Abstract
Sleeping Beauty transposon (SB-Tn) has emerged as an important nonviral vector for integrating transgenes into mammalian genomes. We report here a novel dual fluorescent reporter cis SB-Tn system that permitted nonselective fluorescent-activated cell sorting for SB-Tn-transduced K562 erythroid cells. Using an internal ribosome entry site element, the green fluorescent protein (eGFP) was linked to the SB10 transposase gene as an indirect marker for the robust expression of SB10 transposase. Flourescence-activated cell sorting (FACS) by eGFP resulted in significant enrichment (> 60%) of cells exhibiting SB-Tn-mediated genomic insertions and long-term expression of a DsRed transgene. The hybrid erythroid-specific promoter of DsRed transgene was verified in erythroid or megakaryocyte differentiation of K562 cells. Bisulfite-mediated genomic analyses identified different DNA methylation patterns between DsRed+ and DsRed− cell clones, suggesting a critical role in transgene expression. Moreover, although the host genomic copy of the promoter element showed no CpG methylation, the same sequence carried by the transgene was markedly hypermethylated. Additional evidence also suggested a role for histone deacetylation in the regulation of DsRed transgene. The presence of SB transgene affected the expression of neighboring host genes at distances > 45 kb. Our data suggested that fluorescent reporter cis SB-Tn system can be used to enrich mammalian cells harboring SB-mediated transgene insertions. The epigenetic modification detected on the DsRed transgene demonstrated that transgenes inserted by SB could also be selectively modified by host cellular epigenetic systems. In addition, long-range activation of host genes must now be recognized as a potential consequence of an inserted transgene cassette containing enhancer elements.
The Sleeping Beauty transposon (SB-Tn) is a nonviral gene delivery system based on a Tc1/mariner type transposon, whose activity was restored by genetic correction of accumulated mutations (1). SB-mediated transposition utilizes a “cut and paste” mechanism through SB transposase binding to the inverted repeats (IRs) of the IR/DRs that flank the DNA fragment for integration into a genomic TA dinucleotide. Shortly after its reactivation, the SB-Tn system was successfully applied to the insertion and expression of transgenes into human cells and mammalian somatic tissues (1, 2). The SB-Tn is now widely used as a gene delivery system for human cells and is regarded as a promising nonviral vector for gene therapy (reviewed in ref. 3), particularly as the efficiency of SB insertion is much higher than previously reported with antibiotic selection following simple plasmid DNA delivery (4). Although many significant advances have been made for the SB-Tn system including numerous engineered SB transposases (5, 6), a number of issues have been raised, including the potential impact on expression of adjacent host genes, and inactivation of the transgene by epigenetic modifications such as CpG methylation at the cargo and flanking sequences (4, 7).
β-thalassemia and sickle cell disease (SCD) are severe congenital anemias that result from deficient or altered synthesis of the β-chain of hemoglobin (8, 9). Although the use of retroviral vectors as gene delivery tools has been studied extensively, significant safety issues have yet to be resolved (10). Previously, we reported long-term stable, efficient, and erythroid-specific expression of human β-globin, mediated by the SB-Tn system using a single vector, SB-Tn-IHK-β-globin carrying both the Tn and transposase (11). The hybrid promoter, IHK, is composed of human erythroid 5-aminolevulinate synthase (eALAS) intron 8 enhancer element (12), HS-40 core element from the human αLCR and human ANKYRIN1 promoter sequences (13). A number of challenges remain including the integration efficiency of the SB-Tn-IHK-β-globin in hematopoietic stem cells (HSCs), activity of the IHK promoter expressing β-globin at critical stages in hematopoietic differentiation, modification of the inserted IHK-driven transgene in the transduced cells and the potential effect of transgene insertion on the host genome flanking the insertion loci.
To investigate these issues, we designed and tested a dual SB-Tn reporter system eIF-SB10-IRES-GFP//pT2-IHK-DsRed in K562 erythroid-cells using a nonselective fluorescence-activated cell sorting (FACS) method for identification of SB-Tn-transduced cells. We used an internal ribosome entry site (IRES) (14, 15) to link an eGFP reporter to the constitutively expressed proximal SB transposase gene external to the Tn IR/DRs. The eGFP provided a selectable marker for FACS sorting of transfected cells. We found that initial sorting of cells based on the non-transposable eGFP marker enriched the cells with the SB insertions > 10-fold. When cells were individually sorted, > 60% of the single clones expressed DsRed, with ~ 90% having a single insertion of the IHK-controlled DsRed. DsRed transgene silencing over time was correlated with DNA methylation.
Taken together, these results provide important insight into the SB-Tn system, particularly the advantage of isolating cells using marker expression that is linked to SB transposase expression. Moreover, they provide a means to investigate the tissue/developmental specificity of a transgene promoter following SB-mediated transposition, in addition to a rational basis for SB-Tn vector design for ex vivo therapeutic applications, including primary CD34+ HSCs.
MATERIALS AND METHODS
Dual-reporter SB Transposon Vector
pT2/IHK-β-globin//eIF-SB10 was used as a parental structure to build the pT2/IHK-DsRed//eIF-SB10-IRES-GFP (11). DsRed cDNA was obtained by PCR from pDsRed-Express (BD Biosciences Clontech). The β-globin gene was replaced by cDNA of DsRed, yielding pT2/IHK-DsRed//eIF-SB10. The IRES-GFP element was amplified from pIRES2-EGFP (BD Biosciences Clontech) by PCR, and was inserted into the pT2/IHK-DsRed//eIF-SB10 at EcoRI site which is right on the tail of SB10
Cell Culture, Transfection, FACS Sorting, and Transduced Cell Clone Isolation
The K562 (ATCC # CCL-243) cell line was maintained in RPMI 1640 (Invitrogen) supplemented with 10% FBS (Omega Scientific), 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, and 0.25 μg/ml amphotericin B. Lipofectamine 2000 (Invitrogen) was used to transfect the K562 cell line according to the manufacturer’s instruction. The cells were sorted using the FACSDiva software and FACS Calibur (BD Biosciences).
Erythroid or Megakaryocytic Induction
For erythroid differentiation of K562, 20 μM of hemin (Sigma-Aldrich) was used. Megakaryocytic differentiation was induced by incubation with 50 ng/ml phorbol 12-myristate 13-acetate (PMA, Fisher Scientific). After each induction, the cell clones were harvested for FACS analysis.
Demethylation and Histone Acetylation of Single Clones
5-Aza-2′-deoxycytidine (5-aza, Sigma-Aldrich) at a final concentration of 1 μM and trichostatin A (TSA, Sigma-Aldrich) at a final concentration of 250 nM were used for reversing methylation and histone deacetylation, respectively. 5 × 105 cells were cultured in a 6-well plate with 1.5 ml medium. 5-aza stock solution was prepared daily as previously described (16). Since 5-aza decays after 5 hrs in solution, we replaced the medium every 24 hrs with fresh 1-μM 5-aza containing medium. A similar procedure was used for TSA treatment except that 250 nM of TSA was added to each well at each time point.
Western Blot Analyses
Fifty μg of total proteins were separated by 13.5% SDS-PAGE and eletrophoretically transferred to nitrocellulose membranes. The immunoblots were processed as previously described (11). The primary mouse monoclonal anti-γ-globin 51-7 (Santa Cruz Biotechnology) was used at a dilution of 1:500. The monoclonal anti-β-actin AC-15 (Sigma-Aldrich) was used as a loading control at a dilution of 1:10,000. The in-house polyclonal anti-SB transposase antibody was used at a dilution of 1:200. The secondary horseradish peroxidase-conjugated antibody, goat anti-mouse or anti-rabbit, and the Supersignal West Pico Chemiluminescent substrate were obtained from Pierce Biotechnology, Inc. (Rockford, IL).
PCR Detection of IHK-DsRed and SB10 CDS (coding sequence) in Single Cell Clones
Genomic DNA was purified by DNeasy Tissue Kit (Qiagen) according to the manufacturer’s recommendation. Genomic DNA (0.1 μg) was used as template, and the PCR amplification was performed in 25-μl reaction volume with primers (forward, 5′GAGAACGTCATCACCGAG-TTCA3′ and backward, 5′CAGGCGCTCGGTGGAGGCCT3′) designed specifically for DsRed, using Expand Hi-Fidelity™ PCR system (Roche Molecular Diagnostics). Following 2-min denaturation at 94°C, the DNA was amplified for 26 cycles of 94°C for 30 sec; 57°C for 30 sec; 72°C for 30 sec with a final extension for 7 min at 72°C. The HS3 element (648 bp) in the β-globin locus control region was used as a control for DNA input and amplified using primers (forward, 5′ACTGAGCTCAGAAGAGTCAA3′; reverse, 5′AATAACCCTATGAGATA-GAC3′) by the same thermal cycles as used with DsRed except that the annealing temperature was 52°C.
The SB10 transposase gene was detected using 120 ng of DNA (~ 2 × 104 copies of human diploid genome) isolated from the single cell clones as template with primers SBF 5′-GGACCACGCAGCCGTCATAC-3′ and SBR 5′-CCTGTTTCCTCCAGCATCTTCAC-3′. These were used to amplify a 136 bp region of the SB transposase gene with an initial denaturation step at 95°C for 7 min and subsequent 32 cycles of 95°C 20 sec, 58°C 20 sec, 72°C 25 sec with Expand Hi-Fidelity™ PCR system.
