Abstract
Tumor associated macrophages (TAMs) are implicated in breast cancer progression and metastasis, but relatively little is known about the genes pathways in these cells that contribute to malignant phenotypes. The transcription factor Ets2 is a direct target of signaling pathways involved in regulating macrophage functions during inflammation. To test whether Ets2 in TAMs modulated mouse mammary tumor growth and metastasis a genetic approach was used to conditionally delete Ets2 in TAMs. Ets2 deletion in TAMs decreased the frequency and size of mammary tumor metastases to lung in three different metastatic models. Expression profiling and chromatin immunoprecipitation assays with isolated TAMs established that Ets2 repressed several well characterized inhibitors of angiogenesis. Consistent with these results, Ets2 ablation in TAMs led to decreased tumor angiogenesis and growth. An Ets2-TAM expression signature was identified within human breast cancer expression data and this signature could retrospectively predict overall survival of breast cancer patients in two independent sets of human breast cancer microarray data. In summary, we have identified Ets2 as a critical factor that acts to enhance mammary tumor growth and metastasis by regulating a transcriptional program in TAMs.
Keywords: Tumor Macrophages, Breast Cancer, Ets2, Metastasis, Angiogenesis
INTRODUCTION
Sporadic human cancer results from somatic gene mutations that lead to aberrant growth, survival, genetic instability and increased motility of tumor cells (1). In addition to genetic complexity, it is increasingly apparent that cellular complexity inherent in the tumor stroma plays an active role in promoting all stages of tumor progression (2). Among the many cell types in the tumor stroma, the tumor associated macrophage (TAM) is a broadly defined myeloid cell type that has been implicated in tumor progression (2). TAMs are thought to be a polarized M2 subtype of macrophage that promote tumor growth, invasion, and angiogenesis (3). Alternatively, the pleiotropic effects of macrophages within the tumor microenvironment may be mediated by distinct subpopulations of TAMs that can selectively affect distinct processes such as tumor angiogenesis or invasion (4, 5).
The link between TAMs and tumor progression is especially well established in breast cancer. Human clinical studies have demonstrated that a high focal infiltration of TAMs in primary human breast tumors directly correlates with tumor cell invasion, increased vascularization, axillary lymph node involvement and reduced relapse-free survival of patients (6-9). In a mouse mammary tumor model, genetic ablation of Colony Stimulating Factor-1 (Csf-1), a growth factor critical for macrophage growth, differentiation and survival, results in a reduction in mammary TAMs and a lower incidence of lung metastasis (10).
Ets2, a member of Ets family of transcription factors, is a direct effector of CSF-1 signaling pathways that modulates macrophage functions and survival during inflammation (11, 12). ETS2 activates or represses transcription of target genes in a context-dependent manner (13, 14). Elevated expression of ETS2 has been correlated with human breast cancer (15). However, in mouse mammary tumor models, Ets2 promotes tumor progression from the stroma and not the tumor epithelial cell (16).
In the current study, a genetic approach was used to define the action of Ets2 in mouse mammary TAMs. The results demonstrate that Ets2 in TAMs decreased the growth rate of the primary tumor and tumor metastases and the mechanism involved repressing genes that are inhibitors of angiogenesis. 133 human genes orthologous to the Ets2-TAMs profile could retrospectively predicted disease free survival among patients present in two human breast cancer microarray data sets (17, 18). These results identify an Ets2-regulated transcriptional program in TAMs that regulates growth and spread of breast tumors.
MATERIALS AND METHODS
Mice
The Ets2LoxP allele, Ets2db knockout allele, MMTV-PYMT transgenic mice and Lys-Cre knockin allele were previously described (19-22). The c-fms-YFP construct was identical to the published c-fms-EGFP construct except for the substitution of YFP (23). Transgenic mice were produced by standard DNA microinjection procedures. All alleles utilized were >10 generations FVB/N background. Use and care of mice were approved by the Ohio State University Institutional Animal Care and Use Committee.
Orthotopic and tail vein injection assays
The two breast cancer cell lines used, Met-1 (MMTV-PyMT)) and MVT-1 (MMTV-c-Myc;MMTV-VEGF) were used (24, 25). The cell lines were cultured in Dulbecco’s Modified Eagle Medium containing 10% Fetal Bovine Serum at 37°C in 5% CO2 incubator. Cultured tumor cells were harvested at 80-90% confluence and suspended in filtered cold 0.9% NaCl. Three million Met-1 cells or 200,000 MVT-1 cells were injected into the tail vein or mammary gland, respectively. Tail-vein and orthotopic injected animals were dissected 18 and 35 days post-injection, respectively.
Isolation of TAMs
Minced mammary glands or lungs with metastatic tumors were digested with 20mg Collagenase Type 2 (Worthington), 480 Units DNAseI (Boehringer) and 1mM MgCl2 at 37°C and stroma was enriched by gravity separation (26). The YFP-positive cell population was sorted using Fluorescent Activated Cell Sorting with the BD FACSAria™.
RNA extraction and Quantitative real-time PCR
RNA extraction and cDNA preparation were done as described previously (27). For samples used in microarray analysis, RNA was extracted with the RNeasy Stratagene micro-prep column (Stratagene) as per the manufacturer’s instructions. Two independent sets of RNA isolated from different TAMs/mice than RNA used for the microarrays were used for verification.
