Abstract
Stat5a mediates prolactin-induced differentiation of mammary epithelia, and loss of Stat5 signaling in human breast cancer is associated with undifferentiated histology and poor prognosis. The transcriptional repressor BCL6 shares DNA target sequences with Stat5, disrupts differentiation of breast epithelia, is down-regulated during lactation, and upregulated in poorly differentiated breast cancer. Here we identify prolactin as a potent suppressor of BCL6 protein expression in human breast cancer through a mechanism that requires Stat5a, but not prolactin-activated Stat5b, MEK-ERK, or PI3K-AKT pathways. Prolactin rapidly suppressed BCL6 mRNA in T47D, MCF7, ZR75.1 and SKBr3 breast cancer cell lines, followed by prolonged reduction of BCL6 protein levels within 3h. Prolactin suppression of BCL6 was enhanced by overexpression of Stat5a but not Stat5b, was mimicked by constitutively active Stat5a, but did not require the transactivation domain of Stat5a. Stat5 chromatin immunoprecipitation demonstrated physical interaction with a BCL6 gene regulatory region, and BCL6 transcript repression required histone deacetylase activity based on sensitivity to trichostatin A. Functionally, BCL6 overexpression disrupted prolactin induction of Stat5 reporter genes. Prolactin suppression of BCL6 was extended to xenotransplant tumors in nude mice in vivo and to freshly isolated human breast cancer explants ex vivo. Quantitative immunohistochemistry revealed elevated BCL6 in high grade and metastatic breast cancer compared to DCIS and nonmalignant breast, and cellular BCL6 protein levels correlated negatively with nuclear Stat5a (r=−0.52; P<0.001) but not with Stat5b. Loss of prolactin-Stat5a signaling and concomitant upregulation of BCL6 may represent a regulatory switch facilitating undifferentiated histology and poor prognosis of breast cancer.
Keywords: breast cancer, BCL6, prolactin, Stat5a, Stat5b, transcription
Introduction
Prolactin regulates growth and differentiation of breast epithelia during pregnancy and lactation (1, 2). Prolactin activates the prolactin receptor-associated Jak2 tyrosine kinase and downstream signaling proteins, including signal transducer and activator of transcription-5a (Stat5a) and Stat5b (3). Stat5a and Stat5b have 92% amino acid identity and share many characteristics but are encoded by distinct genes that vary in tissue expression and display subtle differences in phospho-regulation and DNA binding (3). Upon tyrosine phosphorylation of a conserved motif, Stat5 proteins form stable homo and heterodimers that translocate to the nucleus and bind to target DNA sequences. Prolactin activates both Stat5a and Stat5b in mammary epithelia of pregnant and lactating rodents (1, 4) and in many human breast cancer cell lines (5). Nonetheless, Stat5a null mice have a more pronounced lactational deficiency than Stat5b null mice (1, 6).
Studies in mice indicate that prolactin promotes mammary tumor initiation and growth via Stat5a (7–14). Several other prolactin-responsive signaling pathways have also been implicated in breast cancer growth and progression including MEK-ERK, PI3K-AKT, and AP-1 pathways (11) (15–17). Evidence has also implicated prolactin and Stat5 in the maintenance of cell differentiation and suppression of invasive characteristics in breast cancer (18–21). Basal activation of Stat5 in healthy human breast epithelia is frequently lost in invasive and metastatic human breast cancer (19). Indeed, loss of active Stat5 in breast cancer correlated with poorly differentiated histology and poor prognosis (19, 22–24). Thus, a working model has been proposed in which prolactin-Jak2-Stat5 signaling promotes mammary tumor initiation but also maintains differentiation and suppresses progression of established breast cancer (25).
The proto-oncogene B-cell CLL/lymphoma 6 (BCL6) is a master regulator of B-lymphocyte development and facilitates proliferative expansion and blocks differentiation into plasma and memory cells (26). BCL6 is a zinc-finger protein and a potent transcriptional repressor (27). Intriguingly, the BCL6 consensus DNA binding sequence resembles that of Stat5 and BCL6 competes for binding to many Stat5 DNA interaction sites (28–30). Emerging evidence points to a tumor-promoting role of BCL6 in breast cancer. First, BCL6 protein is elevated in human breast cancers, especially in high grade, poorly differentiated cases (31, 32). Second, BCL6 is expressed in mouse mammary epithelia, primarily in virgin and pregnant animals but is completely suppressed during lactation, a terminal differentiation stage which coincides with peak activation of Stat5a and Stat5b (33). Third, overexpression of BCL6 in immortalized mouse mammary EpH4 cells blocked cellular differentiation and promoted proliferation, supporting a differentiation-suppressive role of BCL6 in mammary epithelial cells (32).
Both negative and positive regulation of BCL6 by Stat5 has been reported. Stat5 suppressed BCL6 expression in B-cell lymphomas, adipocytes and hepatocytes (28, 34, 35), but stimulated BCL6 in B lymphocytes (36) and in insulin-producing β-cells during pregnancy (37). A recent gene profiling study of breast cancer cells indicated that prolactin inhibited expression of BCL6 mRNA, an effect that could be mimicked by a constitutively active Stat5a mutant (38). However, the study did not determine whether prolactin affected BCL6 protein levels or whether Stat5b or other prolactin pathways were involved. In fact, exposure of mammary epithelial cells to prolactin-containing differentiation media increased BCL6 mRNA but not protein (32). The present study provides novel evidence that prolactin effectively suppresses BCL6 protein and mRNA levels in human breast cancer through a mechanism that depends on Stat5a but not prolactin signaling via Stat5b, MEK-ERK or AKT pathways. The data are supported by experimental studies of prolactin-responsive human breast cancer cell lines in vitro and in vivo, as well as patient tumors ex vivo. In addition, correlative studies on a progression series of archival human specimens representing normal and malignant breast tissues further supported the conclusions.