Ligation-Mediated PCR for the Identification of Genomic Integration Sites
Genomic DNA (1 μg) from cell clones was digested with either SspI or EcoRV, and purified by QIAquick PCR Purification Kit (Qiagen). The DNA was ligated using 1200 U of T4 DNA ligase (Promega) in a volume of 300 μl at room temperature for 5–6 hr. The ligation products were purified by the QIAquick PCR Purification Kit, and the DNA was eluted in 25 μl of elution buffer. Using 10 μl of the eluted DNA as template, the first inverted PCR was performed in 25 μl reaction volume with RP1 (5′CTGGGATTAAATGTCAGGAATTGTG3′) and LP1 (5′GTGTCATGCACA-AGTAGATGTCC3′) primers using Expand Hi-Fidelity™ PCR system. Following 3 min of denaturation at 94°C, the DNA was amplified for 21 cycles of 93°C for 30 sec; 58°C for 30 sec; 68°C for 5 sec with a final extension for 7 min at 72°C. From this initial PCR reaction, 0.5 μl was removed to serve as the template for a second PCR reaction using nested primers RP2 (5′GTGAGTTTAAATGTATTTGGCTAAG3′) and LP2 (5′ACTGACTTGCCAAAACTAT-TGTTTG3′) in 50 μl. The PCR was performed for 40 cycles using the same cycle parameters. PCR products were excised from 1% agarose gel, purified by QIAquick Gel Extraction Kit (QIAGEN), and sequenced directly by the RP2 or LP2 primer.
Flow Cytometry and FACS Analysis
CD235a-FITC, CD41-APC, FITC-mouse IgG isotype control, and APC-mouse IgG isotype control were purchased from BD Biosciences, Pharmingen. The cells were analyzed by FACS Calibur; and the signal of DsRed was shown in FL-2 channel; CD235a-FITC in FL-1; and CD41 in channel FL-4. Flowjo software was used to analyze the FACS data.
CpG Methylation Analysis
Bisulfite-mediated genomic sequencing was performed as described previously (7, 17, 18) on DNA isolated from clones by phenol/chloroform extraction and ethanol precipitation (19). PCR primers were designed on the basis of sequences converted by Methprimer (20). Bisulfite PCR primers for SB-IHK-DsRed Tn were 5′GGAGGAGGTG-TTTTTGTAATTTG3′ (forward) & 5′CTTCACCTTATAAATAAAACAACC3′ (reverse). For the endogenous host chromosomal ANKYRIN1 promoter, primers were 5′ATAAGTA-GAAGAGGAGATGTTTTG3′ (forward) & 5′AAAATATACAAAACTACTCTTACTC3′ (reverse). PCR amplification used 7 min at 94°C followed by 37 cycles (20 sec at 94°C, 20 sec at 55°C, and 45 sec at 72°C) with final extension of 5 min at 72°C. PCR products were purified by QIAquick® Gel Extraction kit (QIAGEN), cloned into pGEM-T Easy vectors (Promega), and bisulfite PCR plasmid clones were isolated and sequenced using SP6 upstream primers. Combined bisulfite restriction analysis (COBRA) (21) was performed with the PCR products.
Statistical analyses
ANOVA and Bonferroni’s multiple comparison tests for percent DsRed positive cells were performed using InStat version 3.1 (GraphPad Software). The methylation data analysis used paired t-tests based on the number of methylated CpGs at every CpG position between the groups of cell clones calculated using two-tailed tests in Microsoft Excel. Values of P < 0.05 were considered significant.
RESULTS
Highly Efficient Selection of K562 Cells Harboring SB-IHK-DsRed Transposon
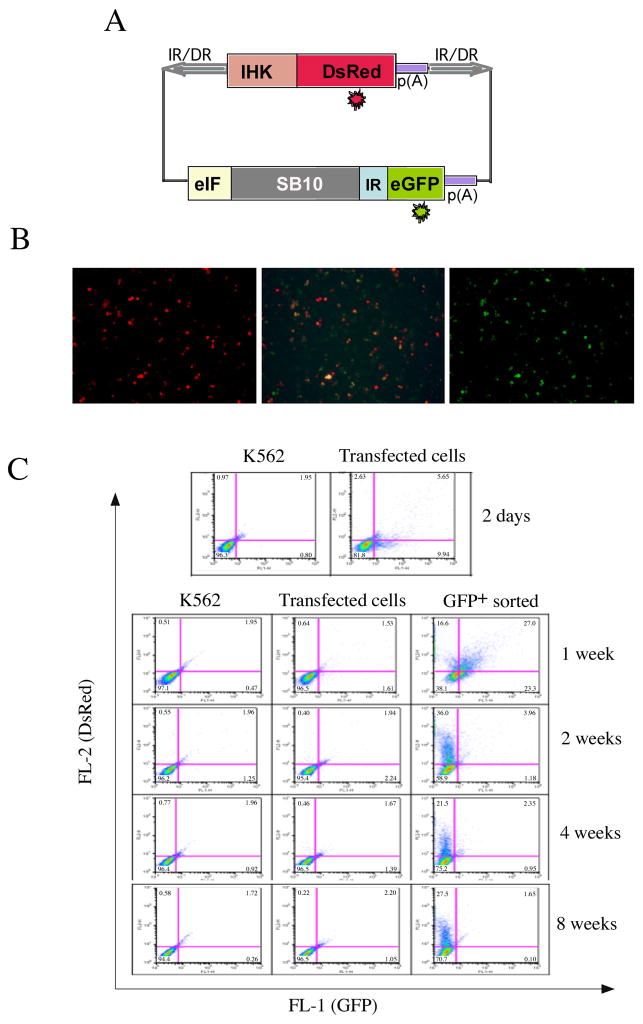
In order to evaluate an alternative approach for transducing cells with cis SB-Tn, we designed a cis SB-Tn dual reporter system, in which a fluorescent reporter gene, DsRed, driven by an erythroid-specific promoter, was placed between the two IR/DRs for transposition. Another reporter gene, GFP expressed by the ubiquitous mouse initiation factor gene (eIF) promoter, was linked to the SB10 transposase gene by an IRES element (14, 15) (Figure 1A). Thus, the expression of GFP indirectly indicates transposase expression as shown in the fluorescence images of K562 cells 2 days post-transfection expressing DsRed (left) as well as GFP (right) (Figure 1B). FACS analysis of the transiently transfected cells (Figure 1C, top panel) indicated that most of the DsRed+ cells were GFP-positive whereas about 65% GFP-positive cells were DsRed negative.
FIGURE 1.
Dual reporter Sleeping Beauty transposon system designed for FACS enrichment of K562 cells with DsRed transgene expression. (A) Schematic of the dual fluorescent cis SB-Tn plasmid showing the erythroid specific IHK hybrid promoter consisting of two small enhancer elements, i8 from the human ALAS2 intron 8 and HS40 from human α-globin locus control region, linked to the human ANKYRIN-1 promoter for the red fluorescent protein, DsRed. On the vector backbone external to the SB-Tn’s IR/DRs, the ubiquitous eukaryotic initiation factor 4A1 (eIF) drives a dual expression cassette of SB10 transposase followed by an internal ribosome entry site (IRES) and enhanced green fluorescent protein (GFP) that was utilized in the selection of the transiently transfected cells by fluorescent activated cell sorting (FACS). (B) Fluorescence images two days after transfection with the eIF-SB10-IRES-GFP//pT2-IHK-DsRed showed K562 cells expressing DsRed (left), GFP (right), respectively, and both (middle) with different shades of yellow color when merged. (C) FACS analysis of the transiently transfected cells at two days post-transfection (top panel) using GFP selection. The thirty thousand GFP+ cells selected by FACS were cultured, passaged under normal conditions, and subjected to FACS analysis for tracking DsRed and GFP expression at 1, 2, 4 and 8 weeks post-sorting (lower panels). The culture time in weeks after initial selection for GFP is shown on the right side, and the identity of the cells analyzed is indicated at top of the FACS panels.
Approximately 30,000 cells positive for GFP were acquired by FACS, and passaged for tracking DsRed and GFP expression by FACS over an 8-week period (Figure 1C, lower panel). In the GFP-sorted group, DsRed-positive cells were ~ 38% at 1 week and remained relatively high at about 28% over 8 weeks in culture. The number of GFP-positive cells was dramatically decreased to ~ 5% after 2 weeks, suggesting the SB10-IRES-GFP element was episomal and gradually degraded or excluded from cells. In contrast to the GFP-sorted population, DsRed/GFP-positive population was significantly diminished (P < 0.001), dropping from 18% at 48 hrs to 3% at 1 week (Figure 1C, lower middle panel) in cells transfected with the same SB construct but not sorted for GFP expression at 48 hrs. The enormous difference (P < 0.001) in DsRed expression between GFP-sorted and non-sorted groups at all later time points is strong evidence that GFP sorting successfully enriched SB-Tn transduced cells and resulted in long-term expression of the transgene.
Table 1 summarizes the DsRed expression levels in the single clones established from those collected either manually or by FACS sorting (with respect to GFP expression). Surprisingly, the percentage of established clones showing DsRed expression above control was 61% and 77% in manually-selected and machine-collected clones, respectively, which was significantly higher (P < 0.001) than ~ 29% observed from GFP-sorted cells at 8 weeks (Figure 1C, lower panel, right column). Tables 2 and 3 show the ratio of DsRed-positive cells for each clone at 2 months after the initiation of individual cell clone cultures.