Real-time quantitative RT-PCR was conducted using the Roche Universal Probe Library system (Roche Diagnostics) in an Icycler iQ Real-Time Detection system (Bio-Rad) as described previously (27). Primer-probe combinations are available on request.
Histology and immunohistochemistry
Tumor tissues were fixed in formalin overnight, processed, paraffin-embedded and 5μM sections were prepared. For immunostaining, rat α-mouse F4/80 (1:40 dilution; Caltag Labs), rat α-mouse CD31 (1:50 dilution, Abcam), mouse α-human THBS1 (1:50 dilution, Abcam), mouse α-mouse THBS2 (1:50 dilution, BD Biosciences), goat α-mouse SPARC (1:100 dilution, B.D Biosciences), and mouse α-mouse BrDU (1:50 dilution, DAKO) primary antibodies were used. Biotinylated goat α-rat, goat α-mouse, or donkey α-goat (BD Biosciences) were the secondary antibodies used for immunohistochemical analysis. Images were acquired using an Axioscope 40 microscope (Zeiss) equipped with an Axiocam HRc camera (Zeiss). Immunohistochemical data was quantified by calculating the area of antibody staining per unit area of tumor using Metamorph 6.0 software. Wholemount hematoxylin staining of lungs was performed as described (28).
For co-localization studies, frozen sections of mammary tumors fixed in 4% paraformaldehyde were double immunostained with α-F4/80 antibody (Alexa-594 secondary antibody, Invitrogen) and either α-THBS2, α-THBS1 or α-SPARC antibody (Alexa-488 secondary antibody, Invitrogen). Nuclei were stained with DRAQ5. Images of stained mammary tumors were acquired using a Zeiss 510 META laser scanning confocal microscope Results are presented as the percentage of F4/80-positive or negative cells that had co-localized staining in or around (extracellular space) for α-THBS2, α-THBS1 or α-SPARC, respectively,.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed as described (27). Immunoprecipitation was carried out with 2.5μg of antibodies. The ETS2 antibody has been previously described (19). Rabbit α-mouse HDAC1 and rabbit-IgG were purchased from SantaCruz Biotechnology and Upstate, respectively. For lung TAMs, the immunoprecipitated chromatin was amplified using an unbiased genome amplification kit (Sigma Aldrich). Samples were analyzed by real-time PCR using the Roche Universal Probe Library (Roche Diagnostics) and the Faststart TaqMan master kit (Roche Diagnostics).
Microarray and survival analysis
Microarrays were performed on the Mouse Affymetrix 130A.2 platform. The primary data was analyzed by a modified RMA method to yield an average gene expression value (29, 30). The detailed description of the experiment and subsequent data analysis is presented in Supplementary Table 1A.
A high confidence 142 probe set (p<0.05) human Ets2-TAM signature was generated by comparing 407 mouse probe sets (357 genes, absolute INT > 1.5) to the 98 lymph-node negative Rosetta cohort (www.rii.com/publications/2002; divided in to 2 groups based on lymphocyte/leukocyte infiltration status (31). For survival analysis, the 142 probe set Ets2-TAM signature was used as a query to retrieve gene-expression data from Stockholm (GSE1456) breast cancer microarrays (downloaded from NCBI-GEO webpage). Similarly, gene-expression data was also extracted from total and lymph node-negative Rosetta microarrays. The resultant data sets were loaded onto BRB-Array Tools as described in Supplementary Table 3. Briefly, unsupervised K-means clustering of each dataset was performed by using Cluster 3.0 (32) and samples were assigned into two groups. Kaplan-Meier survival analysis was performed by using the Survival Analysis module of the software package StatsDirect (StatsDirect Ltd). Significance of survival analyses was performed by using the Log-Rank (Peto) test.
Statistical Analysis
For lung metastases data, a non-parametric Kruskal-Wallis test with no multiplicity adjustment was used to compare medians between experimental and control groups. A repeated measures ANOVA model was used to analyze mammary tumor progression between the genetic groups over a period of 42 days. This approach takes into consideration longitudinal data, and the following terms were included in the model: genetic group, time and interaction (genotype* time). For the statistical analysis of imaging data, an unpaired Student’s t-test was used. All the tests were two sided.
RESULTS
Deletion of Ets2 in TAMs decreases lung metastasis in spontaneous and orthotopic breast tumor models
Cre/LoxP technology was used to conditionally delete Ets2 in TAMs in the PyMT model, a penetrant breast cancer model with a high frequency of lung metastasis (21). The conditional Ets2LoxP allele used for this study contained LoxP sites flanking exon3-exon5 so that Cremediated recombination of the region resulted in the generation of a null allele (19). The well-characterized Lys-Cre knockin allele was used to delete Ets2 specifically in the macrophage compartment (22). However, initial studies revealed that Cre-recombination in Lys-Cre;Ets2LoxP/LoxP mice was only 30-50% efficient (data not shown). To circumvent this problem, we adopted a strategy whereby mice contained one conditional Ets2LoxP allele and one conventional knockout allele, Ets2db (20). In the final cross, PyMT;Lys-Cre;Ets2db/+ males were crossed with Ets2LoxP/LoxP females to generate both the experimental genotype, PyMT;Lys-Cre;Ets2LoxP/db, and the control genotype, PyMT;Ets2LoxP/db (Supplementary Figure 1A). The frequency of Ets2 rearrangement in isolated mammary tumor macrophages varied between 70-90% with this allele configuration (Supplementary Figure 1B).
