Abstract
Dicer or Dicer-like (DCL) protein is a catalytic component involved in microRNA (miRNA) or small interference RNA (siRNA) processing pathway, whose fragment structures have been partially solved. However, the structure and function of the unique DUF283 domain within dicer is largely unknown. Here we report the first structure of the DUF283 domain from the Arabidopsis thaliana DCL4. The DUF283 domain adopts an α-β-β-β-α topology and resembles the structural similarity to the double-stranded RNA-binding domain. Notably, the N-terminal α helix of DUF283 runs cross over the C-terminal α helix orthogonally, therefore, N- and C-termini of DUF283 are in close proximity. Biochemical analysis shows that the DUF283 domain of DCL4 displays weak dsRNA binding affinity and specifically binds to double-stranded RNA-binding domain 1 (dsRBD1) of Arabidopsis DRB4, whereas the DUF283 domain of DCL1 specifically binds to dsRBD2 of Arabidopsis HYL1. These data suggest a potential functional role of the Arabidopsis DUF283 domain in target selection in small RNA processing.
Keywords: NMR structure, Dicer DUF283, miRNA processing, double-stranded RNA-binding fold, protein–protein interaction
INTRODUCTION
RNA silencing is a small regulatory RNA-controlled and revolutionary conserved gene regulation mechanism comprising a set of sequential core reactions (Zamore and Haley 2005). First, Dicer-like enzyme processes primary miRNA transcript (pri-miRNA) or long complementary double-stranded RNA into 21–24 base pairs (bp) small regulatory RNA (Bernstein et al. 2001). Subsequently, the small RNA is loaded into RNA-induced silencing complex (RISC) to pair with target mRNA for degradation or translation repression (Hammond et al. 2000; Nykanen et al. 2001; Liu et al. 2004; Meister et al. 2004; Siomi and Siomi 2009). RNase III enzyme Dicer and RNase H enzyme Argonaute are essential for the initiation and effect of RNA silencing, respectively (Voinnet 2005).
Dicers not only play the catalytic role to process miRNAs/siRNAs, but also load these small regulatory RNAs into the RISC (Lee et al. 2004; Pham et al. 2004). In Arabidopsis thaliana, there are four Dicer-like proteins, namely DCL1–4. DCL1 plays the role for miRNA processing, whereas DCL2–4 proteins generate siRNAs with distinct sizes. DCL2 processes long dsRNAs into 22 bp, whereas DCL3 and DCL4 produce 24 bp siRNA and 21 bp siRNA, respectively (Qi et al. 2005). Therefore the structural information of Dicer could provide the insightful information to understand the molecular mechanism of Dicer initiated RNA silencing pathway.
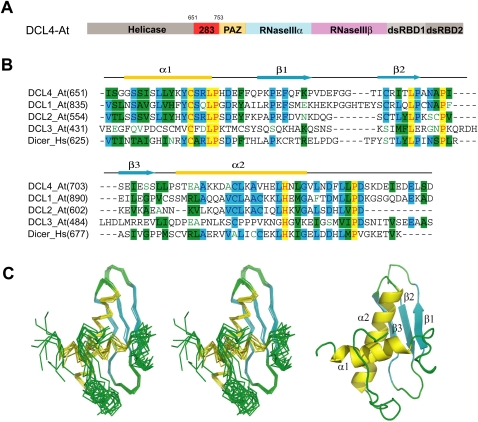
Dicer protein is a multiple-domain protein, which contains the helicase domain, DUF283 domain, PAZ domain, two tandem RNase III domains, and two tandem dsRBD domains from the N-terminus to C-terminus (Fig. 1A). It was proposed that the RNase III domain harbors the catalytic residues for small RNA processing (Zhang et al. 2004). The recent crystal structure of human parasite Giardia Dicer provides the detailed molecular evidence to support the long standing hypothesis that Dicer harbors “one processing center” to cleave the bound pre-miRNA or long dsRNA into ∼19 bp small RNA duplex with 3′-2-nucleotide (nt) overhangs (Macrae et al. 2006). Remarkably, the Giardia Dicer structure adopts a hatchet-like architecture with the PAZ domain recognizing the 3′-2-nt overhangs of bound dsRNAs, whereas the unique connecting α helix functions as a molecular ruler to measure the distance from the dsRNA end (recognized by the PAZ domain) to the cleavage site (provided by the RNase III domains) (Macrae et al. 2006). Although Giardia Dicer contains only the PAZ domain and two RNase III domains, it displays robust dsRNA-processing activity (Macrae et al. 2006). By contrast, the removal of the DUF283 domain, which is ∼100 amino acid positioning at the N-terminus of the PAZ domain (Fig. 1A,B), from either human dicer or Drosophila DCR-1 abolished miRNA processing activity (Lee et al. 2006; Ye et al. 2007). However, a slightly different DUF283 deletion construct in human dicer shows little impact on pre-siRNA or pre-miRNA cleavage activity (Ma et al. 2008). Nevertheless, these data suggest that the DUF283 domain could be an essential functional domain for Dicers from certain higher eukaryotes.