TABLE 1.
Percentage of GFP+ selected single cell clones expressing DsRed at 2 months.
| Method of Clone Isolation |
||
|---|---|---|
| Hand Picked | Machine Sorted | |
| Total Number | 196 | 96 |
| Survived | 26% (51)a | 41% (40) |
| DsRed+ | 61% (31)b | 77% (31) |
Number of single cell clones derived from the total number isolated.
Number of DsRed clones from the single cell clones established.
TABLE 2.
Percentage of DsRed+ cells in clones selected manually at 2 months.a
| clone ID | DsRed % | clone ID | DsRed % | clone ID | DsRed % | clone ID | DsRed % | clone ID | DsRed % |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 82.7 | 11 | 0.05 | 21 | 0.51 | 31 | 4.10 | 41 | 75.5 |
| 2 | 2.66 | 12 | 0.06 | 22 | 0.37 | 32 | 0.14 | 42 | 1.27 |
| 3 | 0.63 | 13 | 0.35 | 23 | 8.36 | 33 | 0.17 | 43 | 2.44 |
| 4 | 0.36 | 14 | 59.1 | 24 | 2.14 | 34 | 0.15 | 44 | 14.4 |
| 5 | 0.11 | 15 | 11.7 | 25 | 8.91 | 35 | 0.11 | 45 | 7.89 |
| 6 | 0.16 | 16 | 0.11 | 26 | no data | 36 | 0.15 | 46 | 59.6 |
| 7 | 2.37 | 17 | 1.30 | 27 | 2.73 | 37 | 0.23 | 47 | 0.84 |
| 8 | 84.8 | 18 | 14.2 | 28 | 0.36 | 38 | 2.03 | 48 | 0.32 |
| 9 | 0.21 | 19 | 0.19 | 29 | 0.33 | 39 | 1.67 | 49 | 67.0 |
| 10 | 0.19 | 20 | 4.46 | 30 | 1.21 | 40 | 10.3 | 50 | 0.23 |
| 51 | 74.3 | ||||||||
| control | 0.11 | ||||||||
Percentage of DsRed+ cells in the single cell clones was determined by FACS analysis. The clones highlighted in gray were considered negative as their values were not significantly above K562.
TABLE 3.
Percentage of DsRed+ cells in machine sorted clones at 2 months.a
| clone ID | DsRed % | clone ID | DsRed % | clone ID | DsRed % | clone ID | DsRed % |
|---|---|---|---|---|---|---|---|
| 1 | 10.0 | 11 | 0.04 | 21 | 0.15 | 31 | 0.13 |
| 2 | 37.1 | 12 | 0.10 | 22 | 84.3 | 32 | 72.2 |
| 3 | 30.4 | 13 | 67.5 | 23 | 70.7 | 33 | 5.41 |
| 4 | 0.61 | 14 | 97.6 | 24 | 98.4 | 34 | 3.01 |
| 5 | 11.1 | 15 | 58.0 | 25 | 68.1 | 35 | no data |
| 6 | 99.6 | 16 | 62.5 | 26 | 0.42 | 36 | 0.81 |
| 7 | 99.8 | 17 | 61.2 | 27 | 39.2 | 37 | 2.62 |
| 8 | 0.40 | 18 | 98.1 | 28 | 66.1 | 38 | 7.69 |
| 9 | 0.18 | 19 | 1.43 | 29 | 1.57 | 39 | 80.5 |
| 10 | 15.5 | 20 | 86.0 | 30 | 32.4 | 40 | 18.1 |
| control | 0.18 | ||||||
Percentage of DsRed+ cells in the single cell clones was determined by FACS analysis. The clones highlighted in gray were considered negative as their values were not significantly above K562.
IHK-DsRed Expression was not Proportional to the Insertion Number, but Likely Affected by Chromosomal Loci in the Host Genome
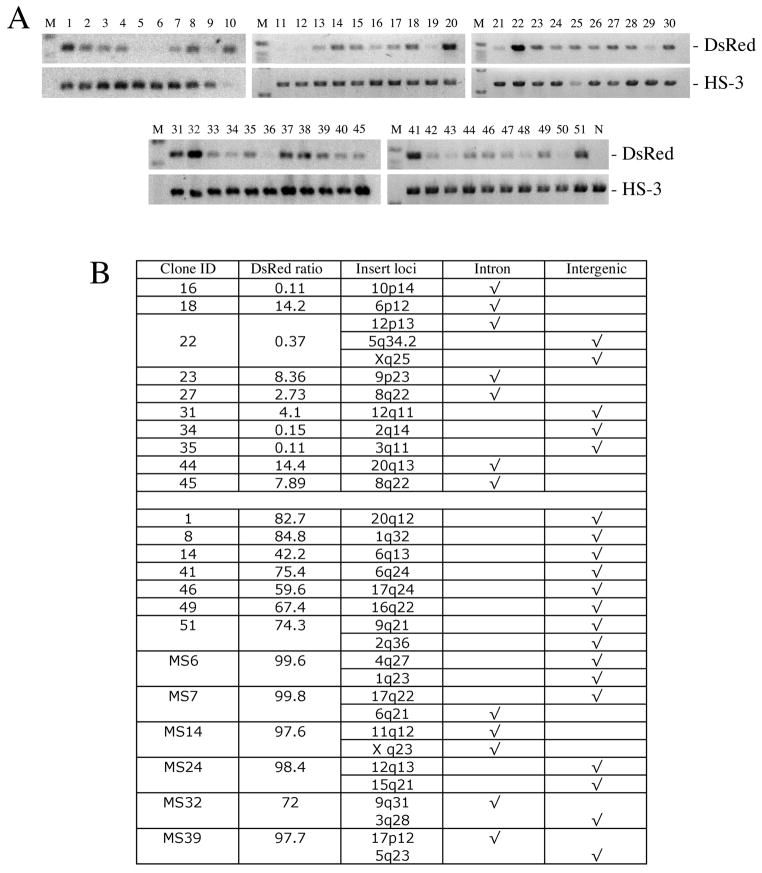
We used semi-quantitative genomic PCR to determine the copy number of the inserted SB transposons in each cell clone. Genomic DNA from each cell clone was subjected to PCR using primers specific to DsRed or host genomic HS3, an internal element in the β-globin locus control region as the autosomal 2-allele endogenous control for copy number comparison. Clones 20, 22, 32 produced significantly stronger signals for DsRed than their respective HS3 control, suggesting more than 2 copies of the SB-Tns had been inserted into the genome (Figure 2A). This was confirmed by inverted-nested PCR, which revealed that clone 22 had 3 insertions (Figure 2B). However, the proportions of DsRed-positive cells in the three clones harboring more than 2 copies of IHK-DsRed transgene were surprisingly low at 4.46%, 0.47%, and 0.37%, respectively. This suggests that expression of DsRed was not simply determined by copy number, but rather the chromosomal environment at the insertion loci may play an important role in regulating the DsRed expression.
FIGURE 2.
Genomic copy number and chromosomal location of SB-IHK-DsRed insertions in individual K562 clones. (A) Genomic DNA isolated from single-cell clones was used as template for PCR using primers specific for DsRed and for the endogenous genomic HS3. (B) Determination of the SB-Tn insertion sites in cell clones. Using ligation-mediated nested PCR of the isolated genomic DNA, the insertion sites of the SB-IHK-DsRed were amplified and sequenced. NCBI BLAST analysis of the recovered flanking sequences was used to determine the genomic insertion sites in the K562 clones. The proportion of DsRed+ cells in each individual clones was determined by FACS analysis. Number of insertion loci identified by ligation-mediated PCR corresponded with the number estimated by semi-quantitative PCR in panel (A). MS prefix represents cell clones isolated by FACS machine whereas other cell clones were isolated manually.
We selected 10 clones with either negative or low DsRed expression and 12 clones with robust expression of DsRed, and conducted inverted-nested PCR to determine the insertion loci (Figure 2B). According to the SB insertion database as well as our previous findings, SB-mediated transgene insertion into the mammalian genome has a random distribution with a slight tendency for TA-rich region (22). In this study using SB10-IRES-GFP//pT2-IHK-DsRed, 62.5% insertions were intergenic whereas 37.5% were intragenic, all of which were located in the introns of the genes. This ratio of inter-/intragenic insertion is quite similar to the previous report of 39% intragenic insertions of SB in human cells, but there were no insertions into exon sequences disrupting host genes. While the expression of transgenes inserted into an intron can be dependent only on the strength of their own promoters (23), the local host gene can also modulate their expression depending on the transcription activity of the local gene. Thus, cell clones with negative or low expression of DsRed could have intronic insertions into transcriptionally inactive host genes while clones with active DsRed expression were shown to have at least one intergenic insertion except clone MS14. However, intergenic insertion did not always lead to high-level expression of the transgene as exemplified by clones 31, 34 and 35, nor was intronic insertion always associated with low level expression as illustrated by clone MS14 (Figure 2, Tables 2 and 3).