Tumor progression was monitored in females of the two genotypes. Tumor initiation was identical between experimental and control mice (data not shown). A small, but statistically significant, decrease in overall tumor growth was observed in the experimental group (Supplementary Figure 1C). This difference in tumor growth was not significant in the early carcinoma stage of progression, but was more pronounced during the late carcinoma stage (days 21-35 post-initiation; Supplementary Figure 1C). However, the final tumor burden and tumor volume were similar in both PyMT;Lys-Cre;Ets2LoxP/db and PyMT; Ets2LoxP/db mice (Supplementary Figure 1D).
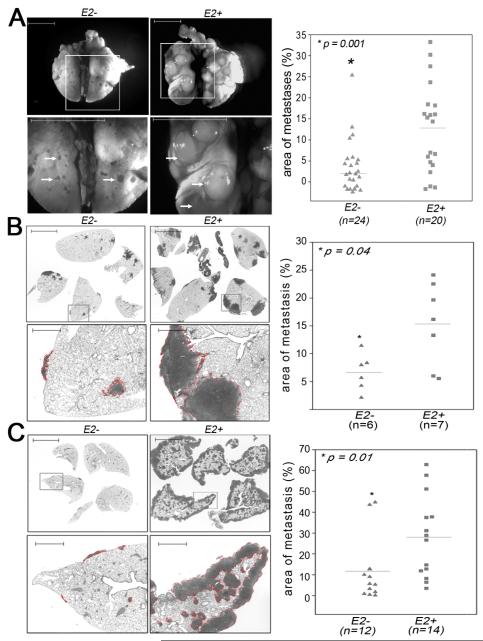
Lung metastasis in both genetic groups was studied by whole-mount analysis (Figure 1A; Supplementary Figure 1E). After image acquisition, the size of the tumors relative to total lung area and the total number of metastases in PyMT;Lys-Cre;Ets2LoxP/db versus PyMT;Ets2LoxP/db mice were quantified. The results showed that both the size and number of lung metastases were significantly reduced in PyMT;Lys-Cre;Ets2LoxP/db mice compared to controls (Figure 1A right panel, size decreased 3-4 fold, p=0.001; Supplementary Figure 2A, number decreased 2-fold, p=0.02).
Figure 1. Deletion of Ets2 in TAMs decreases lung metastasis in spontaneous, orthotopic and tail-vein injection breast cancer models.
A-Wholemount images of lungs obtained from PyMT; Lys-Cre;Ets2LoxP/db (E2−, left) and PyMT;Ets2LoxP/db (E2+, right) mice at late carcinoma stage. B-Analysis of metastatic tumor burden in H&E stained lung sections obtained from Lys-Cre;Ets2LoxP/db (E2−, left) and Ets2LoxP/db (E2+, right) mice in the MVT-1 orthotopic model. C-Analysis of metastatic tumor burden in H&E stained lung sections obtained from E2− (left) and E2+ (right) mice in the Met-1 tail vein injection model. Bottom panels in A, B and C represent high magnification images of areas demarcated by the box in the respective top panels. Lung metastases are indicated by white arrows (A) or outlined with the dotted red line (B, C). Scale Bar = 5mm. Scatter plots on far right of each panel indicates the size of the metastatic tumors in mice with the indicated genotype. Data is presented as the tumor metastases area per unit area. The mean size in each genotype is indicated by the horizontal line. N indicates the number of mice per genetic group. Statistical significance (p-value evaluated by non-parametric Kruskal-Wallis test) is shown.
To confirm and extend the results obtained in the genetic PyMT model, a syngeneic model was employed. The highly metastatic cell line, MVT-1, derived from mice doubly transgenic for MMTV-c-Myc and MMTV-VEGF (25), was injected into mammary fat pads of Lys-Cre;Ets2LoxP/db and control Ets2LoxP/db female mice. After 35 days mice were euthanized and examined. While there was no difference in final tumor burden for the primary tumors (data not shown), the size of metastases per total lung area was three-fold reduced in the experimental Lys-Cre;Ets2LoxP/db group compared to the control group (Figure 1B). These results indicate that the effect of Ets2 is independent of the PyMT oncogene and also demonstrate that haploinsufficieny of Ets2 in the PyMT model is not a confounding factor.
Ets2 in lung macrophages is required for breast tumor metastasis
To firmly establish that the effect of Ets2 in TAMs on metastasis was independent of effects at the primary mammary tumors, a tail-vein injection model was employed. A metastatic PyMT cell line, Met-1 (24), was injected into the circulation via the tail vein in the same two genetic groups as above. After 18 days mice were euthanized and metastases to lungs were quantified in H&E stained sections (Figure 1C). The results demonstrated that the size of lung metastases were significantly reduced more than three-fold in the Lys-Cre;Ets2LoxP/db mice compared to controls.
A potential explanation for the lower levels of metastasis observed in all three models might be that Ets2 regulated genes were required for macrophage survival and/or motility (11, 12). Immunostaining of tumor sections with F4/80 antibody, a marker for mature macrophages, revealed that Ets2 deletion did not result in a decrease in F4/80 positive macrophages associated with either primary or metastatic tumors (Supplementary Figure 2B-C, respectively).