FIGURE 1.
Overall structure of DCL4 DUF283. (A) Domain architecture of Arabidopsis thaliana DCL4. (B) Sequence alignment and secondary structure of Dicer DUF283. The aligned sequences (Swiss Protein ID) are in the order of DCL4_At, DCL1_At, DCL2_At, DCL3_At, and Dicer_Hs. The secondary structure diagram for DCL4_At is shown on top. The α-helices are colored in yellow, β-strands are colored in cyan. Conserved residues are shaded in cyan (80% similarity) and green (60% similarity), whereas essentially invariant residues are shaded in yellow. (C) Stereo view of the ensemble of eight lowest energy NMR structures of the DCL4 DUF283 domain (residues 651–752) and Ribbon representation of the DCL4 DUF283 domain.
In an effort to understand the functional role of the DUF283 domain in Dicer-catalyzed RNA silencing pathway, we report here the first structure of the DUF283 domain of the Arabidopsis Dicer-like 4 (DCL4) protein and show that DUF283 adopts a double-stranded RNA-binding (dsRBD) fold for protein–protein interaction, which is in contrast to bioinformatics prediction that DUF283 is a dsRBD fold for dsRNA binding (Dlakić 2006). We further demonstrate that the DCL4 DUF283 domain selectively binds to its designated partner, DRB4, whereas the DCL1 DUF283 domain selectively binds to its designated partner, HYL1, by in vitro pull-down assay, which suggest that Arabidopsis DUF283 domains probably play significant roles for partner protein selection in small RNA processing.
RESULTS
The DUF283 domain resembles dsRNA-binding fold
We have systematically screened ∼60 different Dicer DUF283 constructs from multiple species with different fragment lengths and tags to assess the expression levels and solubility of the expressed proteins. The Arabidopsis DCL4 DUF283 construct (residues: 651–752) used in our experiments is suitable for NMR determination because of its high expression level, solubility, and monomeric dispersion.
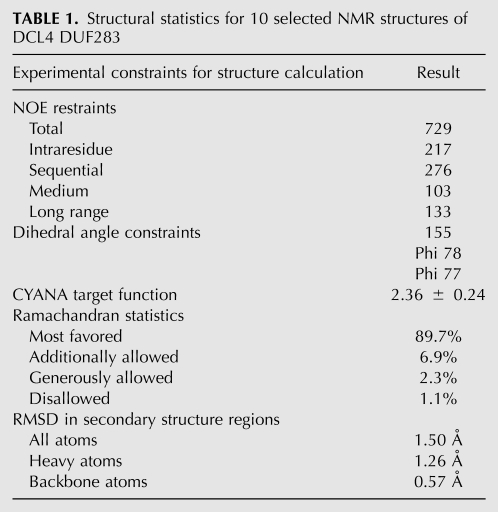
DCL4-DUF283 has a highly dispersed HSQC spectrum in solution characteristic of a well-folded protein. As such, the NMR structure of DUF283 was successfully determined by using NOE distance restraints derived from analyzing 15N- and 13C-edited NOESY spectra; as well as dihedral angle restraints from TALOS prediction based on five chemical shifts. Figure 1C presents the superimposition of 10 selected DUF283 structures in solution with the lowest target functions. As detailed in Table 1, over the secondary structure regions, the 10 structures are very similar, with RMS deviations of 1.50 Å for all atoms, 1.26 Å for heavy atoms and 0.57 Å for backbone atoms. Interestingly, it appears that the C-terminal ∼18 residues (residues: 735–752) are relatively unstructured and poorly defined, having distinct conformations in different structures. This observation is in good agreement with the small chemical shift deviations and lack of the interresidual NOE connectivites over the region (data not shown).
