1694–701. doi: 10.1016/s0006-291x(05)81604-3. [DOI] [PubMed] [Google Scholar]
- Grässer FA, Haiss P, Göttel S, Mueller Lantzsch N. Biochemical characterization of Epstein-Barr virus nuclear antigen 2A. J Virol. 1991;65(7):3779–88. doi: 10.1128/jvi.65.7.3779-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grässer FA, Sauder C, Haiss P, Hille A, K önig S, Göttel S, Kremmer E, Leinenbach HP, Zeppezauer M, Mueller Lantzsch N. Immunological detection of proteins associated with the Epstein-Barr virus nuclear antigen 2A. Virology. 1993;195(2):550–60. doi: 10.1006/viro.1993.1406. [DOI] [PubMed] [Google Scholar]
- Grundhoff AT, Kremmer E, Tureci O, Glieden A, Gindorf C, Atz J, Mueller Lantzsch N, Schubach WH, Grässer FA. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J Biol Chem. 1999;274(27):19136–44. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- Harada S, Yalamanchili R, Kieff E. Epstein-Barr virus nuclear protein 2 has at least two N-terminal domains that mediate self-association. J Virol. 2001;75(5):2482–7. doi: 10.1128/JVI.75.5.2482-2487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265(5168):92–5. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kim S, Merrill BM, Rajpurohit R, Kumar A, Stone KL, Papov VV, Schneiders JM, Szer W, Wilson SH, Paik WK, Williams KR. Identification of N(G)-methylarginine residues in human heterogeneous RNP protein A1: Phe/Gly-Gly-Gly-Arg-Gly-Gly-Gly/Phe is a preferred recognition motif. Biochemistry. 1997;36(17):5185–92. doi: 10.1021/bi9625509. [DOI] [PubMed] [Google Scholar]
- Kremmer E, Kranz B, Hille A, Klein K, Eulitz M, Hoffmann-Fezer G, Feiden W, Herrmann K, Delecluse H-J, Delsol G, Bornkamm GW, Mueller-Lantzsch N, Grässer FA. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B) Virology. 1995;208:336–42. doi: 10.1006/viro.1995.1157. [DOI] [PubMed] [Google Scholar]
- Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocr Rev. 2005;26(2):147–70. doi: 10.1210/er.2004-0008. Epub 2004 Oct 12. [DOI] [PubMed] [Google Scholar]
- Ling PD, Hayward SD. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J Virol. 1995;69(3):1944–50. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S, Santak M, Mantik A, Grabusic K, Kremmer E, Hammerschmidt W, Kempkes B. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J Virol. 2005;79(14):8784–92. doi: 10.1128/JVI.79.14.8784-8792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann KP, Staunton D, Thorley Lawson DA. Epstein-Barr virus-encoded protein found in plasma membranes of transformed cells. J Virol. 1985;55(3):710–20. doi: 10.1128/jvi.55.3.710-720.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger C, Strobl LJ, Marschall G, Bornkamm GW, Zimber-Strobl U. Crucial sequences within the Epstein-Barr virus TP1 promoter for EBNA2-mediated transactivation and interaction of EBNA2 with its responsive element. J Virol. 1994;68(11):7497–506. doi: 10.1128/jvi.68.11.7497-7506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen KA, Tang J, Zhu W, Schurter BT, Shuai K, Herschman HR, David M. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104(5):731–41. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- Paik WK, Kim S. Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem Biophys Res Commun. 1967;29(1):14–20. doi: 10.1016/0006-291x(67)90533-5. [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–45. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo RD, Webb HM, Gunnell A, West MJ. Regulation of transcription by the Epstein-Barr virus nuclear antigen EBNA 2. Biochem Soc Trans. 2008;36(Pt 4):625–8. doi: 10.1042/BST0360625. [DOI] [PubMed] [Google Scholar]
- Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176(2):563–74. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388(Pt 1):379–86. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. Vol. 2. Lippincott-Raven, Philadelphia: 2007. pp. 2655–2700. 2 vols. [Google Scholar]
