Abstract
Background
Creatine is a key intermediate in energy metabolism and supplementation of creatine has been used for increasing muscle mass, strength and endurance. Creatine supplementation has also been reported to trigger the skeletal muscle expression of insulin like growth factor I, to increase the fat-free mass and improve cognition in elderly, and more explorative approaches like transcriptomics has revealed additional information. The aim of the present study was to reveal additional insight into the biochemical effects of creatine supplementation at the protein and metabolite level by integrating the explorative techniques, proteomics and NMR metabonomics, in a systems biology approach.
Methods
Differentiated mouse myotube cultures (C2C12) were exposed to 5 mM creatine monohydrate (CMH) for 24 hours. For proteomics studies, lysed myotubes were analyzed in single 2-DGE gels where the first dimension of protein separation was pI 5-8 and second dimension was a 12.5% Criterion gel. Differentially expressed protein spots of significance were excised from the gel, desalted and identified by peptide mass fingerprinting using MALDI-TOF MS. For NMR metabonomic studies, chloroform/methanol extractions of the myotubes were subjected to one-dimensional 1H NMR spectroscopy and the intracellular oxidative status of myotubes was assessed by intracellular DCFH2 oxidation after 24 h pre-incubation with CMH.
Results
The identified differentially expressed proteins included vimentin, malate dehydrogenase, peroxiredoxin, thioredoxin dependent peroxide reductase, and 75 kDa and 78 kDa glucose regulated protein precursors. After CMH exposure, up-regulated proteomic spots correlated positively with the NMR signals from creatine, while down-regulated proteomic spots were negatively correlated with these NMR signals. The identified differentially regulated proteins were related to energy metabolism, glucose regulated stress, cellular structure and the antioxidative defence system. The suggested improvement of the antioxidative defence was confirmed by a reduced intracellular DCFH2 oxidation with increasing concentrations of CMH in the 24 h pre-incubation medium.
Conclusions
The explorative approach of this study combined with the determination of a decreased intracellular DCFH2 oxidation revealed an additional stimulation of cellular antioxidative mechanisms when myotubes were exposed to CMH. This may contribute to an increased exercise performance mediated by increased ability to cope with training-induced increases in oxidative stress.
Background
Creatine is predominantly situated in skeletal muscle, and originates from both endogenous de novo synthesis and exogenous sources, which are mainly animal products [1]. Creatine and its phosphorylated form are well recognized as key intermediates in the energy metabolism of muscle fibres. Supplementation of creatine has been widely used among athletes as a means for increasing muscle mass and muscle strength and muscle endurance [2-4], but also for elderly people creatine supplementation, seems to enhance muscle strength [5]. The rationale behind CMH supplementation is to increase the content of creatine phosphate in the muscle, and several studies have also shown that the creatine content of the muscle is increased [6], and the majority of this is as creatine phosphate [1,2]. However, creatine supplementation has also been reported to trigger the expression of insulin like growth factor I (IGF-1) in young adults [7], increase fat-free mass [8] and improve cognition in the elderly [9]. Some of these findings have been supported by mechanistic studies in various muscle cell cultures, where IGF-1 [10], myogenesis [11] and protein synthesis [10,12,13] were increased, and also a more explorative approach using microarrays on muscle biopsies from creatine supplemented individuals revealed cytoskeleton remodelling, protein and glycogen synthesis regulation, as well as cell proliferation and differentiation [8]. Other techniques such as proteomics and metabonomics may reveal additional insight into some of the biochemical effects of creatine supplementation at the protein and metabolite level.
High-resolution 1H nuclear magnetic resonance (NMR) spectroscopy is a well-established analytical technique for metabolic fingerprinting of biofluids and various tissues and has also been used for elucidating the metabolic effects of dietary factors in both humans [14-17], animals [18-20], and also in cell cultures [21]. These studies have demonstrated that NMR-based metabonomics is extremely efficient in detecting endogenous and exogeneous metabolic perturbations. However, while being capable of identifying biomarkers and metabolic perturbations, the metabolic network responsible for the perturbations can only be hypothesised. Proteomics displays protein products as a result of gene expression and efficiency of translation, and has been used to separate and identify differentially regulated proteins in response to various treatments of cultured cells [22,23] and muscles [24]. Linking information obtained from metabolic fingerprinting with proteomics would pave the way for obtaining a better understanding of the primary pathways involved in perturbations associated with CMH supplementation. In this study we have for the first time examined and integrated the NMR metabolite profile and the proteomic profile of myotubes in the presence and absence of creatine supplementation in a systems biology approach.
Methods
Muscle Cell Culture
Myotube cultures were established from a mouse myoblast line (C2C12) originally derived from a thigh muscle [25] (American Type Culture Collection, Manassas, VA). A clone from this cell line, which effectively fused and formed myotubes, was isolated [26]. The clone was grown in 80 cm2 culture flask in 10 mL of medium consisting of Dulbecco's modified Eagle's medium (DMEM), 10% (vol/vol) fetal calf serum (FCS), and supplemented with 1% antibiotics giving 100 IU/mL penicillin, 100 μg/mL streptomycin sulfate, 3 μg/mL amphotericin B, and 20 μg/mL gentamycin (growth medium). Cells were maintained in an atmosphere of 95% air and 5% CO2 at 37°C. Prior to confluence, cells were harvested in 0.25% trypsin and sub-cultured into 80 cm2 culture flasks or 96 well plates. Cells were grown to confluence in growth medium, and left to fuse in medium containing 4% FCS (differentiation medium) without phenol red. Following approximately 6 days, the cultures contained differentiated multinuclear myotubes and were ready for experimental use. Culture medium was changed every other day throughout the culture period.