Erythroid-Specific Expression of DsRed by the IHK Promoter in Response to Drug-Induced Erythroid or Megakaryocyte Differentiation
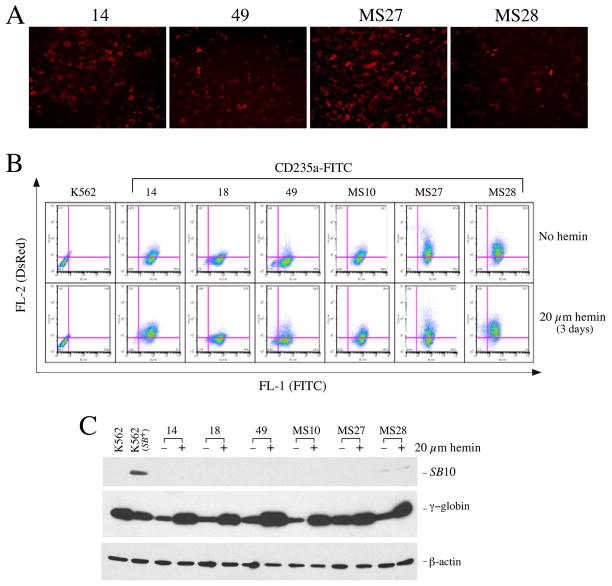
Human K562 cells can be induced to erythroid (24–26) or megakaryocyte differentiation (27–30) by a variety of agents. To test the IHK promoter activity, we chose typical K562 single clones harboring SB-IHK-DsRed, representing low, intermediate and high levels of DsRed expression, respectively (Figure 3A) and monitored DsRed expression following induction of either erythroid or megakaryocytic differentiation. When examined under normal culture conditions, clones 14, 18, 49, MS10, MS27 and MS28 were positive for the erythroid marker, CD235a (glycophorin A) (31) (Figure 3B, top panel). Even though clone 49 has a small portion of CD235a-negative cells, DsRed was expressed only in CD235a-positive cells, suggesting erythroid specificity of the IHK promoter.
FIGURE 3.
The IHK promoter directs erythroid-specific expression of DsRed. SB-IHK-DsRed single clones, representing low, medium and high levels of cells expressing DsRed were used to track the modulation of DsRed expression, following treatment to induce erythroid differentiation. (A) Fluorescent microscopy of the clones 14, 49, MS27 and MS28 showing the DsRed expression prior to treatment. (B) Flow cytometry of the cell clones with DsRed (vertical axis) and erythroid specific marker, CD235a, (horizontal axis), under normal culture condition (upper panels) or under hemin treatment for 3 days to induce erythroid differentiation (lower panels). At far left is the parental K562 cells stained with mouse isotype antibody-FITC as a negative control, while the individual clones are identified above the FACS panels. (C) The western blot analysis of the cell clones against SB10, γ-globin, and β-actin control, respectively. The clone ID number as well as presence or absence of hemin treatment are indicated above the lanes.
When erythroid differentiation was induced by treatment with hemin for 3 days, the DsRed-positive populations were increased in clones 14, 18 and 49 (Figure 3B, bottom panel). The increase of DsRed-positive cells occurred in the fraction of cells that transitioned to CD235a-positive area upon hemin induction (upper right area, Figure 3B, bottom panel). Clones MS10, MS27, and MS28, however, did not respond to hemin treatment, which may have been due to spontaneous erythroid differentiation without induction as indicated by relatively high proportion of DsRed-positive, CD235a-positive cells prior to treatment (Figure 3B, top panel). Since hemin modulates the interactions of the transcription factors NF-E2, Oct-1 and GATA-1, leading to the selective activation of γ-globin (32, 33), we investigated expression of γ-globin by western blot analysis after hemin treatment exhibiting the predicted induction of γ-globin (Figure 3C). In the parallel immunoblot using anti-SB10 antibody, none of the clones expressed detectable SB10 (Figure 3C), suggesting that the SB10-IRES-eGFP cassette outside IR/DR-flanked IHK-DsRed had been eliminated from the cells. This loss of SB10-IRES-eGFP element was confirmed by genomic PCR that indicated no detectable presence of the SB10-coding sequences in most of the clones tested (data not shown).
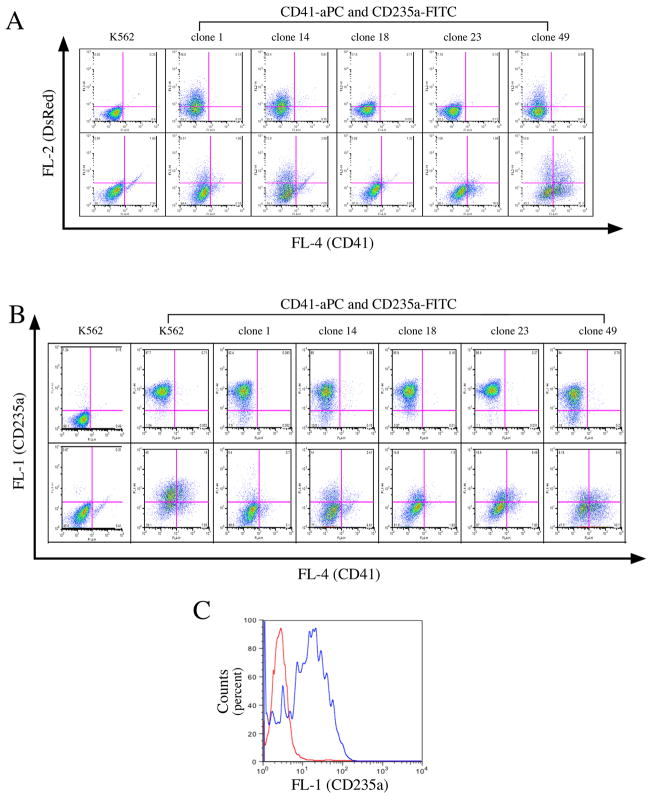
The erythroid specificity of IHK promoter was further tested by megakaryocytic differentiation of clones 1, 14, 18, 23 and 49 by PMA treatment, in which CD41 (platelet membrane glycoprotein IIb-IIIa) was used as a marker for cells of megakaryocytic lineage (34, 35). We monitored the change of DsRed+, CD235a+ and CD41+ population by FACS analysis before and after induction (Figure 4). In contrast to hemin, FACS analysis showed that expression of the IHK promoter-driven DsRed transgene declined after 2 days of PMA treatment, suggesting that IHK promoter activity is decreased in response to megakaryocytic differentiation (Figure 4A, lower panel). In fact, the flow cytometry plot of CD235a versus CD41 confirmed that the number of CD235a+ cells had also dropped (Figure 4B, lower panel, upper left). In clone 49, however, ~ 14% of the cells were DsRed+/CD41+ (Figure 4A, lower panel, right end, upper left) after PMA induction. Overlaying the CD235a signal from this clone with the parental K562 CD235a isotype staining control (CD235a negative control), they still expressed CD235a, suggesting that this DsRed+/CD41+ population originated from DsRed+/CD235a+ and that expression of CD235a persisted following induction of CD41 during megakaryocytic differentiation.
FIGURE 4.
IHK promoter is repressed following megakaryocyte differentiation. Clones 14, 18, 49 and two other clones, 1 and 23 were treated with PMA to induce megakaryocytic differentiation. (A) The clones were stained for CD235a (FITC) and CD41 (APC) as an erythroid-specific and megakaryocytic marker, respectively, and analyzed by flow cytometry against DsRed (vertical axis) and CD41 expression (horizontal axis) before (upper panel) and after PMA induction (lower panel). (B) Flow cytometry plots of CD235a+ (vertical axis) versus CD41+ (horizontal axis) cells on the same set of cell clones shown in (A). Upper panels represent flow cytometry of cell clones without PMA induction, and lower panels are flow cytometry of the cell clones after PMA induction. (C) Plots of CD235a signals of CD41+ and DsRed+ cells in clone 49 (panel A, lower right end) versus that of the parental K562 isotype control. The signal of dual-positive clone 49 is shown in red, the isotype control of K562 is shown in blue.
Long-Term Stable Expression of DsRed in the Single Clones
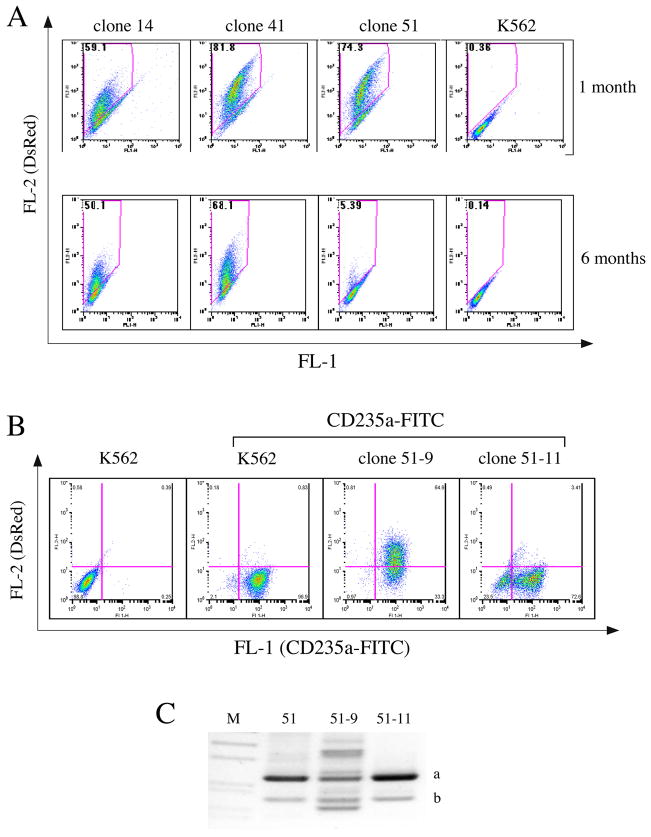
To observe the long-term expression of IHK-DsRed in single clones, we selected three clones with relatively robust expression of DsRed, and passaged them under normal condition. At 1 and 6 months after clone selection, FACS analysis for DsRed was conducted (Figure 5A). DsRed-positive cells exhibited moderate reductions in clones 14 and 41 from 59% to 50%, and from 81.8% to 68.1%, respectively while the DsRed+ ratio in clone 51 decreased dramatically from 74.3% to 5.29%. We also found minor modulations of DsRed expression ranging from 10% to 30% in other clones on long-term culture (data not shown). Taken together, these data imply that persistent long-term expression from the IHK promoter can be maintained in human erythroid cells.