Materials and Methods
Tissue culture
T47D, SKBr3, ZR75.1 and MCF7 cells (ATCC) and surgical human breast tissue explants were cultured in RPMI medium containing 10% FBS and 1mM sodium pyruvate. MDA-MB-231 cells (ATCC) and HEK293 cells (Invitrogen) were grown in DMEM containing 10% FBS and 1mM sodium pyruvate. Recombinant human prolactin (AFP795) was provided by Dr. A.F. Parlow (National Hormone and Pituitary Program). Confluent, serum-starved SKBr3 cells were incubated with DMSO, 10 μM U0126 (Signagen), 10 μM LY294002 (Signagen) or 500 nM of TSA (Sigma) for 1h prior to prolactin stimulation.
Luciferase Assay
BCL6 promoter gene construct (pGL3-BCL6-pr) was generated by PCR using BCL6-pr-f and BCL6-pr-r primers (Table S1) to amplify the BCL6 regulatory Region B of the BCL6 gene (34), digested with KpnI and Hind3 and cloned into pGL3 vector. For BCL6 reporter assays, stably transfected T47D cells (T47D-BCL6-pr) were generated by cotransfecting pGL3-BCL6-pr and pcDNA3 (to provide neomycin selection; 10:1 ratio), and individual cell clones were selected with G418 (500 μg/ml). For Stat5 target gene reporter assays, T47D cells (1.5 × 105) were transiently cotransfected with either β-casein (39) or CIS (40) genomic reporter constructs and pCMV-SPORT6-BCL6 or pCMV-SPORT6 (Open Biosystems). After 12h, cells were serum-starved in RPMI without FBS for 16h and subsequently incubated with or without human prolactin (10 nM) for 24h. MDA-MB-231 cells were seeded at 1 × 105 cells/24-well and transfected with combinations of 0.3 μg of DNA constructs for β-casein reporter (39), CIS reporter (40), pcDNA3-hPRLR (41), pCMV-SPORT-BCL6 and pXM-Stat5a. Transfections were equalized for total DNA with pcDNA3 empty vector. After 24h, cells were incubated with vehicle control or prolactin (10 nM) in DMEM media containing 10% horse serum. Luciferase assays were performed 24h post prolactin stimulation (BMG PolarStar Optima luminescence reader; BMG Technologies).
Lentiviral and Adenoviral vectors
Lentivirus was produced in HEK-293 cells cotransfected with lentiviral vectors carrying shRNAs (Open Biosystems) for non-target control (SHC002), Stat5a (RHS-4533-NM-003142: TRCN0000019305 (5a2), TRCN0000019306 (5a3)) or Stat5b (RHS-4533-NM12448: TRCN0000019356 (5b3), TRCN0000019358 (5b6)) along with pCMV-dR8.2.dvpr and pCMV-VSV-G (Addgene 8454 and 8455) (42). SKBr3 cells (4 × 106/T25) were infected with individual lentivirus and incubated for 48h before exposure to prolactin. Cell lysates were subjected to immunoblot and qRT-PCR analyses. Stat5a, Stat5b, Stat5a-710F and Stat5a-Δ713 adenovirus preparations were prepared using double cesium chloride centrifugation (43) and used for gene delivery into SKBr3 cells (4 × 106/T25; MOI=5). After 24h, cells were incubated with or without prolactin (10 nM) in the absence of FBS for another 24h and subsequently harvested for qRT-PCR analysis.
T47D xenograft tumors
T47D xenotransplants were performed as previously described (39). Briefly, nude mice implanted with 17β-estradiol pellets (0.72 mg; Innovative Research of America) were injected s.c. with 5 × 106 T47D cells into two dorsolateral sites. Once tumors averaging 0.5 cm had formed, mice were injected s.c. with either vehicle control (N=10) or 5 μg/g body mass of human prolactin (N=10) every 12h for 48h. Tumors were harvested and processed for IHC and qRT-PCR.
Chromatin Immunoprecipitation
Confluent SKBr3 cells serum-starved for 16h were treated with or without prolactin (10 nM) for 1h and exposed to 1% formaldehyde for 5 min. Reactions were terminated with 0.125 M glycine. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS) for 1h and subsequently sonicated 10 times on ice. Lysates were incubated with binding buffer (0.1% SDS, 1.1% Triton-X 100, 167 mM NaCl, 16.7 mM Tris-HCl, pH 8.1) with a pan-Stat5 antibody (N20; Santa Cruz) overnight at 4oC, followed by capture with protein A Sepharose (Amersham) for 1h. Samples were washed with binding buffer and resuspended in 100 μl of TE prior to immunoblot and qRT-PCR analyses.
Quantitative reverse transcription polymerase chain reaction
qRT-PCR assays were performed with RNA isolated from cell lines and primary human breast tissues using RNeasy kit (Qiagen). cDNA was generated using Iscript (Bio-Rad). Both cDNA and ChIP DNA were subjected to quantitative PCR using corresponding primers (Table S1).
Immunoblotting
T47D and SKBr3 cell lysates were immunoprecipitated with 4 μl of rabbit Stat5a or Stat5b antibodies as described (39). Proteins were resolved by SDS-PAGE and immunoblotted with mouse pY-Stat5 (AX1, 1:10,000; Advantex), rabbit Stat5a (1:3,000) or rabbit Stat5b (1:1,500) antibodies followed by secondary HRP-conjugated anti-mouse or anti-rabbit antibodies, respectively. BCL6 expression was detected in whole cell lysates with BCL6 antibody (1:1,000; Santa Cruz) followed by secondary HRP-conjugated anti-rabbit antibody. Densitometric analyses were performed using Chemidoc scanner and Quantity One software (Bio-Rad) on three independent experiments.
Immunohistochemistry
Immunohistochemistry and AQUA analyses were performed on sections containing either xenotransplant tissues or a tissue array constructed by cutting edge matrix assembly containing 140 deidentified breast carcinoma specimens (DCIS, primary invasive ductal carcinomas (Grades 1–3), and lymph node metastases) and 40 normal breast tissues (Supplementary Data). Immunohistochemistry was performed as described previously (44) using Stat5a (Advantex, 1:8,000), Stat5b (Advantex, 1:4,000), pY-Stat5 (Epitomics, 1:200), and BCL6 (Santa Cruz, 1:50). AQUA analysis was performed using AQUA/PM2000 (HistoRx) (45). Briefly, slides were scanned and fluorescent images were captured in three channels (FITC/Alexa-488, Cy5, or DAPI). AQUA scores for Stat5a, Stat5b, pY-Stat5 and BCL6 represent average signal intensities within the epithelial cell compartment as defined by cytokeratin-positive mapping.