- Sample J, Brooks L, Sample C, Young L, Rowe M, Gregory C, Rickinson A, Kieff E. Restricted Epstein-Barr virus protein expression in Burkitt lymphoma is due to a different Epstein-Barr nuclear antigen 1 transcriptional initiation site. Proc Natl Acad Sci U S A. 1991;88(14):6343–7. doi: 10.1073/pnas.88.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder C, Gotzinger N, Schubach WH, Horvath GC, Kremmer E, Krebs A, König S, Zimber Strobl U, Mueller Lantzsch N, Grässer FA. Mutational analysis of the Epstein-Barr virus nuclear antigen 2 by far-Western blotting and DNA-binding studies. J Gen Virol. 1996;77(Pt 5):991–6. doi: 10.1099/0022-1317-77-5-991. [DOI] [PubMed] [Google Scholar]
- Sauder C, Haiss P, Grässer FA, Zimber Strobl U, Mueller Lantzsch N. DNA-binding studies of the Epstein-Barr virus nuclear antigen 2 (EBNA-2): evidence for complex formation by latent membrane protein gene promoter-binding proteins in EBNA-2-positive cell lines. J Gen Virol. 1994;75(Pt 11):3067–79. doi: 10.1099/0022-1317-75-11-3067. [DOI] [PubMed] [Google Scholar]
- Stallcup MR, Kim JH, Teyssier C, Lee YH, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. J Steroid Biochem Mol Biol. 2003;85(2-5):139–45. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- Tong X, Yalamanchili R, Harada S, Kieff E. The EBNA-2 arginine-glycine domain is critical but not essential for B-lymphocyte growth transformation; the rest of region 3 lacks essential interactive domains. J Virol. 1994;68(10):6188–97. doi: 10.1128/jvi.68.10.6188-6197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui S, Schubach WH. Epstein-Barr virus nuclear protein 2A forms oligomers in vitro and in vivo through a region required for B-cell transformation. J Virol. 1994;68(7):4287–94. doi: 10.1128/jvi.68.7.4287-4294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss MD, Hille A, Barth S, Spurk A, Hennrich F, Holzer D, Mueller-Lantzsch N, Kremmer E, Grässer FA. Functional cooperation of Epstein-Barr virus nuclear antigen 2 and the survival motor neuron protein in transactivation of the viral LMP1 promoter. J.Virol. 2001;75(23):11781–90. doi: 10.1128/JVI.75.23.11781-11790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human J kappa recombination signal sequence binding protein (RBP-J kappa) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 1994;13(23):5633–8. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J kappa-EBNA2-activated transcription by inhibiting the binding of RBP-J kappa to DNA. J Virol. 1996;70(9):5909–15. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wu DY, Kalpana GV, Goff SP, Schubach WH. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70(9):6020–8. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DY, Krumm A, Schubach WH. Promotor-specific targeting of human SWi-SNF complex by Epstein-Barr virus nuclear protein 2. J.Virol. 2000;74(19):8893–8903. doi: 10.1128/jvi.74.19.8893-8903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, Rowe DT, Greenspan D, Greenspan JS, Rickinson AB, et al. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65(6):2868–74. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Davenport MG, Shackelford J, Pagano JS. Mitosis-specific hyperphosphorylation of Epstein-Barr virus nuclear antigen 2 suppresses its function. J Virol. 2004;78(7):3542–52. doi: 10.1128/JVI.78.7.3542-3552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Gershburg E, Pagano JS. Hyperphosphorylation of EBNA2 by Epstein-Barr virus protein kinase suppresses transactivation of the LMP1 promoter. J Virol. 2005;79(9):5880–5. doi: 10.1128/JVI.79.9.5880-5885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Shackelford J, Pagano JS. cdc2/cyclin B1-dependent phosphorylation of EBNA2 at Ser243 regulates its function in mitosis. J Virol. 2006;80(4):2045–50. doi: 10.1128/JVI.80.4.2045-2050.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber Strobl U, Kremmer E, Grässer F, Marschall G, Laux G, Bornkamm GW. The Epstein-Barr virus nuclear antigen 2 interacts with an EBNA2 responsive cis-element of the terminal protein 1 gene promoter. EMBO J. 1993;12(1):167–75. doi: 10.1002/j.1460-2075.1993.tb05642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimber Strobl U, Strobl LJ. EBNA2 and Notch signalling in Epstein-Barr virus mediated immortalization of B lymphocytes. Semin Cancer Biol. 2001;11(6):423–434. doi: 10.1006/scbi.2001.0409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.