Myotube treatment and sampling for proteomics and metabonomics
For 24 hours the fully differentiated myotubes were cultured in the presence or absence of 5 mM creatine monohydrate (CMH) in the differentiation medium. The treatment and controls were performed in triplicate. Cells were washed in PBS and harvested in 10 ml phosphate buffered saline (PBS) by scraping the flask and mixed thoroughly. The protein content of the cell suspensions was analyzed by the bicinchoninic acid assay (BCA) (BioRad). Five aliquots of 200 μL of each of the triplicates were centrifuged at 6.000 × g for 5 min at 4°C. The cell pellet was kept at -80°C for proteome analysis. The remaining approximately 9 mL was centrifuged at 1000 × g for 10 min at 4°C. The pellet was washed in 1 mL D2O including 0.9% NaCl, centrifuged at 6.000 × g for 5 min and the pellet was kept at -80°C for metabonome analysis.
Two-dimensional gel electrophoresis (2-DGE)
The stored cell pellets were thawed, and 100 μL of lysis buffer (6 M urea, 2 M thiourea, 1.5% (w/v) pharmalyte, 0.8% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propansulfonate (CHAPS), 1% (w/v) dithioerythritol (DTE) in water) was added to triplicate samples. After incubation for 2 h at room temperature, the desired amount of protein from the two aliquots of each sample was combined and further diluted in a rehydration buffer to a final volume of 185 μL. The rehydration buffer consisted of the same substances, in same concentrations as the lysis buffer, but with pharmalyte (5 μL/mL) instead of 1% DTE. For analytical gels subjected to image analysis, a volume of the lysed cell fraction corresponding to 50 μg protein was applied. For preparative gels used for mass spectrometry (MS) analysis a volume corresponding to 125 μg protein was applied. The lysed cells were analyzed in single 2-DGE gel sets consisting of 6 gels representing the three biological replicates of either control cells or CMH treated cells. The first dimension of protein separation was carried out in immobilized 11 cm IPG strips (pH 5-8), whereas 12.5% Criterion gels (BioRad) were used for the second dimension. Running conditions for the 2-DGE gels were essentially as described earlier [27]. Analytical gels were silver stained according to Lametsch and Bendixen [27], whereas preparative gels were stained according to Shevchenko et al.[28].
In gel digestion, desalting and concentration of protein spots
Protein spots of significance were subjected to in-gel digestion by addition of trypsin essentially as described by Jensen et al. [29]. Custom-made chromatographic columns were used for desalting and concentration of the peptide mixture prior to MS analysis as described by Lametsch et al. [30]. The peptides were eluted in 0.5 μL matrix solution (15-20 g/L of α-cyano-4-hydroxycinnamic acid in 70% acetonitrile) directly onto the MALDI target plate.
Identification of myotube proteins by MALDI-TOF mass spectrometry
Mass spectra were obtained using a Bruker Ultraflex MALDI-TOF tandem mass spectrometer in reflection mode. A peptide calibration standard (0.2 μl) containing seven standard peptides ranging in molecular mass from 1046.54 to 3147.47 Da was spotted separately onto the MALDI target plate. The ion accelerating voltage was 25 kV with a delay time of 40 ns. The laser frequency was 50 Hz and 200 laser shots were accumulated for each spectrum. Proteins were identified by peptide mass fingerprinting (PMF) by mass searches in the database Swiss Prot (Swiss Institute of Bioinformatics, Genève, Switzerland) using the search program Mascot (Matrix Science, Boston, USA). In this program the experimental mass value, obtained from MS, is compared with calculated peptide masses from a database. A scoring algorithm is used to identify the closest match. Significant protein identifications (protein scores above 60, P < 0.05) were reported, and manually verified.
Image analysis
The 2-DGE gels were photographed by a Vilber Lourmat digital camera (ImageHouse, Copenhagen, Denmark) equipped with Gel Pro analyzer software. The gel spots were detected and quantified using ImageMaster 2D platimum software (Amersham Pharmacia Biotech, Uppsala, Sweden). After initial analysis using automated spot detection and segmentation, all images were manually checked and the spots were matched by comparing the relative positions of the individual spots on each gel, which reduced the number of spots used in the further analysis. The spots were quantified by adding the pixel intensities within the spot boundary, and the spot volumes were calculated. To overcome gel-to-gel variations in spot intensities due to technical variations related to the staining procedure, the relative spot volumes were calculated for each separate spot on the gels and these values were used in the further data analysis.
NMR measurements
Cells were extracted prior to NMR measurements using a dual methanol/chloroform extraction. The culture dishes were placed on liquid nitrogen and cells were added 2 mL of cold chloroform/methanol (1:1, vol/vol). The cells were homogenized using an electrical homogenizer, and centrifuged for 20 min at 1300 g at 4°C. After centrifugation the supernatants were collected and the pellets were resuspended with 1 mL of chloroform/methanol, centrifuged, and the supernatants were collected. The supernatant was washed with 1 mL ice-cold water, and the water phase was removed and added to the pellet. Two mL of water was added, the pellet was centrifuged, the supernatant was freeze-dried and subsequently dissolved in 0.6 mL D2O containing 0.5 mM sodium trimethylsilyl-[2,2,3,3-2H4]-1-propionate (TMSP), and analyzed by 1H NMR.
The NMR measurements were performed at 300 K on a Bruker Avance 400 NMR Spectrometer (Bruker BioSpin), operating at a 1H frequency of 400.13 MHz, and equipped with a standard 5-mm HX inverse probe. One-dimensional 1H NMR spectra were obtained using a single 90° pulse experiment, solvent suppression was achieved by irradiating the solvent peak during the relaxation delay of 2 s. A total of 128 transients of 8 K data points spanning a spectral width of 24.03 ppm were collected. An exponential line-broadening function of 1 Hz was applied to the free induction decay (FID) prior to Fourier transform (FT). All spectra were referenced in chemical shift value to the TMSP signal at 0 ppm. The 1H NMR spectra in the 10.0-5.0 and 4.5-0.5 ppm regions were subdivided into 0.005 ppm integral regions and integrated, reducing each spectrum into 616 independent variables. The reduced spectra were normalized to total intensity to remove any concentration effects.