Results
Prolactin suppresses BCL6 protein and mRNA levels in breast cancer cell lines
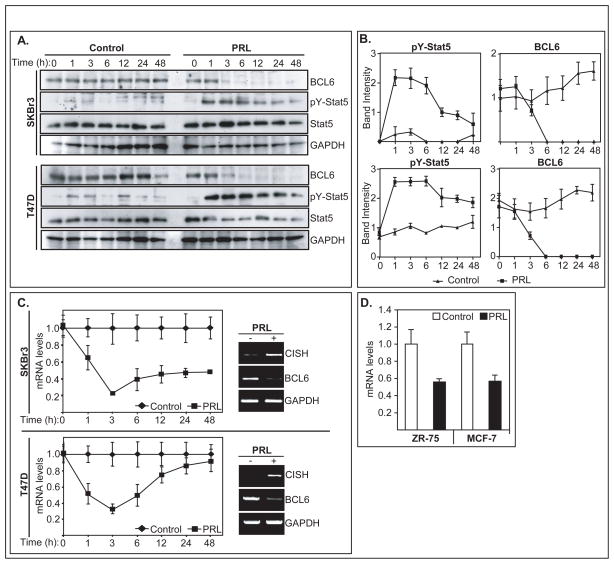
BCL6 protein levels in lysates of SKBr3 and T47D human breast cancer lines fell rapidly within 3h of prolactin stimulation, while levels remained unchanged in untreated cells (Figure 1A). By 6h and throughout the 48h time-course, levels of BCL6 protein in both cell lines were markedly suppressed in the continued presence of prolactin. In parallel, levels of pY-Stat5 rose rapidly following prolactin receptor activation and remained elevated. Densitometry on four independent experiments confirmed the inverse relationship between pY-Stat5 and BCL6 proteins in breast cancer cells (Figure 1B). Prolactin suppression of BCL6 protein levels in T47D and SKBr3 was associated with reduction in mRNA levels as revealed by qRT-PCR. BCL6 transcript levels were repressed as early as 1h of prolactin stimulation and reached maximum repression by 3h in both cell lines (Figure 1C). In contrast, Cytokine-Inducible SH2 (CISH) mRNA, an established prolactin-stimulated gene (46), was markedly induced by prolactin in both SKBr3 and T47D cells (Figure 1C). Marked inhibitory effect by prolactin on BCL6 mRNA levels was also observed in ZR-75.1 and MCF7 cells (Figure 1D), suggesting a broad negative regulation by prolactin of BCL6 expression in human breast cancer lines. The rapid suppression of BCL6 transcript levels by prolactin is consistent with the ~30min half-life of BCL6 mRNA (34).
Figure 1.
Prolactin suppression of BCL6 protein and mRNA in human breast cancer lines. (A) Immunoblot of representative time-course showing protein levels of BCL6, pY-Stat5, Stat5 and GAPDH in T47D and SKBr3 cells treated with or without prolactin for up to 48h. (B) Corresponding densitometry of BCL6 and pY-Stat5 (three experiments) normalized to GAPDH loading controls. (C) Time-course of BCL6 mRNA levels in T47D and SKBr3 cells (three experiments) in response to prolactin by qRT-PCR. Values are normalized to untreated control cells. Inset: prolactin stimulated CISH mRNA and reduced BCL6 mRNA by agarose gel. (D) BCL6 mRNA levels in ZR75.1 and MCF7 human breast cancer lines 24h post prolactin stimulation.
Prolactin suppression of BCL6 is dependent on Stat5a but not Stat5b, MEK/ERK or AKT pathways
BCL6 mRNA and protein expression were examined in SKBr3 cells treated with vehicle or prolactin in the presence of MEK inhibitor U0126 or AKT inhibitor LY294002. Cells exposed to neither inhibitor displayed marked prolactin suppression of BCL6 and stimulation of CISH mRNA, effects that were not affected by MEK or AKT inhibitors (Figure 2A, upper panel). The two inhibitors were effective as judged by inhibition of prolactin induced phosphorylation of ERK or AKT, but did not affect prolactin suppression of BCL6 protein levels as indicated in a representative protein blot and densitometric analyses of repeat experiments (Figure 2A, middle and bottom panels, respectively).
Figure 2.
Prolactin inhibits BCL6 expression via Stat5a but not Stat5b, ERK or AKT pathways. (A) qRT-PCR analyses of BCL6 mRNA in SKBr3 cells treated with or without prolactin with or without ERK (U0126) or AKT (LY294002) inhibitor (top panel). Densitometry (middle panels) and immunoblotting (bottom panel) showing BCL6, pY-Stat5, ERK, p-ERK, AKT, p-AKT protein levels in SKBr3 cells in the absence or presence of prolactin with or without U0126 or LY294002. (B) Stat5a is required for prolactin suppression of BCL6. qRT-PCR assay showing relief of BCL6 mRNA suppression in cells expressing Stat5a shRNA (5a2) but not Stat5b shRNA (5b6) (top panel). Densitometry (three experiments; middle panel) and a representative immunoblot (bottom panel) of BCL6, Stat5a, Stat5b and GAPDH proteins in SKBr3 cells expressing shRNA targeting either Stat5a (5a2 or 5a3) or Stat5b (5b3 or 5b6) or a non-target control shRNA (NT) using lentiviral delivery (* P<0.001 compared to levels in prolactin treated cells exposed to non-target (NT) control shRNA). (C) qRT-PCR analyses of BCL6 or CISH mRNA levels in SKBr3 cells overexpressing Stat5a (top left), Stat5b (top right), Stat5a-Δ713, Stat5a-S710F (bottom) using adenoviral gene delivery and 16h later treated with or without prolactin for 6h.