TABLE 1.
Structural statistics for 10 selected NMR structures of DCL4 DUF283
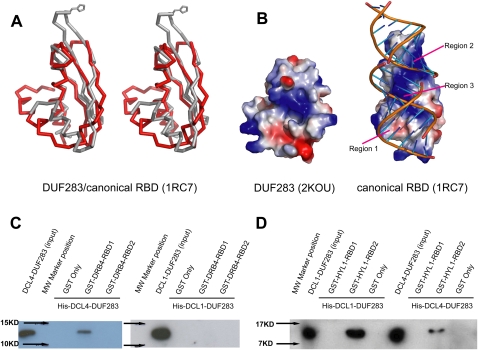
DUF283 consists of three β-strands (β1: residues 675–683; β2: residues 691–697; β3: residues 703–708) and two α-helices (α1: residues 654–667; α2: residues 712–732) and adopts α-β-β-β-α topology with N-terminal α-helix (α1) running cross over the C-terminal α-helix (α2) orthogonally (Fig. 1B). Hence, N-terminal and C-terminal α-helices pack against one surface of the three-stranded antiparallel β-sheet (Fig. 1C). The structural topology search by Dali server (www2.ebi.ac.uk/dali) reveals that the overall structure of the DCL4 DUF283 domain resembles the close structural similarity to the dsRBD domain of Aquifex aeolicus RNase III (PDBID:1RC7, Z score, 4.6, RMSD 3.7 Å, 62 Cα) (Fig. 2A). The comparisons of these two proteins show structural similarity at their β-sheet and C-terminal α-helix regions. However, the N-terminal α-helix of DUF283 swings ∼30° back toward the C-terminal α helix, hence the N- and C-termini of DUF283 are in close proximity, which is different from the bioinformatics prediction (Dlakić 2006).
FIGURE 2.
DCL4 DUF283 resembles dsRBD fold for protein target selection (A) Stereoview diagram of the superimposition of DCL4 DUF283 (red) and the dsRBD domain from Aquifex aeolicus RNase III (1RC7, gray). Invariable histidine side chain of Aquifex aeolicus RNase III dsRBD involved in dsRNA binding shown in stick. For clarification purpose, the C-terminal disordered region (residues: 735–752) was omitted. (B) Electrostatic potential surface view of DCL4 DUF283 and Aquifex aeolicus RNase III dsRBD with the blue, red, and white colors representing the positive, negative, and neutral charges, respectively. Three regions within Aquifex aeolicus RNase III dsRBD involved in dsRNA recognition are also labeled. (C) DCL4 DUF283 selectively binds to DRB4 dsRBD1. Similar amounts of recombinant His-DCL4-DUF283 (left panel) or His-DCL1-DUF283 (right panel) was loaded onto the prebound GST-fused DRB4 fragments (dsRBD1: residues 3–70 and dsRBD2: residues 81–155). His-DCL4-DUF283 or His-DCL1-DUF283 was detected by Western blotting with anti-His antibody. (D) DCL1 DUF283 selectively binds to HYL1 dsRBD2. Similar amount of recombinant His-DCL1-DUF283 (left panel) or His-DCL4-DUF283 (right panel) was loaded onto the prebound GST-fused HYL1 fragments (dsRBD1: residues 11–85 and dsRBD2: residues 100–172). His-DCL4-DUF283 or His-DCL1-DUF283 was detected by Western blotting with anti-His antibody.