Identification of Ets2 target genes in TAMs
To address the mechanism of Ets2 function in TAMs, mammary TAMs were isolated and subjected to gene expression profiling using the Affymetrix platform. To accomplish this, mammary TAMs were tagged using a c-fms-YFP transgene ((23); Supplementary Figure 3A). This transgene was incorporated into the breeding scheme outlined above to produce experimental PyMT;Lys-Cre;Ets2LoxP/db;c-fms-YFP and control PyMT;Ets2LoxP/db;c-fms-YFP mice. YFP-positive cells isolated from collegenase digested tissue by digital high-speed fluorescence activated cell sorting (FACS) represented approximately 10-15% of the total cells from the primary mammary tumor-site (Supplementary Figure 3B). Greater than 90% of these YFP-positive cells co-expressed macrophage markers like F4/80 (Supplementary Figure 3C). Typically, 3-5×105 YFP-positive TAMs could be isolated from a single mouse.
YFP-positive TAMs were isolated from both genetic groups at the stage when early carcinoma was initially detected in the PyMT model (21). The percentage of YFP-positive cells per mammary gland isolated by FACS was similar in both genetic groups, supporting the conclusion that a reduction in tumor macrophages was not responsible for the observed effects (Supplementary Figure 3B). Since macrophages have also been shown to play a central role in tissue remodeling during mammary gland development (33), YFP-positive macrophages were extracted from the mammary gland of Lys-Cre;Ets2LoxP/db;c-fms-YFP and Ets2LoxP/db;c-fms-YFP females approximately 14 days after the onset of puberty. We reasoned that the role of macrophages in tissue remodeling during mammary gland development would provide a useful comparison to unmask the tumor-specific effects of Ets2.
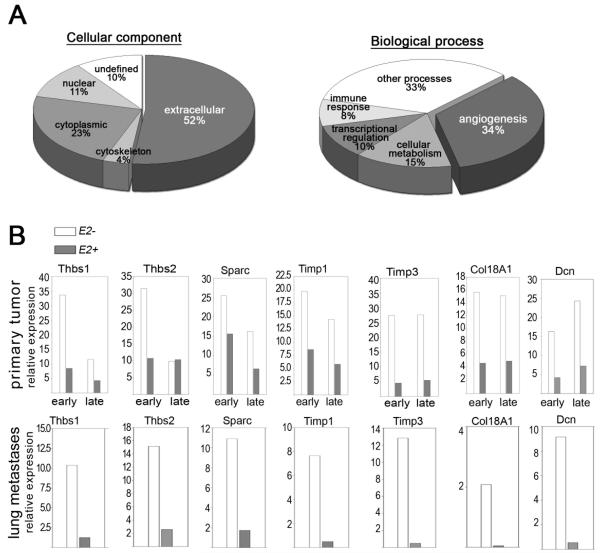
Expression profiling was performed on the resulting four sets of RNA samples. Comparisons between all four sets of expression data were used to identify 357 genes (407 probe sets) whose expression depended on both loss of Ets2 and presence of tumor (see Supplementary Table 1 for details). Approximately 25% of these genes were negatively regulated in the tumor microenvironment and the expression of these genes increased when Ets2 was deleted in TAMs. Gene ontology indicated that genes encoding extracellular components were principally affected by Ets2 deletion (Figure 2A). The major biological process represented was angiogenesis, with 34% of the genes annotated as having a role in this process (Figure 2A). Many of the genes in the angiogenesis class were classified as inhibitors of angiogenesis.
Figure 2. Ets2 represses expression of extracellular-matrix modifying genes in TAMs.
A- Gene ontology based on cellular localization (pie-chart on left) and biological process (pie-chart on right) of the genes differentially regulated in TAMs with or without Ets2. B- Confirmation of genes identified by the microarray analysis using real-time qPCR. RNA was extracted from independently isolated sets of TAMs derived from mammary gland (top panel) or from lungs of mice injected with the Met-1 cell line (bottom panel). Genotypes and stage of tumor development are indicated. Data is represented as average fold induction in samples analyzed in duplicates.
Quantitative RT-PCR using RNA from independently isolated mammary TAMs representing early (first palpable tumor) and late (6 weeks after tumor initiation) carcinoma stages were used to verify the microarray results (Figure 2B). Of 31 genes tested, 25 were confirmed to be differentially expressed in TAMs with or without Ets2 (Supplementary Table 2). Data for fourteen of the genes classified as encoding inhibitors of angiogenesis are shown (Figure 2B and Supplementary Figure 3D). Expression of these genes in both early and late tumors was increased when Ets2 was deleted. In contrast, potential ETS2 targets known to be involved in inflammation like Mmp9 and Tnfα (12), and other genes associated with inflammation like Il6 were not significantly affected by Ets2 deletion in TAMs, emphasizing that the analysis identified tumor-specific targets of ETS2 (Supplementary Figure 3D).
The same 31 genes were also studied in lung TAMs isolated following tail-vein injection of Met-1 cells (bottom panel in Figure 2B and Supplementary Table 2). In these TAMs, 25/31 genes were differentially expressed when Ets2 was deleted, including the angiogenic gene set, indicating the Ets2 targets were similar in mammary or lung TAMs.
ETS2 directly regulates anti-angiogenic genes in isolated TAMs
Examination of 1kb of the proximal promoter regions of four candidate genes not previously reported as ETS2 targets (Thbs1, Thbs2, Timp1, and Tim) revealed conserved ETS binding motifs in their proximal promoter regions (Supplementary Figure 4A). Based on these conserved sequences, chromatin immunoprecipitation (ChIP) experiments were performed on lung TAMs from mice with or without Ets2. For the experiments, approximately 50,000 YFP-tagged, F4/80 positive cells were isolated from lungs containing metastases following tail vein injection of Met-1 cells. Antibodies against ETS2 and its co-repressor HDAC1 (14) were used in the ChIP assays (Figure 3).