To examine the requirement of Stat5 signaling for prolactin-induced BCL6 repression, short hairpin RNA (shRNA) sequences that targeted either Stat5a (shRNA-5a2) or Stat5b (shRNA-5b6) or a non-target control shRNA were introduced into SKBr3 cells by lentiviral delivery, and cells were subsequently treated with prolactin for 6h. Selective knockdown of Stat5a significantly reversed both prolactin-suppression of BCL6 mRNA levels and stimulation of CISH mRNA expression, while selective knockdown of Stat5b did not (Figure 2B, upper panel). Prolactin suppression of BLC6 protein levels was also reversed by shRNA-5a2 as well as to a lesser but statistically significant extent by a second independent Stat5a-targeted shRNA, shRNA-5a3 (Figure 2B, middle and lower panels). Two shRNA constructs, 5b3 and 5b6, targeting Stat5b effectively knocked down Stat5b but did not affect prolactin suppression of BCL6 protein, consistent with mRNA data (Figure 2B, middle and lower panels).
Conversely, overexpression of Stat5a but not Stat5b in SKBr3 cells using adenoviral gene delivery suppressed basal levels of BCL6 and further enhanced prolactin suppression of BCL6 (Figure 2C, upper panels). The lack of efficacy of Stat5b could not be attributed to differences in expression (Figure S1). Furthermore, Stat5a-Δ713, which lacks the transactivation domain, retained the ability to mediate prolactin suppression of BCL6 (Figure 2C, lower left panel). Stat5a-Δ713 acts as a dominant-negative mutant for transactivation function and suppressed both basal and prolactin-induced CISH transcript levels (Figure 2C, lower right panel). Importantly, Stat5a-Δ713 was at least as effective as Stat5a in enhancing prolactin suppression of BCL6 protein levels. Finally, the constitutively active Stat5a-S710F mimicked prolactin suppression of BCL6 in the absence of prolactin and further suppressed BCL6 in response to prolactin (Figure 2C, lower left panel). In conclusion, prolactin suppression of BCL6 could be reversed by knockdown of Stat5a but not Stat5b or by disruption of MEK-ERK or AKT pathways.
Prolactin-activated Stat5 directly binds and functionally inhibits the BCL6 regulatory region by a trichostatin A sensitive mechanism
Stat5 chromatin immunoprecipitation (ChIP) assays in SKBr3 cells revealed prolactin-inducible Stat5 binding to the exon I region of the BCL6 gene (Region B), which contains 4 adjacent canonical GAS sites (Figure 3A) previously shown to be regulated by Stat5 in B-cell lymphoma lines (34). Anti-Stat5 chromatin immunoprecipitation and qPCR revealed that capture of the BCL6 response region almost exclusively occurred when Stat5 was activated although Stat5 protein was captured equally well in cell lysates from prolactin-treated or untreated cells (Figure 3B). In fact, activation of Stat5 was associated with more than 10-fold enrichment for BCL6 DNA (Figure 3C). Likewise, the genomic promoter region of CISH, a known Stat5 target gene, was also significantly enriched upon Stat5 immunoprecipitation in prolactin treated cells, but not the GAPDH negative control DNA.
Figure 3.
Stat5 directly interacts with BCL6 regulatory region in T47D breast cancer cells. (A) Graphic representation of BCL6 regulatory Region B containing four GAS sites. (B) Chromatin immunoprecipitation enriched for CISH and BCL6 regulatory regions in prolactin-stimulated cells, coinciding with increased pY-Stat5 levels. (C) Corresponding qPCR analyses of BCL6 and CISH regulatory sequences purified by anti-Stat5 chromatin immunoprecipitation from T47D cells treated with or without prolactin. (D) Luciferase reporter assays showing reconstitution of prolactin suppression of BCL6 regulatory region upon genomic integration of reporter gene into T47D cells.
To test whether the interaction between Stat5 and the BCL6 regulatory sequence was associated with transcriptional repression of BCL6, a genomic BCL6 luciferase reporter was generated that contained the regulatory Region B with the four GAS sites (34). When tested in transient transfection assays, prolactin consistently stimulated this reporter gene (data not shown) in agreement with previous analysis of this regulatory genomic element in isolation outside of chromatin context (34). However, when stably transfected into T47D cells, two out of ten clones consistently demonstrated prolactin repression of the BCL6 luciferase reporter gene by approximately 50% (Figure 3D) while the other clones did not respond to prolactin (data not shown). This observation suggested that prolactin repression of BCL6 is dependent on chromatin context and may require additional cofactors. In fact, prolactin-induced repression of BCL6 required HDAC activity as revealed by reversal upon pretreatment of cells with Trichostatin A (TSA), a histone deacetylase inhibitor that inactivates HDACs class I and II (47) (Figure S2). In the absence of TSA, prolactin effectively inhibited BCL6 mRNA expression, stimulated expression of CISH, and de-repressed the BCL6 target gene, BLIMP1 (26). TSA effectively blocked prolactin repression of BCL6 but did not affect basal levels of BCL6 (Figure S2A, S2B). Consistent with HDAC requirement for prolactin repression of BCL6, the associated prolactin de-repression of the BCL6-target gene, BLIMP1 (26), was also sensitive to Trichostatin A (Figure S2C). In contrast, prolactin stimulation of CISH mRNA levels remained intact (Figure S2D), a result consistent with the lack of requirement for HDAC for transcriptional activation by Stat5 of CISH. Collectively, ChIP assays and the reporter gene analyses provided evidence of direct involvement of Stat5 in occupying the regulatory region of the BCL6 gene, and suggested critical involvement of HDAC activity for gene suppression.