DCFH2 oxidation analysis
Differentiated myotubes in 96 well plates were analyzed as described earlier [31]. Briefly, myotubes were pre-incubated with different concentrations of CMH (0.04-10 μM) for 24 h. Myotubes were then washed and loaded with 10 μM 2',7'dichlorodihydroflourescein diacetate (Molecular Probes, Inc. Eugene, OR) (H2DCF-DA) for 2 h at 37°C (95% air, 5% CO2) washed again, 100 μM H2O2 was added and intracellular DCFH2 oxidation was determined by fluorescence from 2,7-dichloroflourescein (DCF) at excitation and emission wavelengths of 490 and 515 nm, respectively, at 37°C with a microtiter plate reader (Synergy 2, BioTek Instruments Inc., Vermont, USA). Data is presented as average of 12 replicate wells after background correction.
Data analyses
Multivariate data analysis was performed using the Unscrambler software version 9.2 (Camo, Oslo, Norway). Partial least squares-discriminant analysis (PLS-DA) was performed on the metabonomic and the proteomic data to explore intrinsic biochemical dissimilarities between control cells and CMH treated cells. For the metabonomic data, the NMR signals were used as continuous X-parameters, while the treatment consisted the discriminant regressors (control = 0, treated = 1). For the proteomic data, the relative spot volumes obtained by image analysis of the 2-DGE gels were used as continuous X-parameters. Protein spots contributing least to the PLS-DA models were removed by Jack-knifing [32] through variable selection until an optimal calibrated and validated model was achieved, and based on the remaining spots significant (P < 0.05) regression coefficients were identified using the uncertainty test. For elucidation of correlations between metabonomic and proteomic data, a PLS-2 regression was carried out with NMR variables as X and proteomic spots identified as significant from the D-PLS model as y-variables. A students' t-test was carried out to compare the concentrations of each myotube protein in the triplicate controls and CMH treated C2C12 cells. A two-tailed paired t-test was used with a 0.95% confidence interval.
Results
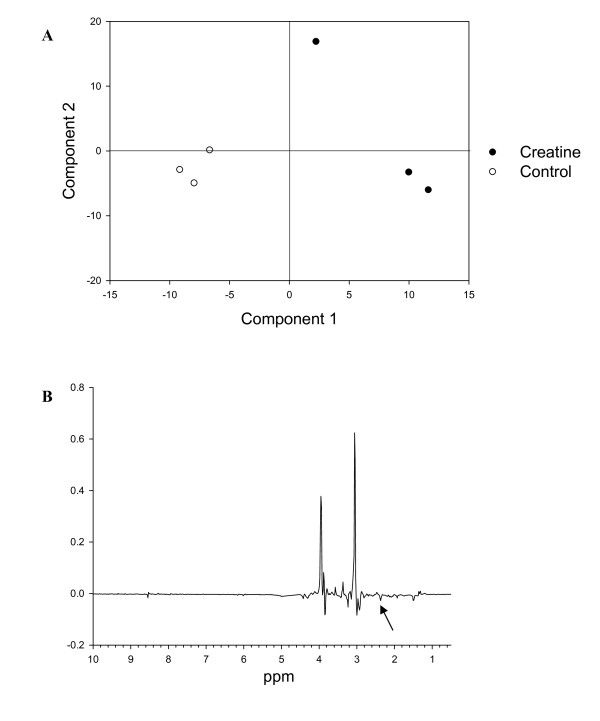
Myotubes exposed to creatine clearly changed their overall metabolic profile as reflected in the PLS-DA on the NMR-based metabonomic data (Figure 1A), where the discrimination between the control myotubes and CMH treated myotubes was evident. The loading plot (Figure 1B) revealed that signals at 3.04 ppm and 3.94 ppm dominates the discrimination, and this can be ascribed to a higher content of creatine in the treated cells, confirming the expected increased incorporation of creatine into the myotubes. The myotube protein expression in response to creatine was analyzed by proteomics using 2-DGE. An obtained proteomic profile of myotube extracts is shown in Figure 2.
Figure 1.
PLS-DA scores plot of NMR-based metabonomic data. (A) PLS-DA scores plot from analysis of NMR-based metabonomic data obtained on extracts of control (open circles) and creatine monohydrate (CMH) treated C2C12 muscle cells (closed circles), (B) the X-loadings of the PLS-DA. The dominating signals at 3.04 and 3.94 ppm are ascribed to CH3 and CH2 in creatine, respectively. The arrow shows a signal at 2.40 ppm, which was also found to be significant in the discrimination of control and CMH-treated cells. The 2.40 ppm signal is tentatively assigned to malate.
Figure 2.
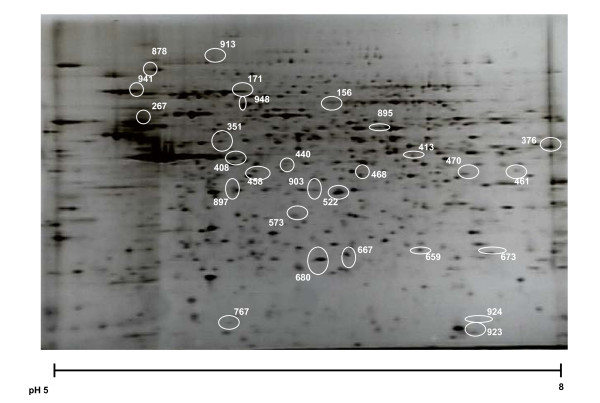
Proteomic profile of myotubes. Proteomic profile of myotubes as analyzed by 2-DGE visualized by silver staining. The positions of protein spots identified to be significantly different in controls and in creatine monohydrate-treated myotubes by PLS-DA of 2-DGE proteomics data are indicated.