The DUF283 domain is a noncanonical dsRBD
In addition to the different orientations of the N-terminal α-helix between DUF283 and dsRBD (Fig. 2A), there are significant structural deviations between them at the putative RNA-binding surfaces (Fig. 2B). All the canonical dsRBDs exampled by Xenopus laevis dsRBD2 have three critical dsRNA-binding regions, referred to as regions 1, 2, and 3 (Ryter and Schiltz 1998). These proteins share a conserved positive electrostatic potential charge in region 3, which is involved in recognizing the major groove of dsRNA. Similar neutral or slightly negative electrostatic potential surfaces in regions 1 and 2, together with a small patch of a positive electrostatic potential surface in region 2, which contains the invariable histidine residue, specifically recognize the dsRNA minor groove (Ryter and Schultz 1998). Unlike canonical dsRBD, the DUF283 domain has a significant different structure at region 1 due to the different orientation of N-terminal α-helix, which further changes the overall charge distribution at the putative dsRNA-binding surface (Fig. 2B). In addition, DUF283 also lacks the invariable His residue at region 2 and adopts short loop architecture with different loop orientation (Fig. 2A,B). The significant structural deviations of the DUF283 domain from other consensus dsRBD at the putative dsRNA-binding surface strongly suggest that the DUF283 domain may not be able to bind to canonical dsRNA.
In order to test whether the DUF283 domain indeed is a noncanonical dsRBD not for dsRNA-binding, we performed electrophoretic mobility shift assay (EMSA), isothermal titration (ITC), and NMR titration methods to detect the binding affinity between the DUF283 domain and a self-complementary siRNA-like duplex (5′-P-AGACAGCAUUAUGCUGUCUUU-3′), whose sequence has been verified as a strong binding partner for several different dsRNA-binding proteins (Chen et al. 2008; Cheng et al. 2009). As expected, EMSA did not show any detectable dsRNA-binding affinity by DUF283 (data not shown), whereas ITC titration showed that no significant heat change was associated with the binding of DUF283 to dsRNA (data not shown). On the other hand, NMR titration revealed that the binding between DUF283 and dsRNA was not saturated, even at a molar ratio of 1:13 (DUF283:dsRNA) (Supplemental Fig. S1). However, attempts to further increase the dsRNA concentration failed because the DUF283 protein precipitated at a higher dsRNA concentration. This result strongly suggests that there is only a weak binding between DUF283 and dsRNA, although a large portion of the DUF283 residues were perturbed (Supplemental Fig. S1). Taken together, these data demonstrated that the DUF283 domain is a noncanonical dsRNA-binding domain.
The DUF283 domain selectively binds to its partner
In addition to dsRNA-binding ability, some dsRBDs have functions in dimerization or nuclear localization (Doyle and Jantsch 2002). Hence, the apparently weak dsRNA-binding affinity displayed by the DUF283 domain strongly suggests that the DUF283 domain may adopt a dsRBD fold for protein–protein interaction.
In Arabidopsis thaliana, four Dicer-like proteins display diversified biological functions within different small RNA processing pathways (Baulcombe 2004; Chapman and Carrington 2007). DCL1 plays the role for miRNA processing, whereas DCL4 plays the primary function for viral RNA processing (Kurihara and Watanabe 2004; Deleris et al. 2006). Notably, DCL1 most strongly interacts with the double-stranded RNA-binding protein HYPONASTIC LEVAES1 (HYL1), whereas DCL4 specifically interacts with HYL1 homolog, DRB4 (Hiraguri et al. 2005). To this end we performed in vitro pull-down assay to test whether the DCL4 DUF283 domain is able to bind to DRB4 in vitro.