BCL6 interferes with Stat5-induced gene transcription
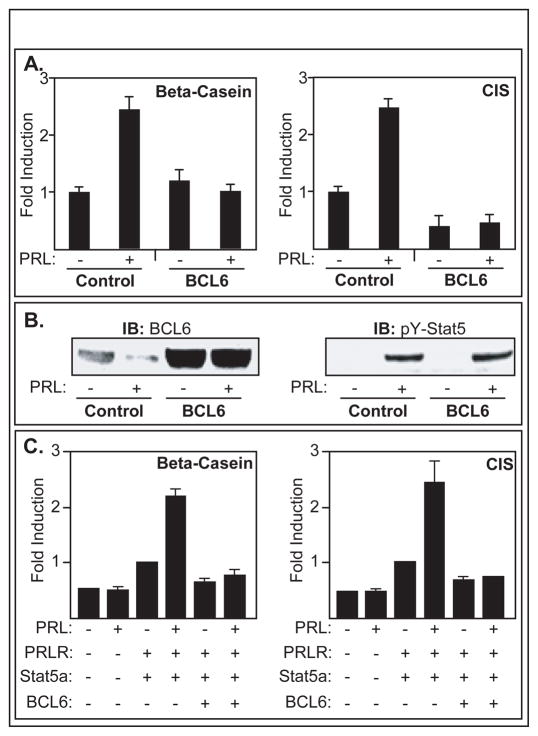
While Stat5a suppressed BCL6 protein expression, BCL6 conversely interfered with prolactin-Stat5 signaling in breast cancer. Overexpression of BCL6 in T47D cells completely blocked prolactin-induced expression of β-casein and CIS reporter gene constructs (Figures 4A, 4B), indicating that BCL6 effectively disrupts at least some of the Stat5-induced genes in breast cancer cells. Stat5a-induction of β-casein and CIS reporter genes was also disrupted by BCL6 in MDA-MB-231 breast cancer cells. Since our MDA-MB-231 cells do not express appreciable levels of prolactin receptor or Stat5, prolactin receptor and Stat5a cDNAs were also cotransfected. Overexpression of BCL6 completely blocked prolactin-induced expression of both Stat5 target genes in MDA-MB-231 cells (Figure 4C). We conclude that BCL6 disrupts prolactin induction of Stat5 regulated reporter genes in both T47D and MDA-MB-231 cells.
Figure 4.
BCL6 blocks prolactin-induced Stat5 target gene expression in breast cancer. (A) Luciferase assays demonstrating that overexpression of BCL6 abolished prolactin-stimulated β-casein and CIS reporter genes in T47D cells. (B) Corresponding immunoblots of BCL6 and pY-Stat5 proteins. (C) Luciferase assays demonstrating that BCL6 overexpression abolished prolactin induction of β-casein and CIS reporters in MDA-MB-231 cells expressing prolactin receptor and Stat5a.
Prolactin inhibits BCL6 expression in human breast cancer in vivo and ex vivo
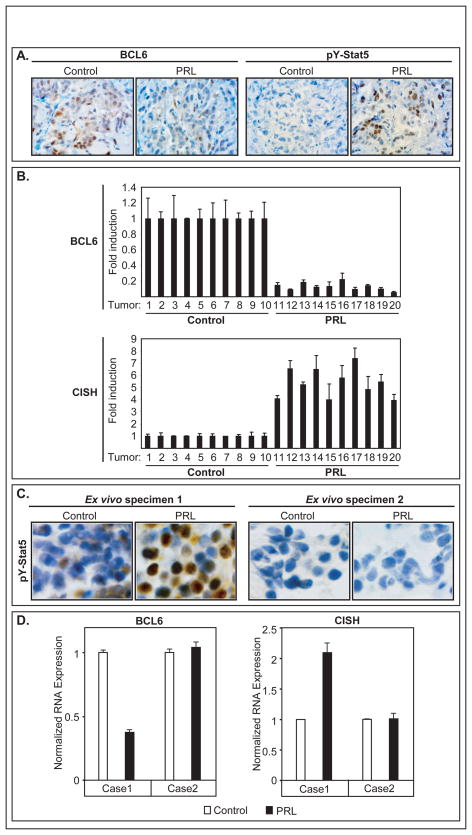
We tested whether BCL6 expression was suppressed by prolactin in vivo using T47D xenotransplants and ex vivo using freshly isolated explant cultures of human surgical breast cancer tissues. For xenotransplant experiments, T47D tumor bearing mice were treated with either PBS control (N=10) or human prolactin (N=10) for 48h. Immunohistochemistry revealed an inverse relationship between levels of nuclear pY-Stat5 and cellular BCL6 protein in xenotransplant tumors (Figure 5A). Without prolactin treatment, Stat5 was inactive and BCL6 expression was detectable in the majority of T47D tumor cells. In contrast, tumors in prolactin-treated animals displayed high levels of nuclear pY-Stat5 and markedly reduced BCL6 protein levels. In addition, qRT-PCR analysis showed high levels of BCL6 transcripts in untreated control tumors and at least 4-fold reduction of BCL6 transcripts in prolactin stimulated tumors, consistent with the observed in vitro prolactin suppression of BCL6 (Figure 5B). Conversely, control tumors expressed low levels of CISH mRNA that were stimulated up to 8-fold by prolactin treatment.
Figure 5.
Prolactin suppression of BCL6 expression in vivo. (A) Representative immunohistochemistry of pY-Stat5 and BCL6 proteins in T47D xenotransplant tumors in mice treated with or without prolactin for 48h. (B) Corresponding qRT-PCR analyses of BCL6 mRNA levels in T47D xenograft tumors. (C) Immunohistochemistry of two surgical human breast cancer tissue explants treated ex vivo with or without prolactin (100 nM) for 1h. Specimen 1 but not Specimen 2 is responsive to prolactin stimulation as measured by pY-Stat5 induction. (D) qRT-PCR quantified CISH and BCL6 mRNA levels in response to prolactin in Specimens 1 and 2.
Human primary breast cancer tissue explants in short-term ex vivo cultures were also examined, extending the effect of prolactin on BCL6 to more clinically relevant conditions. Human breast cancer tissue explants from two patients were exposed ex vivo to either vehicle or prolactin for 1h before subjected to immunohistochemical or qRT-PCR analyses. Specimen 1 responded to prolactin by increased levels of pY-Stat5 (Figure 5C) while Specimen 2 had no detectable pY-Stat5 in response to prolactin. qRT-PCR assays revealed that prolactin suppressed BCL6 expression more than two-fold in prolactin-responsive Specimen 1 but not in prolactin-unresponsive Specimen 2 (Figure 5D). Consistent with Stat5 activation, CISH mRNA was stimulated two-fold by prolactin in Specimen 1 but not in Specimen 2 (Figure 5D). Collectively, these data further extended prolactin suppression of BCL6 expression in human breast cancer to both in vivo and ex vivo conditions.