After the manual check of the automatically assigned number of spots, a total of 584 protein spots were annotated by the image analysis and used in the further statistical analysis. By PLS-DA, 28 proteins were found to be differentially expressed when comparing CMH-treated myotubes with the control myotubes (results not shown). The significance of the spots identified by the PLS-DA was further tested by statistical t-test (Table 1). Of the 28 protein spots in the PLS-DA model, 20 of these were found to be either significantly different (P < 0.05) or exhibited tendency to be significantly different (P < 0.1) by the t-test. Accordingly, the t-test confirms that the intensities of the majority of the spots identified by PLS-DA are considerably affected by CMH treatment. Of these, 13 were up-regulated by CMH treatment, while 7 were down-regulated. This shows, as probably expected, that CMH stimulates the expression of more proteins than it down-regulates. The spots which were identified by the t-test to be differentially expressed in the myotubes in response to CMH treatment were cut out from the gels, and subjected to MALDI-TOF MS analysis using peptide mass fingerprinting. Those protein spots which were identified by MS are listed in Table 2. The identified proteins include vimentin, malate dehydrogenase, peroxiredoxin, thioredoxin dependent peroxide reductase, 75 kDa and 78 kDa glucose regulated protein precursors.
Table 1.
Relative protein spot volumes of spots identified by PLS-DA
| Spot ID | Mean C | SD | Mean CMH | SD | n | C→CMH1 | P-value |
|---|---|---|---|---|---|---|---|
| 156 | 0.140 | 0.042 | 0.271 | 0.005 | 3 | ↑ | 0.028 |
| 171 | 0.182 | 0.027 | 0.138 | 0.022 | 3 | ↓ | 0.004 |
| 267 | 0.309 | 0.248 | 0.811 | 0.233 | 3 | ↑ | 0.019 |
| 376 | 0.362 | 0.169 | 0.109 | 0.010 | 3 | ↓ | 0.120 |
| 408 | 0.400 | 0.072 | 0.380 | 0.165 | 3 | ↓ | 0.828 |
| 413 | 0.058 | 0.011 | 0.0716 | 0.002 | 3 | ↑ | 0.113 |
| 440 | 0.048 | 0.004 | 0.077 | 0.010 | 3 | ↑ | 0.042 |
| 458 | 0.118 | 0.003 | 0.102 | 0.002 | 3 | ↓ | 0.015 |
| 461 | 0.051 | 0.008 | 0.069 | 0.006 | 3 | ↑ | 0.134 |
| 483 | 0.072 | 0.005 | 0.087 | 0.004 | 3 | ↑ | 0.021 |
| 515 | 0.192 | 0.027 | 0.255 | 0.016 | 3 | ↑ | 0.079 |
| 522 | 0.410 | 0.008 | 0.587 | 0.081 | 3 | ↑ | 0.073 |
| 573 | 0.079 | 0.008 | 0.135 | 0.004 | 3 | ↑ | 0.002 |
| 659 | 0.091 | 0.005 | 0.107 | 0.005 | 3 | ↑ | 0.115 |
| 667 | 0.140 | 0.005 | 0.170 | 0.012 | 3 | ↑ | 0.038 |
| 673 | 0.140 | 0.027 | 0.187 | 0.006 | 3 | ↑ | 0.086 |
| 680 | 0.255 | 0.009 | 0.302 | 0.004 | 3 | ↑ | 0.006 |
| 767 | 0.062 | 0.005 | 0.040 | 0.012 | 3 | ↓ | 0.030 |
| 878 | 0.277 | 0.086 | 0.094 | 0.025 | 3 | ↓ | 0.055 |
| 895 | 0.175 | 0.011 | 0.114 | 0.016 | 3 | ↓ | 0.011 |
| 897 | 0.181 | 0.049 | 0.085 | 0.011 | 3 | ↓ | 0.066 |
| 900 | 0.087 | 0.008 | 0.048 | 0.011 | 3 | ↓ | 0.025 |
| 903 | 0.068 | 0.020 | 0.152 | 0.028 | 3 | ↑ | 0.086 |
| 923 | 0.070 | 0.018 | 0.153 | 0.031 | 3 | ↑ | 0.038 |
| 924 | 0.029 | 0.006 | 0.064 | 0.011 | 3 | ↑ | 0.015 |
| 941 | 0.566 | 0.184 | 0.078 | 0.134 | 3 | ↓ | 0.114 |
| 948 | 0.080 | 0.020 | 0.120 | 0.008 | 3 | ↑ | 0.126 |
| 951 | 0.047 | 0.021 | 0.045 | 0.024 | 3 | ↓ | 0.9 |
1, direction of change of relative spot volume in samples in relation to CHM treatment (C, data from control cells; CMH, data from CMH treated cells).
Table 2.
Proteins from myotubes identified by MALDI-TOF MS of spots after 2-DGE.
| Spot id | Protein | Sequence coveragea | Matched peptidesb | Scorec | Theo. pId | Theo. Mwe (kDa) | Access keyf |
|---|---|---|---|---|---|---|---|
| High in CMH | |||||||
| 267 | Vimentin | 37 | 21 | 189 | 4.9 | 54 | P20152 |
| 522 | Malate dehydrogenase - cytoplasmic | 21 | 6 | 65 | 6.2 | 37 | Q6PAB3 |
| 667 | Peroxiredoxin-4 | 26 | 6 | 73 | 6.8 | 31 | O08807 |
| 680 | Thioredoxin dependent peroxide reductase | 45 | 9 | 98 | 5.9 | 28 | P20108 |
| High in Controls | |||||||
| 171 | GRP75, 75 kDa glucose regulated protein precursor | 16 | 10 | 76 | 5.8 | 74 | P38674 |
| 941 | GRP78, 78 kDa glucose regulated protein precursor | 24 | 16 | 120 | 4.9 | 72 | P06761 |
a, The minimum coverage of the matched peptides in relation to the full-length sequence.
b, The number of matched peptides in the database search.
c, Score of the Mascot search.
d, Theoretical pI of the full length protein.
e, Theoretical molecular mass (Mw) of the full length protein.
f, Primary accession key in the SWISS-PROT database.