DRB4 has a similar domain architecture to HYL1 and comprises two tandem dsRBDs at its N-terminus and ∼200 amino acid fragment without structural and functional assignments at its C-terminus. We made two GST-fused DRB4 constructs with the lengths covering the dsRBD1 (DRB4, 3–70) and dsRBD2 (DRB4, 81–155). We used N-terminal GST-fused DRB4 fragment as bait and N-terminal His-fused DCL4-DUF283 as a target. As expected, GST-fused DRB4 dsRBD1 was able to pull-down the His-fused DCL4 DUF283 domain, whereas GST-fused DRB4 dsRBD2 was failed to pull-down the His-fused DCL4 DUF283 domain by Western blotting detection using the antibody against the His tag (Fig. 2C, left panel). Next we tested whether the specific protein–protein interaction displayed by DUF283 determined the protein partner selection. We performed an in vitro pull-down assay by using the same N-terminal GST-fused DRB4 fragment as bait and N-terminal His-fused DCL1 DUF283 (DCL1, 836–942) instead as a target. As expected, none of the DRB4 fragment was able to pull-down DCL1 DUF283 (Fig. 2C, right panel).
We further ask whether DCL1 DUF283 specifically recognizes its own partner, HYL1. To this end, we made two GST-fused HYL1 constructs with the lengths covering the dsRBD1 (HYL1, 11–85) and dsRBD2 (HYL1, 100–172). Indeed, HYL1 RBD2 was able to pull-down a larger amount of the DCL1 DUF283 domain than of the DCL4 DUF283 domain. By contrast, the HYL1 RBD1 domain failed to pull-down either DCL1 DUF283 or DCL4 DUF283 (Fig. 2D). The relatively moderate binding affinity between the DCL4 DUF283 domain and DRB4 probably suggests that other domains of DCL4 also participate in partner protein binding. Nevertheless, these data suggest that HYL1/DRB proteins may assist the recruitment of DCL proteins by targeting the DUF283 domain through heterodimerization at its noncanonical dsRBD domain. Hence, each HYL1/DRB family protein probably individually modulates Dicer function through selectively interacting with one specific partner among the four DCL proteins.
The DUF283 domain binds to Zn ion
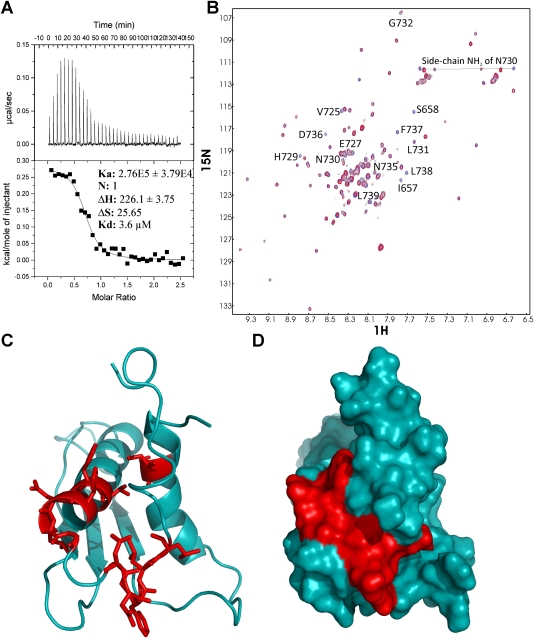
Since DUF283 sequences harbor three Cys residues, therefore, we assessed whether the DUF283 domain was capable of binding Zn ion. ITC experiments showed that DUF283 was able to bind Zn ion of ZnCl2, with a dissociation constant (Kd) of 3.6 μM (Fig. 3A). Next, we performed NMR HSQC titrations to monitor the binding of DUF283 to ZnCl2. As seen in Figure 3B, at a molar ratio of 1:5 (DUF283:Zn), the binding was largely saturated, with several HSQC peaks shifted and some disappeared. The disappearance of peaks implies that the binding provokes conformational exchanges over these residues on the microsecond to millisecond time scale. Notably, the significantly perturbed residues are mainly from three regions; Ile657 and Ser658 located on the N-end of the first helix; Val725, His729-Gly732 on the C-end of the second helix; and Asn735-Leu739 on a loop. It appears that the Zn binding may be involved in a patch formed by residues from three regions of the DUF283 structure (Fig. 3C,D). This result also suggests that although the C-terminal residues are relatively flexible in structure, they are probably essential for biological functions in vivo. However, at this moment, we are not able to link the zinc binding ability of DUF283 with the biological consequences of small RNA processing functions of Dicer.