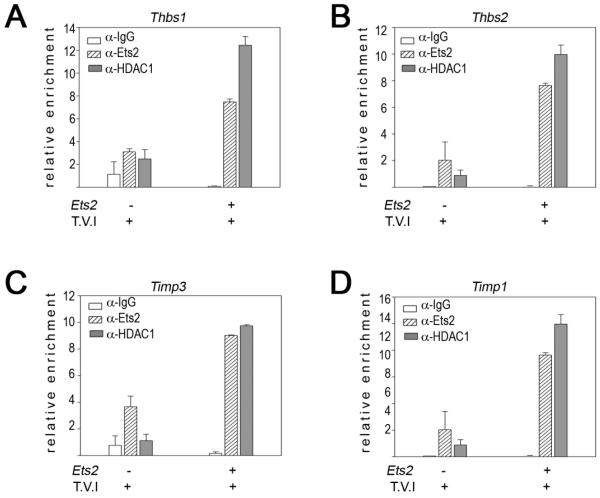
Figure 3. Ets2 represses expression of anti-angiogenic genes in TAMs.
ChIP analysis of the Ets-binding sites in Thbs1 (A, distal Ets site), Thbs2 (B), Timp3 (C) and Timp1 (D) promoters from YFP+ cells extracted from the lung following tail-vein injection of Met-1 cells. ChIP was performed with α-ETS2 and α-HDAC1 antibodies and rabbit IgG control, as indicated. Subsequently, qPCR was performed on the immunoprecipitated chromatin. Results from two independent experiments are represented as mean of relative enrichment of the amplified chromatin ± standard errors of the mean (error bars)
The ChIP experiments revealed that in wild-type cells ETS2 and HDAC1 were both enriched at all four of these promoter sequences (Figure 3). In contrast, when Ets2 was conditionally deleted both the levels of ETS2 and HDAC1 were significantly reduced at each of the four promoters. Similar results were obtained for the Thbs1 promoter in TAMs isolated from the primary mammary tumor (Supplementary Figure 4B).
Expression of angiogenesis inhibitors in TAMs lacking Ets2 correlates with reduced tumor angiogenesis and proliferation
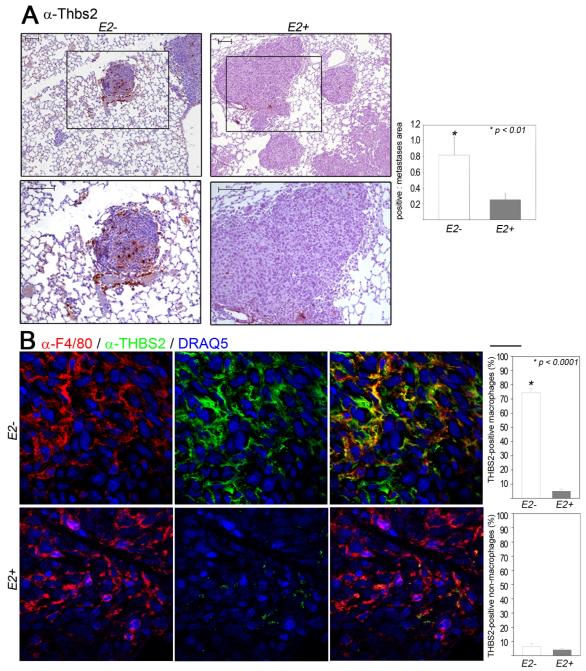
To verify the expression of ETS2 targets in situ, we performed immunohistochemical staining on paraffin-imbedded samples prepared from metastatic lung tumors using commercially available antibodies. This analysis demonstrated robust expression of THBS2, THBS1 and SPARC within tumors from mice with Ets2 deletion in TAMs compared to Ets2+ controls (Figure 4A and Supplementary Figures 5A-B).
Figure 4. Increased expression of Thrombospondin-2 in TAMs that lack Ets2.
A-Images of lung sections from mice injected with the Met-1 cell line in the indicated genotypes, immunostained with α-THBS2. Bottom panel in B represents high magnification image of areas demarcated by the box in the respective top panels. Scale Bar = 100μm. Quantification of antibody staining is presented as the average area of staining per tumor area (graphs at bottom of each panel). Five different tumor areas from five different mice in each group were analyzed. B-Images of mammary tumor sections from mice injected with the MVT-1 cell line harvested 1 week post-injection. Double-immunostained with α-F4/80 (red, left panel) and α-THBS2 (green, middle panel), and merged F4/80-THBS2 images (yellow, right panel).. Quantification of antibody staining is presented as the average percentage of F4/80 positive cells that are also positive for THBS2 in the mammary tumors (graph at right panel). Scale Bar at the top right corner = 20μm. Five different tumor areas from four different tumors in each group were analyzed. Statistical significance (p-value evaluated by unpaired Student’s t-test) is shown.