Cellular levels of BCL6 protein are negatively correlated with levels of nuclear localized Stat5a but not Stat5b in human breast cancer tissues
A breast cancer progression array containing 40 normal and 140 malignant breast tissues, including ductal carcinoma in situ (DCIS), invasive ductal carcinomas (IDC), and metastases, was analyzed by automated quantitative immunohistochemistry (45) for levels within the epithelial compartment of cellular BCL6 protein, nuclear localized pY-Stat5, nuclear localized Stat5a protein, and nuclear localized Stat5b protein (Figures 6A-C). Overall, while cellular levels of BCL6 rose, levels of nuclear localized, pY-Stat5 gradually fell over the progression series from normal breast to metastatic lesions (Figure 6B, left panel). A weak negative correlation between levels of BCL6 and nuclear localized tyrosine phosphorylated Stat5 (r=−0.23, P<0.014; Figure 6B, right panel) was observed in the clinical specimens. However, when Stat5a and Stat5b proteins were analyzed separately, a strong negative correlation was observed between levels of cellular BCL6 and nuclear Stat5a (r=−0.52, P<0.001) but not with nuclear Stat5b (Figure 6C). Instead, a weak positive correlation was noted between cellular BCL6 and nuclear Stat5b (r=0.27; P=0.006). The observed selective negative correlation between cellular BCL6 and nuclear Stat5a but not Stat5b is consistent with the observed selective mechanistic role of Stat5a in prolactin-suppression of BCL6 based on the in vitro cell line data.
Figure 6.
AQUA immunohistochemical quantification of BCL6 and pY-Stat5 protein levels in human breast tissues. (A) Representative immunofluorescent images of normal human breast tissue or primary breast cancer stained for pY-Stat5 (red) or BCL6 (red) and cytokeratin (green) and DNA (blue). Case 1, Case 2 and normal tissue were selected from a progression series to demonstrate a range of BCL6 and pY-Stat5 levels. (B) AQUA quantification of pY-Stat5 and BCL6 levels in a progression array of normal, DCIS and primary invasive breast cancer (grades 1–3), and metastases (left panel). Correlation analyses of levels of cellular BCL6 protein and nuclear localized pY-Stat5 in the human breast tissues (right panel). (C) Correlation analyses between levels of cellular BCL6 protein and nuclear Stat5a protein (left) or nuclear Stat5b protein (right).
Discussion
The present study provides novel evidence of prolactin suppression of BCL6 protein levels in human breast cancer and suggests a mechanism that selectively involves Stat5a despite robust parallel activation of Stat5b, ERK and AKT by prolactin in breast cancer cell lines. Prolactin inhibited BCL6 protein expression through rapid suppression of BCL6 mRNA, an effect that could be reversed by shRNA-mediated suppression of Stat5a but not Stat5b. A strong negative correlation between protein levels of cellular BCL6 and nuclear Stat5a, but not Stat5b in a progression material of normal and malignant breast tissues supported the selective role of Stat5a as a suppressor of BCL6 as suggested by the in vitro data and provided clinical relevance to the observations. Furthermore, several lines of evidence indicated that the effect was mediated by repressor activity and direct binding of Stat5a to regulatory elements in the BCL6 gene based on 1) chromatin immunoprecipitation, 2) rapid reduction of BCL6 mRNA levels within minutes of Stat5a activation, 3) requirement for HDAC activity as determined by sensitivity to TSA, and 4) the lack of requirement for the transactivation domain of Stat5a. Furthermore, a genomic BCL6 DNA fragment containing the Stat5-binding elements coupled to a luciferase reporter gene could restore Stat5-dependent repression when stably transfected into breast cancer cells. However, repression appeared to be chromatin context-dependent since not all of the stably transfected clones revealed repression. Finally, functional involvement of BCL6 as a direct negative regulator of Stat5-induced gene transcription supported the concept that elevated BCL6 may enhance the effects of reduced Stat5 signaling during breast cancer progression.
The observation that shRNA targeting of Stat5a but not Stat5b reversed prolactin suppression of BCL6 suggests a unique repressor capacity of prolactin-activated Stat5a that is not mimicked by prolactin-activated Stat5b in breast cancer cells. While Stat5a and Stat5b generally recognize the same primary GAS sites (3), the greater ability of Stat5a to form N-domain-dependent tetramers on tandem GAS sites (48) provides one possible Stat5a-selective mechanism. Further analyses are therefore needed to map in detail which of the regulatory GAS sites of the BCL6 gene are required for Stat5a-mediated repression of BCL6, whether Stat5a preferentially binds to this region, and to identify the exact molecular complexes responsible for prolactin suppression of BCL6 mRNA. The transcriptional co-repressor, SMRT, can bind to the coiled-coil domain of Stat5a and Stat5b activated by interleukin-3 and provide repressor functions (49), and future work will determine the requirement for SMRT for prolactin-dependent repression of BCL6.
Previously, the observed loss of nuclear localized, tyrosine phosphorylated Stat5 protein in invasive breast cancer was associated with poor prognosis and raised the possibility that Stat5 inhibits cell invasion and metastasis. In established breast cancer, active Stat5 is associated with more differentiated histology and experimental evidence suggests that Stat5a promotes differentiation and suppresses invasive features (19, 20, 23). Our present results demonstrating that prolactin-activated Stat5a negatively regulates BCL6 expression provide a new mechanism by which Stat5 may control differentiation of normal and malignant breast epithelia. BCL6 functions as a tumor promoting factor by blocking differentiation and stimulating cell cycle progression in lymphomas. Consistent with this action, poorly differentiated and rapidly growing breast cancers exhibited elevated levels of BCL6 protein. Overexpression of BCL6 in the mammary cell line EpH4 inhibited cellular differentiation and promoted growth by increasing cell proliferation and reducing apoptosis (32, 50). Correspondingly, in the present study, overexpression of BCL6 in breast cancer cell lines abolished prolactin induced expression of Stat5 reporter genes, β-casein and CIS. Ongoing studies will address the impact of BCL6 for prolactin regulation of breast cancer cell biology in vitro and in vivo.