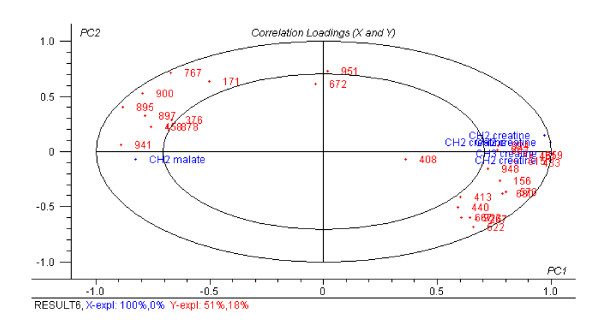
Moreover, in order to investigate the relationship between the proteomic spots, identified by the PLS-DA model and the metabolite profile of the myotubes, a PLS2 regression was carried out between the NMR metabolite profile and the 28 differentially regulated spots. Figure 3 shows the correlations loadings for variables selected from jack-knifing of this PLS-2 regression. Proteomic spots found to be up-regulated with CMH are clearly positively correlated with the NMR signals from creatine (3.05; 3.95-3.99 ppm), while proteomic spots found to be down-regulated with CMH are negatively correlated with these NMR signals. Noticeably, a NMR signal at 2.39 ppm, which is tentatively assigned to malate, is positively correlated with the proteomic spots down-regulated by CMH.
Figure 3.
Correlation loading plot of PLS2 (X: NMR variables, Y: selected proteomic spots). Correlation loading plot (1st and 2nd PLS component) of PLS2 using NMR variables as X and selected proteomic spots as Y. Jack knifing has been applied to eliminate insignificant variables. The inner and outer ellipses refer to 50 percent and 100 percent explained variance in X and Y, respectively. The validated explained variances are 100%/0% for X and 51%/18% for Y, the 1st and the 2nd component, respectively.
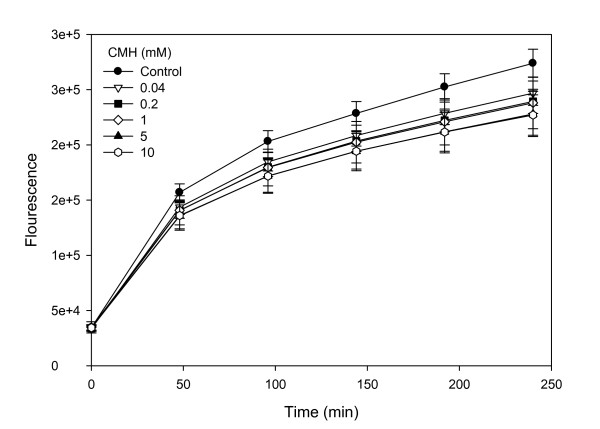
The results from the proteomic data indicate an antioxidative effect of CMH on the cells as two thioredoxin reductases (peroxiredoxin-4 and thioredoxin dependent peroxide reductase) were up-regulated. On the basis of this, the overall intracellular antioxidative capacity was analyzed in myotubes after pre-incubation with CMH for 24 h. The protective effect of CMH pre-incubation was supported by a reduced intracellular DCFH2 oxidation with increasing concentrations of CMH (Figure 4).
Figure 4.
Intracellular oxidation of 2,7-dichloroflourescein. Oxidation of intracellular 2,7-dichloroflourescein in myotube cultures exposed to 100 μM H2O2 after pre-incubation with increasing amounts of creatine monohydrate (CMH) for 24 h.
Discussion
The identified differentially regulated proteins (Table 2) are related to different cellular functions. Malate dehydrogenase is central in the energy metabolism, GRP75 and GRP78 are glucose regulated stress proteins, the filament protein vimentin is involved in maintaining cell integrity, and perturbation of the antioxidant defence system is indicated by peroxiredoxin-4 and thioredoxin dependent peroxide reductase.
The reason why malate dehydrogenase is elevated in creatine treated cultures is not known. However, we speculate that increased re-synthesis of glycogen is involved following treatment with CMH. This is based on the following considerations. In muscle creatine phosphate is an available energy source for muscle contraction during anaerobic conditions:
This reaction is under the control of creatine phosphokinase. Addition of CMH increases intra cellular concentrations of creatine (Figure 1) and this in turn will force the equilibrium to the right resulting in a higher level of creatine phosphate and ADP. Reduced ATP and increased ADP will increase the ratio of ADP:ATP which increases the rate of glycogenolysis. Thus, to restore ATP glycogen is degraded causing an elevated intracellular glucose level, which in the present study was indicated by down regulation of the glucose regulated protein precursors GRP75 and GRP78, both of which has been shown to increase with glucose starvation in the cell [33].
Following ATP restoration, glyconeogenesis is stimulated (by ATP). The substrate for the re-synthesis of glycogen is oxaloacetate and in the mitochondria oxaloacetate is converted to malate in order to enable the transport to the cytoplasm. Malate is then oxidised to oxaloacetate in the cytoplasm and this oxidation process is catalyzed by malate dehydrogenase. Oxaloacetate is then available as substrate for glycogen re-synthesis. Increased expression of malate dehydrogenase in CMH supplemented myotubes together with reduced intracellular content of the reaction substrate malate as detected by the NMR signal at 2.39 ppm. (Figure 3) support the assumptions above.