FIGURE 3.
DCL4 DUF283 is a zinc-binding module. (A) ITC titration of DCL4 DUF283 by zinc chloride. (B) Chemical shift perturbation after addition of zinc chloride to a 15N-labeled DCL4 DUF283. The chemical shifts are monitored by 1H–15N heteronuclear single quantum correlation (HSQC) spectra. The spectrum of the free protein is shown in blue and the spectrum after addition of a fivefold molar excess of zinc chloride is shown in red. (C) Ribbon representation of the DCL4 DUF283 domain colored according to chemical shift perturbation induced following the addition of zinc chloride. Residues that are affected/not affected by the zinc addition are shown in red/cyan. (D) Surface representation of the DCL4 DUF283 domain colored according to chemical shift perturbation induced following the addition of zinc chloride. The same orientation as in C.
DISCUSSION
Dicers are large proteins of ∼220 kDa, which only adopts a “single processing center” that measures and cleaves ∼21 bp miRNA/siRNA duplex from the end of a pre-miRNA or from long dsRNA to dsRNA with 3′-2-nt overhangs (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003; Ma et al. 2004). Within this model, a unique long α helix connecting the PAZ domain to the RNase III domains functions as a molecular ruler to measure the distance from the characteristic 3′-2-nt overhang of dsRNA end (recognized by the PAZ domain) to the cleavage site (provided by the RNase III domains) (Lingel et al. 2003; Song et al. 2003; Yan et al. 2003; Ma et al. 2004). This model is strongly supported by the Giardia Dicer structure and the mutation data on human dicer (Macrae et al. 2006). However, this model does not shed light on the structural mechanism of the unique DUF283 domain and the helicase domain, which positions at the N-terminus of the PAZ domain. Importantly, recent function analysis on Drosophila DCR-1 showed that the DUF283 domain is essential for pre-miRNA processing because the removal of the PAZ and/or the DUF283 domains completely abolished the ability of DCR-1 to generate miRNA, although the N-terminal helicase domain and the C-terminal dsRBD domain are dispensable (Ye et al. 2007). Therefore, our experimental results on the structure and function characterization of the DUF283 domain could provide a basis to further fine-tune the “single processing center model” by the addition of the DUF283 domain as a target for Dicer partner protein selection.
Although, the truncated dicer comprising only the PAZ domain and two RNase III domains, such as Giardia Dicer, could cleave pre-miRNA or long dsRNA with 3′-2-nt overhangs efficiently in vitro (Macrae et al. 2006), Dicer requires a dsRNA-binding partner for efficient processing of pre-miRNA in vivo (Liu et al. 2003; Tomari et al. 2004; Han et al. 2006; Dong et al. 2008). The structural discovery that the DUF283 domain is a dsRNA-binding domain for selectively protein–protein interactions strongly suggests that the DUF283 domain serves as a heterodimerization domain for specific dsRNA-binding (DRB) protein binding.
We therefore propose that DRB family protein probably plays a role to selectively recruit specific dicer to its native target by primary targeting the DUF283 domain within dicer. The high degree of structural and surface charge compatibility between different DRB family members and the individual Dicer DUF283 domains could help to guide the specific Dicer into its designated small RNA processing pathway. Therefore, in Arabidopsis, the HYL1/DCL1 pair probably determines the miRNA processing fate of DCL1, whereas the DRB4/DCL4 pair probably determines the viral RNA processing fate of DCL4.