In summary, the present work has revealed a novel negative regulatory interaction placing the proto-oncogene BCL6 within the prolactin-Jak2-Stat5a signaling network in human breast cancer. Resulting up-regulation of BCL6 may exacerbate the biological consequences associated with loss of Stat5a-signaling in breast cancer due to the suppressive effect of BCL6 on Stat5 target gene induction. Further work is now warranted to determine the diagnostic and therapeutic implications for human breast cancer of the mutually negative crosstalk between Stat5a and BCL6 signaling.
Supplementary Material
Acknowledgments
We thank Marja Nevalainen and Ayush Dagvadorj for Stat5b adenovirus, and Albert Kovatich and Arthur Gutierrez-Hartmann for helpful suggestions and discussions.
Supported by NIH grants, R01-CA101841 and R01-CA118740 (H.R.), NIH fellowship T32-CA09678 (T.H.T.), and NCI Support Grant 1P30CA56036 to the Kimmel Cancer Center. Project is funded, in part, under a Commonwealth University Research Enhancement Program grant with the Pennsylvania Department of Health (H.R.). The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Hennighausen L, Robinson GW, Wagner KU, Liu W. Prolactin signaling in mammary gland development. J Biol Chem. 1997;272:7567–9. doi: 10.1074/jbc.272.12.7567. [DOI] [PubMed] [Google Scholar]
- 3.Grimley PM, Dong F, Rui H. Stat5a and Stat5b: fraternal twins of signal transduction and transcriptional activation. Cytokine Growth Factor Rev. 1999;10:131–57. doi: 10.1016/s1359-6101(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 4.Cella N, Groner B, Hynes NE. Characterization of Stat5a and Stat5b homodimers and heterodimers and their association with the glucocortiocoid receptor in mammary cells. Mol Cell Biol. 1998;18:1783–92. doi: 10.1128/mcb.18.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neilson LM, Zhu J, Xie J, et al. Coactivation of janus tyrosine kinase (Jak)1 positively modulates prolactin-Jak2 signaling in breast cancer: recruitment of ERK and signal transducer and activator of transcription (Stat)3 and enhancement of Akt and Stat5a/b pathways. Mol Endocrinol. 2007;21:2218–32. doi: 10.1210/me.2007-0173. [DOI] [PubMed] [Google Scholar]
- 6.Udy GB, Towers RP, Snell RG, et al. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci U S A. 1997;94:7239–44. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wennbo H, Gebre-Medhin M, Gritli-Linde A, Ohlsson C, Isaksson OG, Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. J Clin Invest. 1997;100:2744–51. doi: 10.1172/JCI119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose-Hellekant TA, Arendt LM, Schroeder MD, Gilchrist K, Sandgren EP, Schuler LA. Prolactin induces ERalpha-positive and ERalpha-negative mammary cancer in transgenic mice. Oncogene. 2003;22:4664–74. doi: 10.1038/sj.onc.1206619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manhes C, Kayser C, Bertheau P, et al. Local over-expression of prolactin in differentiating mouse mammary gland induces functional defects and benign lesions, but no carcinoma. J Endocrinol. 2006;190:271–85. doi: 10.1677/joe.1.06829. [DOI] [PubMed] [Google Scholar]
- 10.Ren X, Zhang X, Kim AS, et al. Comparative genomics of susceptibility to mammary carcinogenesis among inbred rat strains: role of reduced prolactin signaling in resistance of the Copenhagen strain. Carcinogenesis. 2008;29:177–85. doi: 10.1093/carcin/bgm224. [DOI] [PubMed] [Google Scholar]
- 11.Clevenger CV, Furth PA, Hankinson SE, Schuler LA. The role of prolactin in mammary carcinoma. Endocr Rev. 2003;24:1–27. doi: 10.1210/er.2001-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barash I. Stat5 in the mammary gland: controlling normal development and cancer. J Cell Physiol. 2006;209:305–13. doi: 10.1002/jcp.20771. [DOI] [PubMed] [Google Scholar]
- 13.Humphreys RC, Hennighausen L. Signal transducer and activator of transcription 5a influences mammary epithelial cell survival and tumorigenesis. Cell Growth Differ. 1999;10:685–94. [PubMed] [Google Scholar]
- 14.Ren S, Cai HR, Li M, Furth PA. Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene. 2002;21:4335–9. doi: 10.1038/sj.onc.1205484. [DOI] [PubMed] [Google Scholar]
- 15.Das R, Vonderhaar BK. Activation of raf-1, MEK, and MAP kinase in prolactin responsive mammary cells. Breast Cancer Res Treat. 1996;40:141–9. doi: 10.1007/BF01806209. [DOI] [PubMed] [Google Scholar]
- 16.Das R, Vonderhaar BK. Prolactin as a mitogen in mammary cells. J Mammary Gland Biol Neoplasia. 1997;2:29–39. doi: 10.1023/a:1026369412612. [DOI] [PubMed] [Google Scholar]
- 17.Gutzman JH, Rugowski DE, Nikolai SE, Schuler LA. Stat5 activation inhibits prolactin-induced AP-1 activity: distinct prolactin-initiated signals in tumorigenesis dependent on cell context. Oncogene. 2007;26:6341–8. doi: 10.1038/sj.onc.1210454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nouhi Z, Chughtai N, Hartley S, Cocolakis E, Lebrun JJ, Ali S. Defining the role of prolactin as an invasion suppressor hormone in breast cancer cells. Cancer Res. 2006;66:1824–32. doi: 10.1158/0008-5472.CAN-05-2292. [DOI] [PubMed] [Google Scholar]
- 19.Nevalainen MT, Xie J, Torhorst J, et al. Signal transducer and activator of transcription-5 activation and breast cancer prognosis. J Clin Oncol. 2004;22:2053–60. doi: 10.1200/JCO.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 20.Sultan AS, Xie J, LeBaron MJ, Ealley EL, Nevalainen MT, Rui H. Stat5 promotes homotypic adhesion and inhibits invasive characteristics of human breast cancer cells. Oncogene. 2005;24:746–60. doi: 10.1038/sj.onc.1208203. [DOI] [PubMed] [Google Scholar]