Thus, the data related to cellular energy metabolism broadly confirm previously described effects of CMH, but CMH supplementation has also been associated with cytoskeleton remodelling [8]. In the present study, structural perturbations were only indicated by an up regulation of the intermediate filament protein vimentin, which may just reflect maintenance of cellular integrity. Other studies have shown that neither muscle hypertrophy nor performance of rat skeletal muscle was augmented by creatine, and the authors argued that positive findings in relation to performance may rather be due to an enhanced ability to train [34].
Other effects of creatine support the hypothesis of creatine-induced improved ability to train through a direct antioxidant effect of creatine [35] on DNA molecules [36] or through activation of some of the cellular antioxidative systems.
The intracellular protection mechanisms against reactive oxygen species are very delicately balanced and, when exposed to stressors, adjustments in the defense mechanisms may be induced [37]. In various cell cultures including murine myoblasts an increased creatine level was associated with general cytoprotective effects towards oxidative agents [38,39]. However, the activities of the antioxidative enzymes catalase and glutathionperoxidase were not affected by creatine treatment [38,39], and the authors ascribed the cytoprotective effect to scavenging dependent antioxidative mechanisms [38]. In the present study on murine myotubes, we revealed an additional antioxidant effect of creatine, i.e. its capacity to induce up-regulation of one of the cellular antioxidative systems the thiol redox system, which consists of the glutathione and thioredoxin pathways [40]. Two thioredoxin reductases situated in the mitochondria and cytoplasm, respectively, were increased in creatine treated cells (Table 1); peroxiredoxin-4, a type 2 peroxiredoxin, and thioredoxin dependent peroxide reductase. These systems catalyse thiol-disulfide exchange reactions and thereby control the redox state of cytoplasmic cysteine residues, thus protecting e.g. radical sensitive enzymes from oxidative damage. An up-regulation of these very universally important redox systems as well as reduced intracellular DCFH2 oxidation (Figure 4) is an indication of an improved resistance towards oxidative challenges in cells exposed to CMH. Improvement of the intracellular antioxidative mechanisms will enhance the ability to cope with the increased levels of reactive oxygen species inevitably following increased exercise.
Conclusions
CMH supplementation increases the muscle cell energy metabolism, but the direct effect on performance is still debated and investigated. The explorative approach of this study, combined with the finding of a decreased intracellular DCFH2 oxidation, revealed an additional stimulation of cellular antioxidative mechanisms when exposed to CMH. This may contribute to an improved performance through increased ability to cope with training-induced increases in oxidative stress. Combined effects of increased energy load and improved antioxidative defences may thus be the key to the performance improvement experienced by some athletes following creatine supplement, but this approach needs further investigation [41].
List of abbreviations used
CHAPS: 3-[(3-cholamidopropyl) dimethylammonio]-1-propansulfonate; CMH: creatine monohydrate; DCFH2: 2',7'dichlorodihydroflourescein; 2-DGE: 2 dimentional gel electrophoresis; DTE: dithioerythritol; GRP75 and GRP78: glucose regulated protein precursors of 75 kDa and 78 kDa, respectively; MALDI-TOF: Matrix Assisted Laser Desorption Ionization - Time of Flight; MS: mass spectrometry; NMR: nuclear magnetic resonance; PLS: partial least squares; PLS-DA: PLS-discriminant analysis; TMSP: sodium trimethylsilyl-[2,2,3,3-2H4]-1-propionate.
Declaration of competing interests
The authors declare that they have no competing interests.
Authors' contributions
JFY and NO participated in the design of the cellular studies and JFY drafted the manuscript. IKS carried out the cellular experiments for proteomics and metabonomics studies. LBL designed the proteomics analysis and HCB, AM and NCN designed and carried out the metabonomics experiments. LBL, HCB and JFY carried out data and statistical analysis on proteomics, metabonomics and DCFH2 oxidation analysis, respectively. All authors contributed in the drafting of the manuscript and approved the final manuscript.
Contributor Information
Jette F Young, Email: JetteF.Young@agrsci.dk.
Lotte B Larsen, Email: LotteBach.Larsen@agrsci.dk.
Anders Malmendal, Email: anders@chem.au.dk.
Niels Chr Nielsen, Email: ncn@chem.au.dk.
Ida K Straadt, Email: Ida.Straadt@agrsci.dk.
Niels Oksbjerg, Email: Niels.Oksbjerg@agrsci.dk.
Hanne C Bertram, Email: HanneC.Bertram@agrsci.dk.
Acknowledgements
The authors wish to thank Hanne S. Møller, Inge Lise Sørensen and Anne-Grete Dyrvig Petersen for excellent technical assistance. The Danish Technology and Production Research Council (FTP) is thanked for financial support through the project "NMR-based metabonomics on tissues and biofluids" (project no. 274-05-339). The Danish National Research Foundation and the Danish Biotechnological Instrument Centre (DABIC) is acknowledged.