Notably, the heterodimers formed between Dicers and dsRBD proteins are observed from Drosophila to human and ranged from miRNA/siRNA processing to subsequent RISC loading and assembly (Tomari et al. 2004; Chendrimada et al. 2005; Förstemann et al. 2005; Haase et al. 2005; Hiraguri et al. 2005; Curtain et al. 2008). Although there is no evidence suggesting the direct interactions between dsRBD-containing proteins and Dicers through the DUF283 domains in the human and Drosophila systems so far, the discovery that the DUF283 domain adopts a noncanonical dsRBD fold for protein–protein interaction could provide the important insights to further study these molecular events in depth.
MATERIALS AND METHODS
Construction of Escherichia coli expression vectors and protein expression
DNA fragments coding for the DCL4-DUF283 domain (residues 651–752) and DCL1-DUF283 domain (residues 836–942) were amplified by PCRs with designed primers from Arabidopsis thaliana DCL1 and DCL4 cDNAs and inserted into pET-28b with an N-terminal His-tag, respectively. HYL1 dsRBD1 (residues 11–85), HYL1 dsRBD2 (residues 100–172), DRB4 dsRBD1 (residues 3–70), and DRB4 dsRBD2 (residues 81–155) were cloned into pGEX6p-1 vector with N-terminal GST tag.
Recombinant proteins were expressed in E. coli (BL21/DE3 strain) overnight at 20°C induced by 0.4 mM isopropyl β-d-thiogalactoside. The harvested cells were sonicated in lysis buffer containing 150 mM sodium chloride, 20 mM sodium phosphate at pH 7.2. The extracted proteins were purified through Ni2+ affinity column by in-gel digestion followed by HiLoad Superdex S-75 26/60 column.
The generation of the isotope-labeled proteins for NMR studies followed a similar procedure, except that the bacteria were grown in M9 medium with the addition of (15NH4)2SO4 for 15N labeling and (15NH4)2SO4/[13C] glucose for 15N/13C double labeling, respectively. The purified DUF283 domain proteins were further dialyzed in 10 mM phosphate buffer with 1 mM DTT under pH 6.3.
For different fusion proteins from DRB4 and HYL1, cells were resuspended in 1 mM EDTA, 1 mM dithiothreitol (DTT), complete proteinase inhibitor (Roche, www.roche.com), 1.0 M NaCl, 50 mM Tris (pH 7.4), and lysed by cell disruptor. After centrifugation (40,000 g, 1 h), the supernatant was loaded onto a GST affinity column equilibrated in 50 mM Tris (pH 8.0) with 500 mM NaCl. Nonspecific-binding proteins were washed out by the same buffer and target proteins were eluted with GSSH followed by gel filtration purification. All fusion proteins were dialyzed at 500 mM NaCl, 4 mM DTT, and 25 mM Tris-HCl at pH 7.4.
NMR experiments and structure calculation
For NMR experiments, the DCL4 samples were prepared in 10 mM phosphate buffer at pH 6.3, with 10% D2O added for NMR spin-lock. For the samples used for collecting NMR spectra for structural calculation, 20 mM DTT was included to prevent the oxidation of the free cysteines, which was observed to trigger severe protein precipitation at a high protein concentration of ∼1 mM.
All heteronuclear NMR experiments used for assignments and structure determination were collected on an 800 MHz Bruker Avance spectrometer equipped with shielded cryoprobe at 25°C as previously described (Ran and Song 2005; Qin et al. 2008). The NMR spectra acquired for both backbone and side chain assignments included 15N- and 13C-edited HSQC-TOCSY, HSQC-NOESY, and HCCHTOCSY HCCHTOCSY, as well as triple-resonance experiments HNCACB, CBCA(CO)NH, HNCO, (H)CC(CO)NH, and H(CCO)NH. NOE restraints were derived from both 15N- and 13C-edited NOESY spectra.
For structure determination, a set of manually assigned unambiguous NOE restraints, dihedral angle restraints predicted by TALOS program based on five chemical shift values (15N, Cα, Cβ, CO, and Hα) was used to calculate initial structures of DCL4 by CYANA program as previously described (Ran and Song 2005). After several rounds of refinement, the final set of unambiguous NOE, dihedral angle restraints were input for structure determination by CYANA. The structures were displayed and manipulated by use of the graphic software MolMol and PyMol (DeLano Scientific LLC).