To confirm that the tumor macrophages were expressing these proteins, frozen mammary tumor sections were analyzed by double immunofluorescent staining using F4/80 to identify TAMs. The MVT-1 orthotopic mammary fat pad injection model was used for this analysis. Staining with α-F4/80-and α-THBS2 showed extensive overlap between the two proteins in sections obtained from tumors with Ets2 deletion (Figure 4B, top panels, Supplementary Movie 1). Since THBS2 is an extracellular protein, expression was found both intracellularly and in the adjacent extracellular space in approximately 75% of F4/80 positive cells, as clearly evident in confocal reconstructions of 15μM sections (see Supplementary Movie 1). In contrast, co-expression of THBS2 in F4/80 positive cells was 10-fold lower in tumors with Ets2 (Figure 4B, bottom panels). Importantly, expression of THBS2 in F4/80 negative cells was not affected by deletion of Ets2 in TAMs (Figure 4B, lower bar graph). Identical results were obtained for THBS1 and SPARC (Supplementary Figure 6A-B respectively and Supplementary Movies 2-3 respectively).
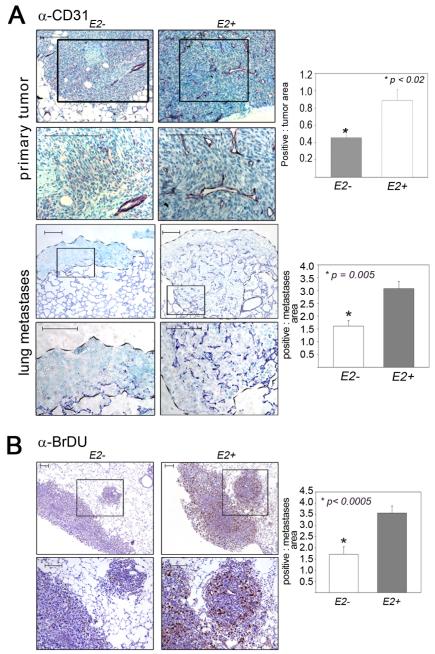
Since many of the tumor-specific Ets2 targets detected, including THBS1, THBS2 and SPARC, have been implicated in angiogenesis, blood vessel density was analyzed in experimental and control mice using α-CD31 immunostaining of paraffin imbedded tumor sections. For these experiments, both primary MVT-1 tumors and lung tumors formed by tail-vein injection of Met-1 cells were studied (Figure 5A). A significant 2-3 fold reduction in tumor vasculature was observed in both primary mammary tumors and lung metastases (Figure 5A).
Figure 5. Impaired tumor angiogenesis and proliferation with Ets2 deletion in TAMs.
A - Images of mammary (top two panels) and lung (bottom two panels) sections from mice injected with the MVT-1 and Met-1 cells respectively; genotypes as indicated. Immunostainedwith α-CD31. B-Micrographs of lung sections from mice injected with the Met-1 cell line in the indicated genotypes, immunostained with α-BrDU. Panels 2 and 4 in A and bottom panel in B represent high magnification image of areas demarcated by the box in the respective top panels. Scale Bar = 100μm. Quantification of antibody staining is presented as the average area of staining per tumor area (graphs at right of each panel). Five different tumor areas from five different mice in each group were analyzed. Statistical significance (p-value evaluated by unpaired Student’s t-test) is shown.
BrDU incorporation was used to measure cell proliferation in lung metastases in the Met-1 tail vein injection model (Figure 5B). The analysis demonstrated a significant 2.5-fold decrease in BrDU-labeled tumor cells in mice with Ets2-deficient TAMs compared to controls. Tumor cell apoptosis, measured by staining with activated caspase-3 antibody, was not significantly affected by Ets2 deletion (Supplementary Figure 2D).
The Ets2-TAM gene expression signature predicts survival of breast cancer patients
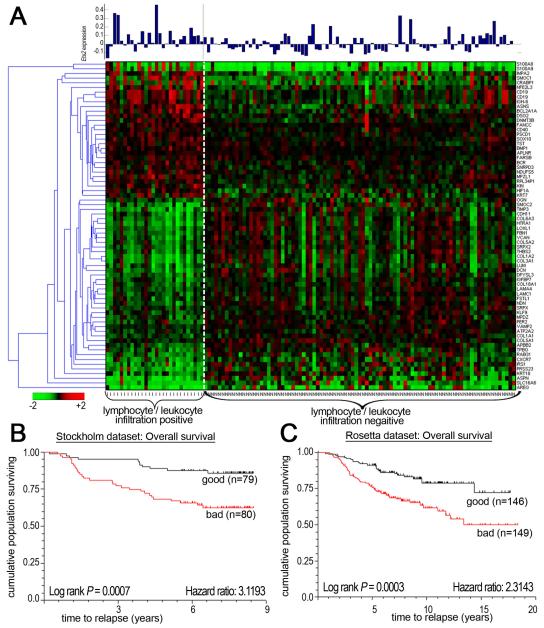
In order to determine if the mouse genetic studies were relevant to human disease, the mouse expression data was compared to the Rosetta human breast cancer data set (31). Initially, 407 mouse probe sets that were differentially expressed in mouse TAMs with or without Ets2 were compared to the Rosetta array platform and 341 homologous human probe sets were identified (see Supplementary Table 3 for details). These 341 probe sets were compared to 2856 probe sets that represented genes differentially expressed in 117 human samples annotated as with or without lymphocyte/leukocyte infiltration (31). This comparison showed that 142 of the mouse Ets2-TAM probe sets, representing 133 genes, were significantly differentially expressed in lymphocyte/leukocyte infiltration-positive versus -negative human breast cancers (p<0.05, see Supplementary Table 3 and Supplementary Figure 7A). Gene ontology analysis of these human genes showed that extracellular matrix components and angiogenesis were predominantly affected, just as for the mouse Ets2-TAMs genes (Supplementary Figure 7B). A subset of 70 genes differentially expressed with high significance (p<0.001) is represented in the heat map presented in Figure 6A. Interestingly, Ets2 expression itself was on average 8-fold higher inlymphocyte/leukocyte infiltration-positive patients when compared to the negative group (Figure 6A, bar graph, p = 0.0002).