References
- Balsom PD, Söderlund K, Ekblom B. Creatine in humans with special reference to creatine supplementation. Sports Med. 1994;18:268–280. doi: 10.2165/00007256-199418040-00005. [DOI] [PubMed] [Google Scholar]
- Casey A, Constantin-Teodosiu D, Howell S, Hultman E, Greenhaff PL. Creatine ingestion favorably affects performance and muscle metabolism during maximal exercise in humans. Am J Physiol. 1996;271:E31–E37. doi: 10.1152/ajpendo.1996.271.1.E31. [DOI] [PubMed] [Google Scholar]
- Dentowski ABH, Opaszowski D, Blachnio D, Polanowski B. Effect of creatine supplementation on the performance in supramaximal, intermittent exercise. Biol Sport. 1997;14:291–298. [Google Scholar]
- Mujika I, Padilla S. Creatine supplementation as an ergogenic acid for sports performance in highly trained athletes: a critical review. Int J Sports Med. 1997;18:491–496. doi: 10.1055/s-2007-972670. [DOI] [PubMed] [Google Scholar]
- Gotshalk LA, Kraemer WJ, Mendonca MAG, Vingren JL, Kenny AM, Spiering BA, Hatfield DL, Fragala MS, Volek JS. Creatine supplementation improves muscular performance in older women. Eur J Appl Physiol. 2008;102:223–231. doi: 10.1007/s00421-007-0580-y. [DOI] [PubMed] [Google Scholar]
- Balsom PD, Söderlund K, Sjödin B, Ekblom B. Skeletal muscle metabolism during short duration high-intensity exercise: influence of creatine supplementation. Acta Physiol Scand. 1995;154:303–310. doi: 10.1111/j.1748-1716.1995.tb09914.x. [DOI] [PubMed] [Google Scholar]
- Burke DG, Candow DG, Chilibeck PD, MacNeil LG, Roy BD, Tarnopolsky MA, Ziegenfuss T. Effect of creatine supplementation and resistance-exercise training on muscle insulin-like growth factor in young adults. Int J Sport Nutr Exerc Metab. 2008;18:389–398. doi: 10.1249/01.mss.0000322659.59003.62. [DOI] [PubMed] [Google Scholar]
- Safdar A, Yardley NJ, Snow R, Melov S, Tarnopolsky MA. Global and targeted gene expression and protein content in skeletal muscle of young men following short-term creatine monohydrate supplementation. Physiol Genomics. 2008;32:219–228. doi: 10.1152/physiolgenomics.00157.2007. [DOI] [PubMed] [Google Scholar]
- McMorris T, Mielcarz G, Harris RC, Swain JP, Howard A. Creatine supplementation and cognitive performance in elderly individuals. Neuropsychol Dev Cogn B Aging Neuropsychol Cogn. 2007;14:517–528. doi: 10.1080/13825580600788100. [DOI] [PubMed] [Google Scholar]
- Louis M, Van Beneden R, Dehoux M, Thissen JP, Francaux M. Creatine increases IGF-I and myogenic regulatory factor mRNA in C(2)C(12) cells. FEBS Lett. 2004;557:243–247. doi: 10.1016/S0014-5793(03)01504-7. [DOI] [PubMed] [Google Scholar]
- Vierck JL, Icenoggle DL, Bucci L, Dodson MV. The effects of ergogenic compounds on myogenic satellite cells. Med Sci Sports Exerc. 2003;35:769–776. doi: 10.1249/01.MSS.0000065005.96298.01. [DOI] [PubMed] [Google Scholar]
- Ingwall JS, Weiner CD, Morales MF, Davis E, Stockdale FE. Specificity of creatine in the control of muscle protein synthesis. J Cell Biol. 1974;62:145–151. doi: 10.1083/jcb.62.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JF, Bertram HC, Theil PK, Petersen A-GD, Poulsen KA, Rasmussen M, Malmendal A, Nielsen NC, Vestergaard M, Oksbjerg N. In vitro and in vivo studies of creatine monohydrate supplementation to Duroc and Landrace pigs. Meat Sci. 2007;76:342–351. doi: 10.1016/j.meatsci.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Daykin CA, Van Duynhoven JPM, Groenewegen A, Dachtler M, Van Amelsvoort JMM, Mulder TPJ. Nuclear magnetic resonance spectroscopic based studies of the metabolism of black tea polyphenols in humans. J Agric Food Chem. 2005;53:1428–1434. doi: 10.1021/jf048439o. [DOI] [PubMed] [Google Scholar]
- Wang YL, Tang HR, Nicholson JK, Hylands PJ, Sampson J, Holmes E. A metabonomic strategy for the detection of the metabolic effects of chamomile (Matricaria recutita L.) ingestion. J Agric Food Chem. 2005;53:191–196. doi: 10.1021/jf0403282. [DOI] [PubMed] [Google Scholar]
- Solanky KS, Bailey NJC, Beckwith-Hall BM, Davis A, Bingham S, Holmes E, Nicholson JK, Cassidy A. Application of biofluid H-1 nuclear magnetic resonance-based metabonomic techniques for the analysis of the biochemical effects of dietary isoflavones on human plasma profile. Anal Biochem. 2003;323:197–204. doi: 10.1016/j.ab.2003.08.028. [DOI] [PubMed] [Google Scholar]
- Bertram HC, Duarte IF, Gil AM, Knudsen KEB, Laerke HN. Metabolic profiling of liver from hypercholesterolemic pigs fed rye or wheat fiber and from normal pigs. High-resolution magic angle spinning H-1 NMR spectroscopic study. Anal Chem. 2007;79:168–175. doi: 10.1021/ac061322+. [DOI] [PubMed] [Google Scholar]
- Solanky KS, Bailey NJ, Holmes E, Lindon JC, Davis AL, Mulder TP, Van Duynhoven JP, Nicholson JK. NMR-based metabonomic studies on the biochemical effects of epicatechin in the rat. J Agric Food Chem. 2003;51:4139–4145. doi: 10.1021/jf025677f. [DOI] [PubMed] [Google Scholar]
- Bertram HC, Hoppe C, Petersen BO, Duus JO, Molgaard C, Michaelsen KF. An NMR-based metabonomic investigation on effects of milk and meat protein diets given to 8-year-old boys. Br J Nutr. 2007;97:758–763. doi: 10.1017/S0007114507450322. [DOI] [PubMed] [Google Scholar]