ITC characterization on the binding of DCL4 to RNA or Zn
Isothermal titration calorimetry (ITC) experiments were performed using a Microcal VP isothermal titration calorimetry machine (Qin et al. 2008). Titration was conducted in 10 mM phosphate buffer at pH 6.3, at 25°C. The powder of ZnCl2 or RNA was dissolved in 10 mM phosphate buffer with the final pH value adjusted to 6.3. The DCL4 protein was placed in a 1.8-mL sample cell, while ZnCl2 or RNA was loaded into a 300 μL syringe. The samples were degassed for 15 min to remove bubbles before the titration was initiated. Control experiment with the same parameter settings was also performed for ZnCl2 and RNA without DCL4, to subtract the effects resulting from dilution. To obtain thermodynamic binding parameters, the titration data after subtracting the values obtained from the control experiment were fit using the built-in software ORIGIN version 5.0 (Microcal Software, Inc.).
NMR characterization on the binding of DCL4 to RNA or Zn
To NMR characterize the binding of DCL4 to RNA or Zn, two-dimensional 1H-15N HSQC spectra were acquired on the 15N-labeled DCL4 at a protein concentration of 100 μM in the absence or presence of RNA at molar ratios: 1:2, 1:5, 1:7, 1:10, and 1:13 (DCL4/RNA), or in the absence or presence of ZnCl2 at molar ratios: 1:0.5, 1:1, 1:5, and 1:10 (DCL4: ZnCl2). By superimposing HSQC spectra, the shifted HSQC peaks could be identified and further mapped back to the corresponding residues on the DCL4 structure (Qin et al. 2008).
GST pull-down assay
One hundred micrograms of GST fused proteins were bound to glutathione sepharose beads (GE healthcare) in binding buffer containing 25 mM Tris at pH 7.4, 1 mM EDTA, 0.01% NP-40, and 2 M NaCl. After 2–4 h of incubation, the beads were washed with the same buffer to remove unbound proteins, and incubated with His6-tagged DCL1 DUF283 and DCL4 DUF283 proteins in binding buffer overnight at 4°C with rotation. The beads were washed 10–12 times in binding buffer. The bound proteins were eluted using 2X SDS PAGE loading dye at 100°C for 5–7 min, and detected by Western blot analysis using anti-poly-histidine monoclonal antibody (Sigma).
Accession code
The atomic coordinates for the NMR structures of the DUF283 domain have been deposited in the Protein Data Bank with the accession code 2KOU.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank N.-H. Chua at the Rockefeller University for the DCL1, DCL4, HYL1, and DRB4 cDNAs. This work was supported by Singapore Ministry of Education (T208A3124 to Y.A.Y.; R-154-000-388-112 to J.S.) and intramural research funds from Temasek Life Sciences Laboratory (Y.A.Y.). The authors declare that they have no competing financial interests.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1965310.
REFERENCES
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chen HY, Yang J, Lin C, Yuan YA. Structural basis for RNA-silencing suppression by Tomato aspermy virus protein 2b. EMBO Rep. 2008;9:754–760. doi: 10.1038/embor.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Watson JM, Smith NA, Eamens AL, Blanchard CL, Waterhouse PM. The roles of plant dsRNA-binding proteins in RNAi-like pathways. FEBS Lett. 2008;582:2753–2760. doi: 10.1016/j.febslet.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;7:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Dlakić M. DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold. Bioinformatics. 2006;22:2711–2714. doi: 10.1093/bioinformatics/btl468. [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Jantsch MF. New and old roles of the double-stranded RNA-binding domain. J Struct Biol. 2002;140:147–153. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J Mol Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon G. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008;283:29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran X, Song J. Structural insight into the binding diversity between the Tyr-phosphorylated human ephrinBs and Nck2 SH2 domain. J Biol Chem. 2005;280:19205–19212. doi: 10.1074/jbc.M500330200. [DOI] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA–protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Induction and suppression of RNA silencing: Insights from viral Infections. Nature. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]