FIGURE 5.
Long-term expression of DsRed in individual clones. (A) Flow cytometry analysis of DsRed expression in clones at 1 month (upper panel) and at 6 months (lower panel). The clone number is indicated above and the percent DsRed+ cells indicated in the upper left corner of each plot. (B) Analysis of DsRed expression by flow cytometry in two individual subclones 51-9 (65.7% DsRed+) and 51-11 (4.0% DsRed+), both of which were derived at 6 months from the same DsRed-positive cell clone 51 which had dropped from 74% of the cells expressing DsRed at 1 month to 5% at 6 months. Clones were stained for the erythroid specific marker CD235a, and the percentage of single or dual positive cells was determined by FACS analysis. The percentages of the population positive or negative for DsRed (vertical axis), and/or CD235a (horizontal axis) are indicated in the upper or lower corner of the relevant quadrant of the FACS plot. (C) Ligation-mediated PCR analysis of the genomic insertion sites in the parental clone 51 and the two subclones 51-9 and 51-11. Clone 51, sub-clones 51-9, and 51-11 gave the same product bands, a and b, which when sequenced were determined to be the same flanking host genomic sequences as those flanking the dual insertion sites in the parental clone 51 (Figure 2A). M, DNA ladder.
To investigate the potential mechanism for such an extreme change in DsRed expression in clone 51, we derived 31 new subclones from clone 51. Among them, a highly DsRed-positive subclone 51-9 was compared by flow cytometry with a subclone 51-11 that had predominately silenced the expression of DsRed (Figure 5B). Similar to the parental K562 control, subclone 51-9 was 98% positive for CD235a while clone 51-11 had lost the CD235a marker in 24% of the population (Figure 5B). This indicated that a significant proportion of subclone 51-11 had differentiated from the original clone 51, leading to altered phenotypes related to expression of both DsRed and erythroid markers. To test the possibility that the decrease in DsRed expression was due to the loss or genomic rearrangement of the inserted IHK-DsRed-Tn, we conducted inverted-nest PCR analysis (Figure 5C). Both subclones exhibited the identical “a” and “b” bands as the parental clone 51, and by direct sequencing revealed the same insertion site loci in 51-9 and 51-11 as the parental clone suggesting no further transposition/movement of the inserted Tns. As the “self-ligations” of genomic DNA for the inverted-nested PCR are random events, the multiple bands in subclone 51-9 other than “a” and “b” most likely resulted from nonspecific PCR amplification of ligated products.
This possibility is supported by the presence of the predicted PCR products from the original insertion loci in subclone 51-9 which should have disappeared if the nonspecific bands were derived from genomic rearrangements/hopping of the Tns from their initial insertion sites. Moreover, we used PCR to detect any potential SB10 transposase gene integration into the host genome by random insertion mechanisms, and thus if expressed, mobilized the genomic inserted SB Tns. Of the 7 cell clones subjected to PCR analyses, only one cell clone, MS14 exhibited a randomly inserted SB10 transposase gene. All the other clones including 51-9 and 51-11 showed no SB10 transposase gene by semi-quantitative genomic PCR (data not shown). In addition, all the cell clones tested by immunoblot analyses demonstrated no detectable SB10 transposase after the establishment of single clones (Figure 3C). Thus, the loss of DsRed expression in subclone 51-11 during long-term culture did not originate from major genetic rearrangement of the inserted SB-Tns.
IHK-DsRed Expression was Regulated by DNA Methylation and Histone Acetylation
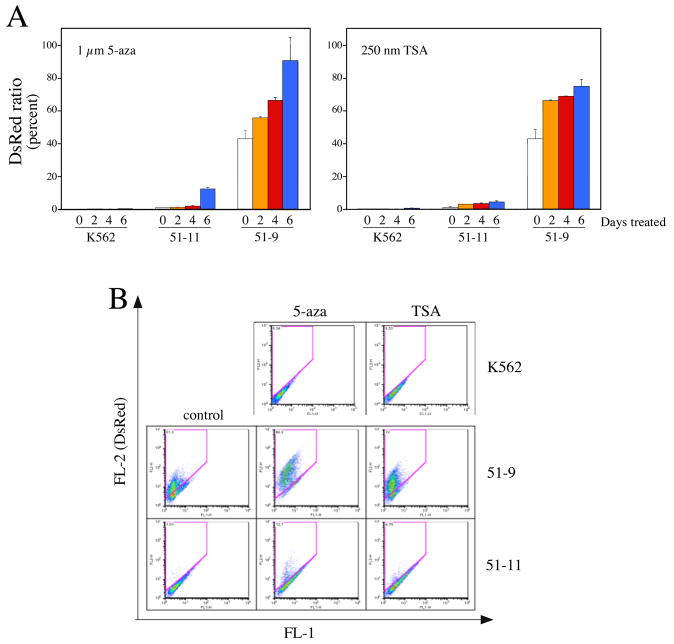
In order to determine whether the silencing of IHK-DsRed in cell clones was due to epigenetic mechanisms such as CpG methylation and/or histone deacetylation (reviewed in ref. 36), we treated subclone 51-9 and 51-11 with 5-aza-2′-deoxycytidine (5-aza), an inhibitor of DNA methyltransferases (Figure 6A, left panel), or with trichostatin A (TSA) which blocks histone deacetylation (Figure 6A, right panel) (16, 37). By day 6 of 5-aza treatment, the percentage of cells expressing DsRed increased substantially in 51-9 and 51-11 (Figure 6A, left panel, 6B middle column), indicating expression of IHK-DsRed was suppressed by DNA methylation in a considerable proportion of both subclones. TSA treatment also led to an increase of DsRed expression in 51-9 and 51-11 (Figure 6A, right panel, 6B, far left) as predicted due to the linkage between CpG methylation and histone deacetylation in transcriptional repression (38). Reactivation of DsRed expression occurred in more cells following 6 days of 5-aza treatment than with TSA treatment, suggesting that DNA methylation may be the primary mechanism for the repression of IHK-DsRed, followed by supplementary histone deacetylation (39).
FIGURE 6.
Increased DsRed expression following treatments to reverse epigenetic modifications of genomic DNA. FACS analyses of clones 51-11 and 51-9 following treatment with 5-Aza-2′-deoxycytidine (5-aza) to inhibit DNA methylation (A, left panel) or with Trichostatin A (TSA) to block histone deacetylation of the chromatin (A, right panel) showed the increase of DsRed expression. The data (mean ± 1 SD) represent results from three independently treated cultures with each reagent. The clones and the number of treatment days with each chemical reagent are indicated below the horizontal axis. (B) Representative FACS analyses are shown that were performed after treatment for 6 days with either 5-aza or TSA, and the percentage of DsRed+ cells is indicated in the upper left of each plot.
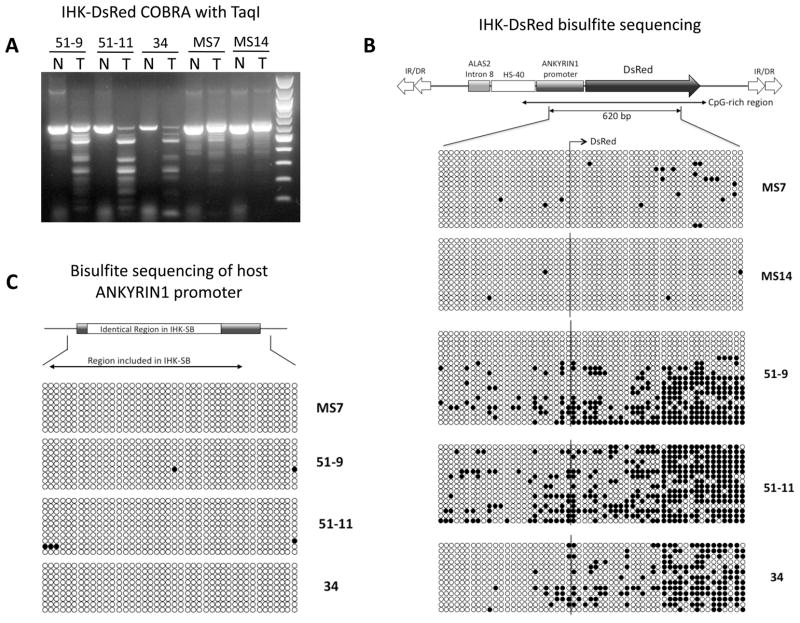
To confirm and further characterize the DNA methylation of the transposed IHK-DsRed, bisulfite-mediated genomic analyses were performed with cell clones exhibiting various levels of DsRed expression. PCR products amplified using IHK-DsRed specific primers from bisulfite-treated genomic DNA were subjected to COBRA analyses using the restriction endonuclease TaqI, in which the CpG of its recognition site (TCGA) must be methylated for cleavage to occur (21). Nearly complete TaqI digestion of the bisulfite PCR products indicated heavy CpG methylation of IHK-DsRed in 51-11 (0.5–1.9% DsRed+) and clone 34 (0.15% DsRed+); while no detectable cleavage in clones MS7 (99.8% DsRed+) and MS14 (97.6% DsRed+) implied a lack of CpG methylation of the same region (Figure 7A). Clone 51-9 (24–61% DsRed+) demonstrated an intermediate level of TaqI cleavage, suggesting heterogeneous CpG methylation of the IHK-DsRed Tns in the cell population. These data support a strong negative correlation between the level of DNA methylation at IHK-DsRed and the percentage of cells in the population expressing DsRed determined by FACS analysis.