Figure 6. The Ets2-TAM gene signature predicts survival in human breast cancer patients.
A- Heat map of differential expression of the Ets2-TAM 70 gene profile (p<0.001, see text) in 98 breast cancer samples distinguished by lymphocyte/leukocyte infiltration. The bar graph on the top indicates the level of Ets2 expression in each of the 98 cancer samples. B&C- Kaplan Meier analysis of overall survival in Stockholm and Rosetta breast tumor cohorts, respectively.
To determine if the TAM gene signature correlated with clinical outcome of patients, the 133 human Ets2-TAM gene signature was used for unsupervised clustering of expression data obtained from 159 sporadic breast cancer patients in the Stockholm data set ((18), see Supplementary Table 3). Expression of the Ets2-TAM signature predicted overall survival in this group with high confidence (Figure 6B, p = 0.0007, Hazard ratio of 3.1). Similar results were obtained with the entire Rosetta 295 patient sample set (Figure 6C, p=.0.0003, Hazard ratio of 2.31).
DISCUSSION
The influence of the microenvironment, particularly macrophages, on tumor growth and metastasis have long been recognized, but relatively little is known of the gene pathways and mechanisms macrophages use to promote tumor malignancy (34). The results presented here demonstrate that in mouse models Ets2 in tumor macrophages promotes angiogenesis and growth of both primary tumors and lung metastases. The mechanism of action of ETS2 in TAMS involved direct repression of genes encoding predominantly extracellular products, including well-characterized inhibitors of angiogenesis. Recently, an independent report of global gene profiling in TAMs also observed expression of several anti-angiogenic genes along with well-known positive regulators like Vegf-a, results consistent with our data (35). However, the anti-angiogenic effect of TAMs lacking Ets2 is dominant even in the context of MVT-1 tumor cells that overexpress VEGF-A. Additionally, the presumed role of VEGF-A produced by TAMs in triggering the angiogenic switch have been challenged by recent finding demonstrating that deletion of VEGF-A in TAMs actually results in increased tumor growth (36, 37). Thus, Ets2 has previously unappreciated role in TAMs in controlling the balance between positive and negative regulators of angiogenesis necessary for tumor metastasis.
Ets2 in TAMs increased the growth of primary and metastatic tumors. Ets2 could indirectly effect tumor growth by modulating angiogenesis, or directly through paracrine mechanisms. The Ets2 targets identified would favor the former possibility, as obvious paracrine candidates like Il6 or Egf were not differentially expressed. In either case, the results are consistent with the Ets2 pathway playing a role in some activities associated with the alternatively-activated M2 macrophage population (2, 3). M2 macrophages are believed to modulate inflammatory response and to promote tissue remodeling and angiogenesis; in the context of tumor progression, M2-like cells are believed to promote immune suppression, as well as tumor angiogenesis, invasion and metastasis (2, 3). Extracellular function and angiogenesis are the major Ets2 targets identified in our studies, providing a molecular mechanism by which M2-like tumor macrophages modulate the extracellular microenvironment to promote tumor growth and angiogensis at both primary and tumor sites.
A key finding is that a portion of the mouse Ets2-TAM gene expression signature was present in human breast cancer expression data and that it could retrospectively predict overall survival in two independent cohorts of sporadic breast cancer patients. This 133 gene signature is independent of other breast tumor signatures capable of predicting patient outcome, including stromal gene signatures (38, 39). While further efforts will be required to fully implement these findings and determine their significance to human disease, the results validate the relevance of our hypothesis-driven mouse modeling approach for dissecting TAM functions in tumor growth and metastasis.
Dispersed tumor cells are present in many breast cancer patients and may be the mediators of tumor recurrence (40). Breast tumor micrometastases are genetically distinct from the primary tumor indicating that they are disseminated early in tumor progression (41, 42). Results obtained in the PyMT and Her2/Neu mouse models demonstrate an early spread of mammary epithelial cells before the carcinoma stage, providing experimental verification of the human data (42). Thus, understanding how dispersed dormant cells progress to growing metastases is a problem with considerable clinical relevance. Further studies on Ets2 and its downstream targets could provide unique insights in understanding how the microenvironment modulates the growth of tumor cells at metastatic sites.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexander Borowsky and Michael Johnson for the Met-1 and MVT-1 cell lines, respectively, Robert Oshima for the Ets2db mice, Karl Kornacker for microarray data analysis, Kartic Krishnamurthy for image analysis software, and Lisa Rawahneh for histology support. We acknowledge the Cancer Center Microscopy/Imaging, Microarray, Genomic, Transgenic/Knockout, Histology, and Flow Cytometry Shared Resources. TZ was supported by a DOD Pre-doctoral Fellowship. This work was supported by NCI Grants P01 CA097189 (MCO, GL, TJR) and R01 CA053271 (MCO) and the Evelyn Simmer’s Charitable Trust (MCO).