- 21.Sultan AS, Brim H, Sherif ZA. Co-overexpression of Janus kinase 2 and signal transducer and activator of transcription 5a promotes differentiation of mammary cancer cells through reversal of epithelial-mesenchymal transition. Cancer Sci. 2008;99:272–9. doi: 10.1111/j.1349-7006.2007.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamashita H, Nishio M, Ando Y, et al. Stat5 expression predicts response to endocrine therapy and improves survival in estrogen receptor-positive breast cancer. Endocr Relat Cancer. 2006;13:885–93. doi: 10.1677/erc.1.01095. [DOI] [PubMed] [Google Scholar]
- 23.Cotarla I, Ren S, Zhang Y, Gehan E, Singh B, Furth PA. Stat5a is tyrosine phosphorylated and nuclear localized in a high proportion of human breast cancers. Int J Cancer. 2004;108:665–71. doi: 10.1002/ijc.11619. [DOI] [PubMed] [Google Scholar]
- 24.Strauss BL, Bratthauer GL, Tavassoli FA. STAT 5a expression in the breast is maintained in secretory carcinoma, in contrast to other histologic types. Hum Pathol. 2006;37:586–92. doi: 10.1016/j.humpath.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Wagner KU, Rui H. Jak2/Stat5 signaling in mammogenesis, breast cancer initiation and progression. J Mammary Gland Biol Neoplasia. 2008;13:93–103. doi: 10.1007/s10911-008-9062-z. [DOI] [PubMed] [Google Scholar]
- 26.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 27.Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci U S A. 1996;93:6947–52. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer RD, Laz EV, Su T, Waxman DJ. Male-Specific Hepatic Bcl6: Growth Hormone-Induced Block of Transcription Elongation in Females and Binding to Target Genes Inversely Coordinated with STAT5. Mol Endocrinol. 2009 doi: 10.1210/me.2009-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez de Mattos S, Essafi A, Soeiro I, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol. 2004;24:10058–71. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda N, Arima M, Tsuruoka N, et al. Bcl6 is a transcriptional repressor for the IL-18 gene. J Immunol. 2003;171:426–31. doi: 10.4049/jimmunol.171.1.426. [DOI] [PubMed] [Google Scholar]
- 31.Bos R, van Diest PJ, van der Groep P, et al. Protein expression of B-cell lymphoma gene 6 (BCL-6) in invasive breast cancer is associated with cyclin D1 and hypoxia-inducible factor-1alpha (HIF-1alpha) Oncogene. 2003;22:8948–51. doi: 10.1038/sj.onc.1206995. [DOI] [PubMed] [Google Scholar]
- 32.Logarajah S, Hunter P, Kraman M, et al. BCL-6 is expressed in breast cancer and prevents mammary epithelial differentiation. Oncogene. 2003;22:5572–8. doi: 10.1038/sj.onc.1206689. [DOI] [PubMed] [Google Scholar]
- 33.Nevalainen MT, Xie J, Bubendorf L, Wagner KU, Rui H. Basal activation of transcription factor signal transducer and activator of transcription (Stat5) in nonpregnant mouse and human breast epithelium. Mol Endocrinol. 2002;16:1108–24. doi: 10.1210/mend.16.5.0839. [DOI] [PubMed] [Google Scholar]
- 34.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–33. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Lin G, Huo JS, et al. Computational and functional analysis of growth hormone-regulated genes identifies the transcriptional repressor Bcl6 as a participant in GH-regulated transcription. Endocrinology. 2009 doi: 10.1210/en.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheeren FA, Naspetti M, Diehl S, et al. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat Immunol. 2005;6:303–13. doi: 10.1038/ni1172. [DOI] [PubMed] [Google Scholar]
- 37.Karnik SK, Chen H, McLean GW, et al. Menin controls growth of pancreatic beta-cells in pregnant mice and promotes gestational diabetes mellitus. Science. 2007;318:806–9. doi: 10.1126/science.1146812. [DOI] [PubMed] [Google Scholar]
- 38.Walker SR, Nelson EA, Zou L, et al. Reciprocal Effects of STAT5 and STAT3 in Breast Cancer. Mol Cancer Res. 2009 doi: 10.1158/1541-7786.MCR-08-0238. [DOI] [PubMed] [Google Scholar]
- 39.Utama FE, LeBaron MJ, Neilson LM, et al. Human prolactin receptors are insensitive to mouse prolactin: implications for xenotransplant modeling of human breast cancer in mice. J Endocrinol. 2006;188:589–601. doi: 10.1677/joe.1.06560. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto A, Masuhara M, Mitsui K, et al. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–54. [PubMed] [Google Scholar]
- 41.Yamashita H, Xu J, Erwin RA, Farrar WL, Kirken RA, Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J Biol Chem. 1998;273:30218–24. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- 42.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dagvadorj A, Kirken RA, Leiby B, Karras J, Nevalainen MT. Transcription factor signal transducer and activator of transcription 5 promotes growth of human prostate cancer cells in vivo. Clin Cancer Res. 2008;14:1317–24. doi: 10.1158/1078-0432.CCR-07-2024. [DOI] [PubMed] [Google Scholar]
- 44.Plotnikov A, Li Y, Tran TH, et al. Oncogene-mediated inhibition of glycogen synthase kinase 3 beta impairs degradation of prolactin receptor. Cancer Res. 2008;68:1354–61. doi: 10.1158/0008-5472.CAN-07-6094. [DOI] [PubMed] [Google Scholar]
- 45.Dolled-Filhart M, McCabe A, Giltnane J, Cregger M, Camp RL, Rimm DL. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006;66:5487–94. doi: 10.1158/0008-5472.CAN-06-0100. [DOI] [PubMed] [Google Scholar]
- 46.LeBaron MJ, Xie J, Rui H. Evaluation of genome-wide chromatin library of Stat5 binding sites in human breast cancer. Mol Cancer. 2005;4:6. doi: 10.1186/1476-4598-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard WJ. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol Cell Biol. 2000;20:389–401. doi: 10.1128/mcb.20.1.389-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima H, Brindle PK, Handa M, Ihle JN. Functional interaction of STAT5 and nuclear receptor co-repressor SMRT: implications in negative regulation of STAT5-dependent transcription. Embo J. 2001;20:6836–44. doi: 10.1093/emboj/20.23.6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alenzi FQ. BCL-6 prevents mammary epithelial apoptosis and promotes cell survival. J Pak Med Assoc. 2008;58:494–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.