FIGURE 7.
DNA methylation at the inserted SB-Tns and endogenous host copy of ANKYRIN1 promoter. (A) COBRA analyses to establish overall CpG methylation levels at the investigated regions. Bisulfite PCR products digested with TaqI restriction endonuclease (T) were compared with uncut controls (N) for each samples of different SB clones. (B) Methylation at each CpG dinucleotide in the 620 bp region indicated under the schematic map was profiled by bisulfite-mediated genomic sequencing. Open circles represent unmethylated CpGs while filled circles are methylated CpGs. Each horizontal line of circles is derived from a single bisulfite PCR amplicon. Bisulfite-PCR amplicons derived from a cell clone were organized into each block with the clone number indicated at right. The end of the ANKYRIN1 promoter sequence and start of the DsRed CDS are indicated by the solid line through the blocks, with the above arrow indicating the direction of transcription. (C) CpG methylation profile at the endogenous host copy of ANKYRIN1 promoter. The region identical with ANKYRIN1 promoter sequence in the SB-IHK-DsRed is indicated in the schematic representation above the blocks of CpG methylation data derived from individual bisulfite-PCR amplicons. The cell clone from which genomic DNA was isolated is indicated at right, and methylated CpGs indicated by filled circles. The absence of CpG methylation at this region was also verified in the cell clone MS14 (data not shown).
PCR products from the bisulfite-treated DNA were cloned and sequenced to establish the precise CpG methylation profile (Figure 7B, 7C). Overall, DNA methylation patterns were consistent with the TaqI-COBRA analyses, revealing the highest level of CpG methylation at IHK-DsRed in 51-11 (0.5–1.9% DsRed+) and clone 34 (0.15% DsRed+). As predicted from an intermediate level of cleavage by TaqI, the bisulfite-mediated sequencing data corroborated heterogeneity of DNA methylation at IHK-DsRed in 51-9 (24–61% DsRed+), while clones MS7 (99.8% DsRed+) and MS14 (97.6% DsRed+) were unmethylated (Figure 7B) in agreement with COBRA analysis. Differences in CpG methylation status between the clones were more pronounced in the DsRed-coding region than in the IHK promoter with the DsRed-negative clones, 51-11 and 34, exhibiting dense CpG methylation in the distal portion of the DsRed CDS. Taken together, these data suggest that methylation in the DsRed CDS plays a key role in regulating its expression as well as in the potential for this region to trigger methylation-dependent silencing. When every CpG is compared between DsRed+ (MS7 and MS14) and DsRed− (51-11 and 34) groups, the difference in the level of methylation was highly significant in the DsRed CDS (p = 2.09×10−12) as well as in the ANK promoter (p = 2.42×10−5). Similar statistical analyses of subclones 51-11 and 51-9 also revealed a significant difference (p = 1.77×10−9) when the promoter region and DsRed CDS were combined.
The endogenous genomic copies of ANKYRIN1 promoter were completely devoid of CpG methylation in all the clones analyzed (Figure 7A) despite the significant level of CpG methylation at the identical human ANKYRIN1 promoter sequences within the transposed IHK-DsRed (Figure 7B). This data suggested that there is a mechanism(s) that can specifically discriminate and methylate CpGs in identical DNA sequences depending on whether they are native or foreign to the host genome.
Perturbation in Expression of Host Genes Neighboring the SB-Tn Insertions
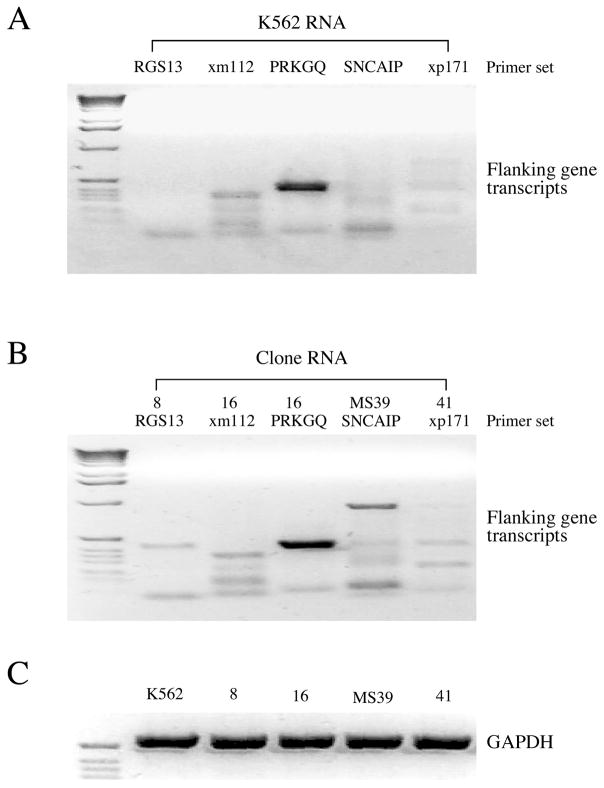
The IR/DRs and their adjacent regions in SB-Tn have been reported to function as mild cis-acting regulatory elements for the expression of nearby genes, although the effect is significantly weaker than that observed with the retroviral long terminal repeat (40, 41). Furthermore, the enhancer element of cargo transgene cassette could also affect the expression of host genes located close to the SB insertion (41). To investigate potential effects of SB-IHK-DsRed insertion on the expression of flanking host genes, we selected clones 8, 16, MS39 and 41, in which SB insertions are relatively close to neighboring host genes: RGS 13, 45 kb downstream of SB-Tn insertion in clone 8; XM_001128367.1 (hypothetical gene), 13.5 kb downstream of SB-Tn insertion in clone 16; SNCAIP, 84 kb downstream of SB-Tn insertion in clone MS39; and XP_001717916.1 (hypothetical protein), 4 kb downstream of SB-Tn insertion in clone 41.
Semi-quantitative RT-PCR analyses of the neighboring genes using total RNA purified from each clones were compared with RT-PCR analysis of control K562. The transcripts from XM_001128367.1 and XP_001717916.1 in clone 16 and 41, respectively, were not affected by the SB insertion relative to the K562 control (Figure 8A, 8B). The PRKGQ gene, which has an SB insertion in its first intron, was also unaffected in clone 16. In contrast, the presence of additional RT-PCR products in clones 8 and MS39 indicated transcriptional activation of SNCAIP and RGS13 loci (Figure 8A, 8B). Since SNCAIP and RGS13 genes are 84 and 45 kb away from the insertion sites in clone MS39 and 8, respectively, their up-regulation is most likely attributable to enhancer elements in the IHK promoter, i8 or HS40. Additional RT-PCR analysis of clone 8 indicated that SNX2 the adjacent downstream gene from SNCAIP exhibited the same transcript level as in K562 control cells (data not shown). Taken together, a transgene cassette harboring enhancer elements could potentially activate neighboring host genes at long range depending on its insertion loci, while the perturbation of SB-Tn insertion on expression of neighboring genes will not jump over the intervening gene.
FIGURE 8.
Effect of SB-IHK-DsRed insertion on the expression of adjacent endogenous host genes. (A) Total RNA purified from K562 control cells was used as template for RT-PCR with primer sets correspondent to RGS13, XM_001128367.1, PRKGQ, SNCAIP and XP_001717916.1, respectively. The RT-PCR products were subjected to agarose gel electrophoresis. (B) Total RNA was purified from cell clone 8, 16, MS39 and 41, respectively, and used as template for RT-PCR of the closest downstream host genes: RGS13 for clone 8; XM_001128367.1 and PRKGQ for clone 16; SNCAIP for clone MS39; and XP_001717916.1 for clone 41. XM_001128367.1 and XP_001717916.1 are abbreviated as xm112 and xp171, respectively. (C) The control RT-PCR of GAPDH was carried out to verify equal amount of template for each sample.
DISCUSSION
In this study, we developed and tested a novel dual fluorescent reporter cis SB-Tn system to select cell clones that expressed an IHK-driven DsRed transgene. By applying this system to a human erythroid K562 cell line coupled with FACS-mediated enrichment of transiently transfected cells via the constitutively expressed GFP, we achieved 61–77% successful transposition and long-term expression of IHK-DsRed. We characterized the involvement of epigenetic modifications such as CpG methylation and histone deacetylation in the regulation of the IHK-driven DsRed after genomic insertion of the transgene. In the clones analyzed, transcriptional activation of host genes located at relatively distant regions from the SB insertions (> 45 kb) was observed, possibly due to enhancer elements present in the IHK-DsRed transgene carried by the SB-Tn. The same IHK-DsRed SB-Tn did not perturb transcription of close host neighboring genes (< 13.5 kb) nor the PRKGQ gene in which the SB insertion site was located in the first intron.