REFERENCES
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 4.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 5.Wyckoff JB, Wang Y, Lin EY, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 6.Leek RD, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–9. [PubMed] [Google Scholar]
- 7.Bolat F, Kayaselcuk F, Nursal TZ, Yagmurdur MC, Bal N, Demirhan B. Microvessel density, VEGF expression, and tumor-associated macrophages in breast tumors: correlations with prognostic parameters. J Exp Clin Cancer Res. 2006;25:365–72. [PubMed] [Google Scholar]
- 8.Tsutsui S, Yasuda K, Suzuki K, Tahara K, Higashi H, Era S. Macrophage infiltration and its prognostic implications in breast cancer: the relationship with VEGF expression and microvessel density. Oncol Rep. 2005;14:425–31. [PubMed] [Google Scholar]
- 9.Valkovic T, Dobrila F, Melato M, Sasso F, Rizzardi C, Jonjic N. Correlation between vascular endothelial growth factor, angiogenesis, and tumor-associated macrophages in invasive ductal breast carcinoma. Virchows Arch. 2002;440:583–8. doi: 10.1007/s004280100458. [DOI] [PubMed] [Google Scholar]
- 10.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–40. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sevilla L, Aperlo C, Dulic V, et al. The Ets2 transcription factor inhibits apoptosis induced by colony-stimulating factor 1 deprivation of macrophages through a Bcl-xL-dependent mechanism. Mol Cell Biol. 1999;19:2624–34. doi: 10.1128/mcb.19.4.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei G, Guo J, Doseff AI, et al. Activated Ets2 is required for persistent inflammatory responses in the motheaten viable model. J Immunol. 2004;173:1374–9. doi: 10.4049/jimmunol.173.2.1374. [DOI] [PubMed] [Google Scholar]
- 13.Baker KM, Wei G, Schaffner AE, Ostrowski MC. Ets-2 and components of mammalian SWI/SNF form a repressor complex that negatively regulates the BRCA1 promoter. J Biol Chem. 2003;278:17876–84. doi: 10.1074/jbc.M209480200. [DOI] [PubMed] [Google Scholar]
- 14.Wei G, Schaffner AE, Baker KM, Mansky KC, Ostrowski MC. Ets-2 interacts with corepressor BS69 to repress target gene expression. Anticancer Res. 2003;23:2173–8. [PubMed] [Google Scholar]
- 15.Turner DP, Findlay VJ, Moussa O, Watson DK. Defining ETS transcription regulatory networks and their contribution to breast cancer progression. J Cell Biochem. 2007;102:549–59. doi: 10.1002/jcb.21494. [DOI] [PubMed] [Google Scholar]
- 16.Man AK, Young LJ, Tynan JA, et al. Ets2-dependent stromal regulation of mouse mammary tumors. Mol Cell Biol. 2003;23:8614–25. doi: 10.1128/MCB.23.23.8614-8625.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei G, Srinivasan R, Cantemir-Stone C, et al. Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood. 2009;114:1123–30. doi: 10.1182/blood-2009-03-211391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamamoto H, Flannery ML, Kupriyanov S, et al. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–26. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin EY, Jones JG, Li P, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol. 2003;163:2113–26. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 23.Sasmono RT, Oceandy D, Pollard JW, et al. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–63. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 24.Borowsky AD, Namba R, Young LJ, et al. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis. 2005;22:47–59. doi: 10.1007/s10585-005-2908-5. [DOI] [PubMed] [Google Scholar]
- 25.Pei XF, Noble MS, Davoli MA, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In Vitro Cell Dev Biol Anim. 2004;40:14–21. doi: 10.1290/1543-706X(2004)40<14:ECOPMT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 26.Soule H, McGrath C. A simplified method for passage and long-term growth of human mammary epithelial cells. In Vitro Cell Dev Biol. 1986;22:6–12. doi: 10.1007/BF02623435. [DOI] [PubMed] [Google Scholar]
- 27.Hu R, Sharma SM, Bronisz A, Srinivasan R, Sankar U, Ostrowski MC. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol Cell Biol. 2007;27:4018–27. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessen KA, Liu SY, Tepper CG, et al. Molecular analysis of metastasis in a polyomavirus middle T mouse model: the role of osteopontin. Breast Cancer Res. 2004;6:R157–69. doi: 10.1186/bcr768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auer H, Newsom D, Nowak N, et al. Gene-resolution analysis of DNA copy number variation using oligonucleotide expression microarrays. BMC Genomics. 2007;8:111. doi: 10.1186/1471-2164-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry R, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 32.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–4. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 33.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–82. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 34.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–6. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Ojalvo L, King W, Cox D, Pollard J. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol. 2009;174:1048–64. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin EY, Li JF, Bricard G, et al. VEGF Restores Delayed Tumor Progression in Tumors Depleted of Macrophages. Mol Oncol. 2007;1:288–302. doi: 10.1016/j.molonc.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockmann C, Doedens A, Weidemann A, et al. Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature. 2008 doi: 10.1038/nature07445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 39.Ma X, Dahiya S, Richardson E, Erlander M, Sgroi D. Gene expression profiling of the tumor microenvironment during breast cancer progression. Breast Cancer Res. 2009;11:R7. doi: 10.1186/bcr2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braun S, Vogl FD, Naume B, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt-Kittler O, Ragg T, Daskalakis A, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–42. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hüsemann Y, Geigl J, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.