- Bertram HC, Knudsen KEB, Serena A, Malmendal A, Nielsen NC, Frette XC, Andersen HJ. NMR-based metabonomic studies reveal changes in the biochemical profile of plasma and urine from pigs fed high-fibre rye bread. Br J Nutr. 2006;95:955–962. doi: 10.1079/BJN20061761. [DOI] [PubMed] [Google Scholar]
- Lamers RJ, Wessels EC, van de Sandt JJ, Venema K, Schaafsma G, van der Greef J, van Nesselrooij JH. A pilot study to investigate effects of inulin on Caco-2 cells through in vitro metabolic fingerprinting. J Nutr. 2003;133:3080–3084. doi: 10.1093/jn/133.10.3080. [DOI] [PubMed] [Google Scholar]
- Lin WY, Song CY, Pan TM. Proteomic analysis of Caco-2 cells treated with monacolin K. J Agric Food Chem. 2006;54:6192–6200. doi: 10.1021/jf061060c. [DOI] [PubMed] [Google Scholar]
- Purup S, Larsen LB. Integrated proteomics and biological assays for characterisation of non-nutritional effects of butyric acid on human intestinal cancer cells. J Anim Feed Sci. 2007;16S:163–171. [Google Scholar]
- Laville E, Sayd T, Terlouw C, Chambon C, Damon M, Larzul C, Leroy P, Glenisson J, Cherel P. Comparison of sarcoplasmic proteomes between two groups of pig muscles selected for shear force of cooked meat. J Agric Food Chem. 2007;55:5834–5841. doi: 10.1021/jf070462x. [DOI] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Oksbjerg N, Petersen JS, Sorensen IL, Henckel P, Vestergaard M, Ertbjerg P, Moller AJ, Bejerholm C, Stoier S. Long-term changes in performance and meat quality of Danish Landrace pigs: a study on a current compared with an unimproved genotype. Anim Sci. 2000;71:81–92. [Google Scholar]
- Lametsch R, Bendixen E. Proteome analysis applied to meat science: Characterizing post mortem changes in porcine muscle. J Agric Food Chem. 2001;49:4531–4537. doi: 10.1021/jf010103g. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Jensen ON, Larsen MR, Roepstorff P. Mass spectrometric identification and microcharacterization of proteins from electrophoretic gels: Strategies and applications. Proteins. 1998. pp. 74–89. [DOI] [PubMed]
- Lametsch R, Roepstorff P, Bendixen E. Identification of protein degradation during post-mortem storage of pig meat. J Agric Food Chem. 2002;50:5508–5512. doi: 10.1021/jf025555n. [DOI] [PubMed] [Google Scholar]
- Young JF, Christensen LP, Theil PK, Oksbjerg N. The polyacetylenes falcarinol and falcarindiol affect stress responses in myotube cultures in a biphasic manner. Dose-Response. 2008;6:239–251. doi: 10.2203/dose-response.08-008.Young. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens H, Martens M. Modified Jack-knife estimation of parameter uncertainty in bilinear modelling by partial least squares regression (PLSR) Food Qual Pref. 2000;11:5–16. doi: 10.1016/S0950-3293(99)00039-7. [DOI] [Google Scholar]
- Flores-Diaz M, Higuita JC, Florin I, Okada T, Pollesello P, Bergman T, Thelestam M, Mori K, Alape-Giron A. A cellular UDP-glucose deficiency causes overexpression of glucose/oxygen-regulated proteins independent of the endoplasmic reticulum stress elements. J Biol Chem. 2004;279:21724–21731. doi: 10.1074/jbc.M312791200. [DOI] [PubMed] [Google Scholar]
- Young JC, Young RE. The effect of creatine supplementation on glucose uptake in rat skeletal muscle. Life Sci. 2002;71:1731–1737. doi: 10.1016/S0024-3205(02)01941-0. [DOI] [PubMed] [Google Scholar]
- Lawler JM, Barnes WS, Wu G, Song W, Demaree S. Direct antioxidant properties of creatine. Biochem Biophys Res Commun. 2002;290:47–52. doi: 10.1006/bbrc.2001.6164. [DOI] [PubMed] [Google Scholar]
- Guidi C, Potenza L, Sestill P, Martinelli C, Guescini M, Stocchi L, Zeppa S, Polidori E, Annibalini G, Stocchi V. Differential effect of creatine on oxidatively-injured mitochondrial and nuclear DNA. Biochim Biophys Acta - General Subjects. 2008;1780:16–26. doi: 10.1016/j.bbagen.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Free Radicals and Antioxidants: A Personal View. Nutr Rev. 1994;52:253–265. doi: 10.1111/j.1753-4887.1994.tb01453.x. [DOI] [PubMed] [Google Scholar]
- Sestili P, Martinelli C, Bravi G, Piccoli G, Curci R, Battistelli M, Falcieri E, Agostini D, Gioacchini AM, Stocchi V. Creatine supplementation affords cytoprotection in oxidatively injured cultured mammalian cells via direct antioxidant activity. Free Radic Biol Med. 2006;40:837–849. doi: 10.1016/j.freeradbiomed.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Sestili P, Barbieri E, Martinelli C, Battistelli M, Guescini M, Vallorani L, Casadei L, D'Emilio A, Falcieri E, Piccoli G, Agostini D, Annibalini G, Paolillo M, Gioacchini AM, Stocchi V. Creatine supplementation prevents the inhibition of myogenic differentiation in oxidatively injured C2C12 murine myoblasts. Mol Nutr Food Res. 2009;53:1187–1204. doi: 10.1002/mnfr.200800504. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Hong SM, Kim BC, Park EH, Ahn K, Lim CJ. Effects of heterologous expression of thioredoxin reductase on the level of reactive oxygen species in COS-7 cells. Mol Cells. 2006;22:113–118. [PubMed] [Google Scholar]
- de Souza TP, Pereira B. Creatine: ergogenic aid with antioxidant potential? Revista de Nutricao-Brazilian Journal of Nutrition. 2008;21:349–353. [Google Scholar]