The unique design of the cis eIF-SB10-IRES-GFP//pT2-IHK-DsRed vector is a significant improvement over SB-Tn systems containing a single fluorescent reporter gene in the SB-Tn itself (4). As the expression of GFP was tightly linked via an IRES to the constitutively expressed SB10 transposase that mediates the insertion of the SB-Tn, we were able to enrich transfected cells that were actively expressing the transposase, which definitely enhanced the probability of transgene insertion into the host genome. Although we used DsRed, replacement with a therapeutic transgene would enable the eIF-SB10-IRES-GFP//pT2-system to be readily applied for therapeutic gene delivery. Moreover, the high frequency of successful transposition events resulting in persistent long-term expression was achieved using the original SB10 version of the transposase. Thus, incorporation of recently developed hyperactive SB transposase genes (42–45) may further enhance the efficiency of transposition using the cis IRES-linked fluorescent reporter SB-Tn vector.
Even with the high efficiency of successful transposition events, the number of SB-Tn insertions in this study was generally 1 or 2, but never more than 3 (a single case, clone 22) (Figure 2B). In contrast, despite dependence on gene transfer efficiency, up to 9 retroviral vectors on average could be inserted into the genome of a single K562 cell (46). The smaller number of transgene insertions mediated by this cis SB-Tn system is advantageous over retroviral vector systems in that it could reduce the risk of insertional mutagenesis. Moreover, no insertions were detected within exons disrupting normal host genes, although we have not extensively elucidated the possible influence of intron insertions of SB-Tn on the host gene expression except PRKGQ. Finally, alterations in expression of the transgene over time in clones with multiple insertions did not result from re-mobilization of the IHK-DsRed from its original insertion site. Quantitative genomic PCR to detect SB10 transposase gene showed no detectable SB10 transposase in the host genome of subclones 51-9 and 51-11. The results are consistent with long-term stable expression of transgenes in both induced pluripotent and cord blood-derived CD34+ stem cells (47, 48).
Slight modifications in the expression of some host genes flanking the SB insertions were detected in this study. This transactivation of the host gene seems to be mediated by enhancer elements, such as i8 or HS40, carried in the SB-IHK-DsRed, since endogenous host genes located relatively far from the insertion (84 kb and 45 kb) were affected. In particular, HS40 has been shown to act as an enhancer element over ~ 40 kb distance (49). This type of long-range transactivation was previously reported with respect to the insertion of viral LTRs (50, 51). The long-range enhancer activity of i8 and HS40 in the context of SB-Tn insertion should be investigated in a more comprehensive manner, as it is possible that the distal versus proximal effect observed is due to erythroid responsive DNA sequences in the gene promoters. Introduction of insulator elements such as cHS4 (52) flanking the transgene should prevent transactivation of host genes by transgenic enhancer elements. In addition, balancing the promoter/enhancer properties of a transgene should ensure the robust expression of the transgene as well as to minimize the potential activation of nearby host genes (53).
In the clones derived, expression of DsRed did not appear to be dependent merely on copy number or if the insertion site was either intronic or intergenic. However, we observed a negative correlation between the expression of IHK-DsRed and its CpG methylation consistent with numerous reports linking DNA methylation and consequent silencing of foreign DNA element(s) inserted in the mammalian host genome (reviewed in ref. 54, 55). High levels of CpG methylation was observed in cell clones, 51-11 and 34, where most cells silenced DsRed expression; whereas clones MS7 and MS14 (> 97% DsRed+) were almost free of transgene methylation. The level of methylation was greatest in the CpG-rich area of the DsRed CDS in all the clones analyzed, suggesting that this sequence, which is both non-mammalian and CpG-rich, could play a role in the recognition and nucleation of DNA methylation machinery at the SB-IHK-DsRed (56).
The DNA methylation of exogenous DNA elements is considered to be a host defense mechanism against the potential harmful activity of such DNA elements in mammalian cells (57). The host endogenous genomic copy of ANKYRIN1 promoter was actually free of CpG methylation, which concurs with a previous report of the complete lack of DNA methylation at the endogenous copy of ROSA26 sequence despite the heavy methylation in the exogenous counterpart in mice (7). However, the Agouti transgene derived from the mouse Agouti gene revealed a negligible level of CpG methylation in the mouse genome when introduced and inserted by SB transposon system (58). It should be noted that transgene silencing could be mediated by mechanisms other than CpG methylation as shown in a recent report (59). Thus, more extensive studies are required to identify the precise sequence elements most susceptible/responsible for triggering DNA methylation or other epigenetic modifications.
The heterogeneity of the expression and DNA methylation demonstrated in 51-9 might have originated from the fact that the subclone actually had two insertions of the SB-Tn at two different chromosomal loci (human chromosome 2 and 9). The local epigenetic status of those two loci could be quite different from each other, presumably affecting CpG methylation and expression of the inserted IHK-DsRed. Bisulfite-mediated sequencing with primers that encompass the neighboring host genomic region as well as the DsRed cDNA would be able to discriminate the two insertions from each other, whereas long-range PCR with bisulfite-treated genomic DNA represents a significant technical challenge. The differences in expression and methylation of DsRed between 51-9 and 51-11 suggest that unknown stochastic processes could make notable molecular and phenotypic changes even in the population of a single cell clone (clone #51) when passaged through long-term culture. This could be compensated for, by sorting cells with the desired phenotype immediately prior to use.
The potential to deliver a wild-type β-globin gene expression cassette to autologous hematopoietic stem cells presents a highly attractive alternative to the management and/or resolution of disorders such as β-thalassemia, sickle cell disease, severe combined immune deficiency (SCID), and other congenital immuno-deficiencies (8). Using nucleofection, over 50% of CD34+ cells can be transduced with SB-Tns (60). However, the survival rate and engraftment in an animal is still problematic most likely due to cytotoxicity, unknown side effects of the ex vivo process as well as epigenetic suppression of transgene expression illustrated in the present study. Although the dual fluorescent reporter system may need to be optimized for other cell types by replacing the promoter driving DsRed, the use of the ubiquitous mouse eIF promoter for transposase expression, the broad range of transposase levels that provide equivalent levels of transposition in the cis configuration (61) illustrates the utility of application of this vector system in multiple cell-types.
In conclusion, an SB-Tn system (eIF-SB10-IRES-GFP//pT2-IHK-DsRed) was designed and tested for the enrichment of transfected human cells according to the expression of the SB transposase and a linked GFP reporter. The initial screening based on the GFP expression greatly enriched the cells that had SB-mediated insertions of DsRed transgene, with up to 60% of the GFP+ cells resulting in DsRed+ cell clones with long-term expression maintained in the majority of the cell clones. These findings indicate that this enrichment system could greatly enhance the efficiency of obtaining genetically modified cells with the SB transposon system, defining mechanism(s) by which modulation of transgene expression occurs in different cell types to enhance long-term persistent expression.
Acknowledgments
This work was supported in part by NHF/NHLBI grant HL082591-01 to R.P. Hebbel and C.J. Steer; and NIH grant R01 HL081582-01 to C.J. Steer.
We would like to acknowledge the assistance of the Flow Cytometry Core Facility of the University of Minnesota Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598. We also thank Phillip Y.P. Wong for assistance in image processing, Dr. Yixin Chen for tissue culture and Dr. Lisa H. Brauer for manuscript editing.
Abbrevations
- DsRed
red fluorescent protein
- FACS
fluorescence-activated cell sorting
- eGFP
green fluorescent protein
- HSCs
hematopoietic stem cells
- hrs
hours
- IHK
human eALAS2 intron 8 enhancer, HS-40 core element and ANKYRIN1 promoter
- IRES
internal ribosome entry site
- IR/DR
inverted repeat, direct repeat
- SCD
sickle cell disease
- SB
Sleeping Beauty
- SB10
Sleeping Beauty transposase version 10
- Tn
transposon
References
- 1.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 2.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvák Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 3.Ivics Z, Izsvák Z. Transposons for gene therapy! Curr Gene Ther. 2006;6:593–607. doi: 10.2174/156652306778520647. [DOI] [PubMed] [Google Scholar]
- 4.Garrison BS, Yant SR, Mikkelsen JG, Kay MA. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yant SR, Huang Y, Akache B, Kay MA. Site-directed transposon integration in human cells. Nucleic Acids Res. 2007;35:e50. doi: 10.1093/nar/gkm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 7.Park CW, Park J, Kren BT, Steer CJ. Sleeping Beauty transposition in the mouse genome is associated with changes in DNA methylation at the site of insertion. Genomics. 2006;88:204–213. doi: 10.1016/j.ygeno.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Sorrentino BP, Nienhuis AW. Gene therapy for hematopoietic diseases. In: Stamatoyannopoulous G, Majerus PW, Perlmutter RM, Varmus H, editors. The Molecular Basis of Blood Diseases. 3. Saunders; Philadelphia, PA: 2001. pp. 969–1003. [Google Scholar]
- 9.Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–328. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Kren BT, Park CW, Bilgim R, Wong PY, Steer CJ. Erythroid-specific expression of β-globin by the Sleeping Beauty transposon for sickle cell disease. Biochemistry. 2007;46:6844–6858. doi: 10.1021/bi6024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau-Gaudry F, Xia P, Jiang G, Perelman NP, Bauer G, Ellis J, Surinya KH, Mavilio F, Shen CK, Malik P. High-level erythroid-specific gene expression in primary human and murine hematopoietic cells with self-inactivating lentiviral vectors. Blood. 2001;98:2664–2672. doi: 10.1182/blood.v98.9.2664. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher PG, Romana M, Tse WT, Lux SE, Forget BG. The human ankyrin-1 gene is selectively transcribed in erythroid cell lines despite the presence of a housekeeping-like promoter. Blood. 2000;96:1136–1143. [PubMed] [Google Scholar]
- 14.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 15.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michalowsky LA, Jones PA. Differential nuclear protein binding to 5-azacytosine-containing DNA as a potential mechanism for 5-aza-2′-deoxycytidine resistance. Mol Cell Biol. 1987;7:3076–3083. doi: 10.1128/mcb.7.9.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:e65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross-Bellard M, Oudet P, Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973;36:32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 20.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michalkiewicz M, Michalkiewicz T, Geurts AM, Roman RJ, Slocum GR, Singer O, Weihrauch D, Greene AS, Kaldunski M, Verma IM, Jacob HJ, Cowley AW., Jr Efficient transgenic rat production by a lentiviral vector. Am J Physiol Heart Circ Physiol. 2007;293:H881–H894. doi: 10.1152/ajpheart.00060.2007. [DOI] [PubMed] [Google Scholar]
- 24.Rutherford TR, Clegg JB, Weatherall DJ. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature. 1979;280:164–165. doi: 10.1038/280164a0. [DOI] [PubMed] [Google Scholar]
- 25.Lozzio CB, Lozzio BB, Machado EA, Fuhr JE, Lair SV, Bamberger EG. Effects of sodium butyrate on human chronic myelogenous leukaemia cell line K562. Nature. 1979;281:709–710. doi: 10.1038/281709b0. [DOI] [PubMed] [Google Scholar]
- 26.Singh MK, Yu J. Accumulation of a heat shock-like protein during differentiation of human erythroid cell line K562. Nature. 1984;309:631–633. doi: 10.1038/309631a0. [DOI] [PubMed] [Google Scholar]
- 27.Villeval JL, Pelicci PG, Tabilio A, Titeux M, Henri A, Houesche F, Thomopoulos P, Vainchenker W, Garbaz M, Rochant H, Breton-Gorius J, Edwards PA, Testa U. Erythroid properties of K562 cells. Effect of hemin, butyrate and TPA induction. Exp Cell Res. 1983;146:428–435. doi: 10.1016/0014-4827(83)90145-3. [DOI] [PubMed] [Google Scholar]
- 28.Whalen AM, Galasinski SC, Shapiro PS, Nahreini TS, Ahn NG. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mignotte V, Vigon I, Boucher de Crevecoeur E, Romeo PH, Lemarchandel V, Chretien S. Structure and transcription of the human c-mpl gene (MPL) Genomics. 1994;20:5–12. doi: 10.1006/geno.1994.1120. [DOI] [PubMed] [Google Scholar]
- 30.Meshkini A, Yazdanparast R. Induction of megakaryocytic differentiation in chronic myelogenous leukemia cell K562 by 3-hydrogenkwadaphnin. J Biochem Mol Biol. 2007;40:944–951. doi: 10.5483/bmbrep.2007.40.6.944. [DOI] [PubMed] [Google Scholar]
- 31.Jokinen M, Gahmberg CG, Andersson LC. Biosynthesis of the major human red cell sialoglycoprotein, glycophorin A, in a continuous cell line. Nature. 1979;279:604–607. doi: 10.1038/279604a0. [DOI] [PubMed] [Google Scholar]
- 32.Chénais B, Molle I, Trentesaux C, Jeannesson P. Time-course of butyric acid-induced differentiation in human K562 leukemic cell line: rapid increase in γ-globin, porphobilinogen deaminase and NF-E2 mRNA levels. Leukemia. 1997;11:1575–1579. doi: 10.1038/sj.leu.2400755. [DOI] [PubMed] [Google Scholar]
- 33.Chénais B, Andriollo M, Guiraud P, Belhoussine R, Jeannesson P. Oxidative stress involvement in chemically induced differentiation of K562 cells. Free Radic Biol Med. 2000;28:18–27. doi: 10.1016/s0891-5849(99)00195-1. [DOI] [PubMed] [Google Scholar]
- 34.Williams MJ, Du X, Loftus JC, Ginsberg MH. Platelet adhesion receptors. Semin Cell Biol. 1995;6:305–314. doi: 10.1006/scel.1995.0040. [DOI] [PubMed] [Google Scholar]
- 35.van Pampus EC, Denkers IA, van Geel BJ, Huijgens PC, Zevenbergen A, Ossenkoppele GJ, Langenhuijsen MM. Expression of adhesion antigens of human bone marrow megakaryocytes, circulating megakaryocytes and blood platelets. Eur J Haematol. 1992;49:122–127. doi: 10.1111/j.1600-0609.1992.tb00915.x. [DOI] [PubMed] [Google Scholar]
- 36.Whitelaw E, Sutherland H, Kearns M, Morgan H, Weaving L, Garrick D. Epigenetic effects on transgene expression. Methods Mol Biol. 2001;158:351–368. doi: 10.1385/1-59259-220-1:351. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 38.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 40.Moldt B, Yant SR, Andersen PR, Kay MA, Mikkelsen JG. Cis-acting gene regulatory activities in the terminal regions of Sleeping Beauty DNA transposon-based vectors. Hum Gene Ther. 2007;18:1193–1204. doi: 10.1089/hum.2007.099. [DOI] [PubMed] [Google Scholar]
- 41.Walisko O, Schorn A, Rolfs F, Devaraj A, Miskey C, Izsvák Z, Ivics Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol Ther. 2008;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- 42.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 43.Zayed H, Izsvák Z, Walisko O, Ivics Z. Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol Ther. 2004;9:292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, Vandendriessche T, Ivics Z, Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 46.Kustikova OS, Wahlers A, Kuhlcke K, Stahle B, Zander AR, Baum C, Fehse B. Dose finding with retroviral vectors: correlation of retroviral vector copy numbers in single cells with gene transfer efficiency in a cell population. Blood. 2003;102:3934–3937. doi: 10.1182/blood-2003-05-1424. [DOI] [PubMed] [Google Scholar]
- 47.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung H-K, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue X, Huang X, Nodland SE, Mátés L, Ma L, Izsavák Z, Ivics Z, LeBien TW, McIvor RS, Wagner JE, Zhou X. Stable gene expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- 49.Higgs DR, Wood WG, Jarman AP, Sharpe J, Lida J, Pretorius IM, Ayyub H. A major positive regulatory region located far upstream of the human α-globin gene locus. Genes Dev. 1990;4:1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- 50.Morishita K, Parker DS, Mucenski ML, Jenkins NA, Copeland NG, Ihle JN. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell. 1988;54:831–840. doi: 10.1016/s0092-8674(88)91175-0. [DOI] [PubMed] [Google Scholar]
- 51.Lazo PA, Lee JS, Tsichlis PN. Long-distance activation of the Myc protooncogene by provirus insertion in Mlvi-1 or Mlvi-4 in rat T-cell lymphomas. Proc Natl Acad Sci U S A. 1990;87:170–173. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitoh N, Bell AC, Recillas-Targa F, West AG, Simpson M, Pikaart M, Felsenfeld G. Structural and functional conservation at the boundaries of the chicken β-globin domain. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber EL, Cannon PM. Promoter choice for retroviral vectors: transcriptional strength versus trans-activation potential. Hum Gene Ther. 2007;18:849–860. doi: 10.1089/hum.2007.067. [DOI] [PubMed] [Google Scholar]
- 54.Doerfler W, Schubbert R, Heller H, Kammer C, Hilger-Eversheim K, Knoblauch M, Remus R. Integration of foreign DNA and its consequences in mammalian systems. Trends Biotechnol. 1997;15:297–301. doi: 10.1016/S0167-7799(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 55.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 56.Chevalier-Mariette C, Henry I, Montfort L, Capgras S, Forlani S, Muschler J, Nicolas JF. CpG content affects gene scilencing in mice: evidence from novel transgenes. Genome Biol. 2003;4:R53. doi: 10.1186/gb-2003-4-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 58.Park CW, Kren BT, Largaespada DA, Steer CJ. DNA methylation of Sleeping Beauty with transposition into the mouse genome. Genes Cells. 2005;10:763–77610. doi: 10.1111/j.1365-2443.2005.00875.x. [DOI] [PubMed] [Google Scholar]
- 59.Chen ZY, Rui E, He CY, Xu H, Kay MA. Silencing of episomal expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–556. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- 60.von Levetzow G, Spanholtz J, Beckmann J, Fischer J, Kogler G, Wernet P, Punzel M, Giebel B. Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev. 2006;15:278–285. doi: 10.1089/scd.2006.15.278. [DOI] [PubMed] [Google Scholar]
- 61.Mikkelson JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA. Helper-independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]