Abstract
RNase MRP is a ribonucleoprotein endoribonuclease found in three cellular locations where distinct substrates are processed: the mitochondria, the nucleolus, and the cytoplasm. Cytoplasmic RNase MRP is the nucleolar enzyme that is transiently relocalized during mitosis. Nucleolar RNase MRP (NuMRP) was purified to homogeneity, and we extensively purified the mitochondrial RNase MRP (MtMRP) to a single RNA component identical to the NuMRP RNA. Although the protein components of the NuMRP were identified by mass spectrometry successfully, none of the known NuMRP proteins were found in the MtMRP preparation. Only trace amounts of the core NuMRP protein, Pop4, were detected in MtMRP by Western blot. In vitro activity of the two enzymes was compared. MtMRP cleaved only mitochondrial ORI5 substrate, while NuMRP cleaved all three substrates. However, the NuMRP enzyme cleaved the ORI5 substrate at sites different than the MtMRP enzyme. In addition, enzymatic differences in preferred ionic strength confirm these enzymes as distinct entities. Magnesium was found to be essential to both enzymes. We tested a number of reported inhibitors including puromycin, pentamidine, lithium, and pAp. Puromycin inhibition suggested that it binds directly to the MRP RNA, reaffirming the role of the RNA component in catalysis. In conclusion, our study confirms that the NuMRP and MtMRP enzymes are distinct entities with differing activities and protein components but a common RNA subunit, suggesting that the RNA must be playing a crucial role in catalytic activity.
Keywords: RNase MRP, ribonucleoprotein, endoribonuclease, mitochondrial DNA replication, rRNA processing, cell cycle
INTRODUCTION
RNase mitochondrial RNA processing (RNase MRP) is a ribonucleoprotein endoribonuclease. RNase MRP was initially isolated from mouse cell mitochondria, as an in vitro enzymatic activity that cleaves RNA transcripts complementary to the origin of replication consistent with it, forming the RNA primers for the leading strand of mitochondrial DNA replication (Chang and Clayton 1987; Chang et al. 1987). Despite its mitochondrial function, the majority of RNase MRP is found in the nucleolus (Reimer et al. 1988; Gold et al. 1989). In Saccharomyces cerevisiae, nucleolar RNase MRP (NuMRP) was shown to play a direct role in processing the rRNA precursor at the A3 site, leading to the generation of the mature 5′-end of the 5.8S rRNA (Schmitt and Clayton 1993; Chu et al. 1994; Lygerou et al. 1996; Cai et al. 2002).
During mitosis in S. cerevisiae, RNase MRP localizes to temporal asymmetric MRP (TAM) bodies where it is believed to cleave the mRNA for a B-type cyclin (Clb2) to promote the end of mitosis (Cai et al. 2002; Gill et al. 2006). Mutations in the gene for the RNA component of human RNase MRP cause the autosomal recessive genetic disease cartilage hair hypoplasia (Ridanpää et al. 2001), which is characterized by short-limb dwarfism, brittle sparse hair, and immunodeficiency. In addition, human RNase MRP mutations lead to changes in cyclin mRNA abundance and rRNA processing (Thiel et al. 2005, 2007).
The composition of the NuMRP has been well-established in S. cerevisiae where a complex containing a single RNA and at least 10 protein components has been identified (Schmitt and Clayton 1992; Salinas et al. 2005). Eight of the proteins (Pop1p, Pop3p, Pop4p, Pop5p, Pop6p, Pop7p, Pop8p, and Rpp1p) are shared with the yeast nucleolar RNase P (Chamberlain et al. 1998), a closely related ribonucleoprotein endoribonuclease that has multiple activities in the cell, including processing cytoplasmic tRNA precursors to generate mature 5′-termini. Two unique protein components of NuMRP are Snm1p and Rmp1p. Snm1p and Rmp1p bind to the RNase MRP RNA but not the RNase P RNA (Schmitt and Clayton 1994; Salinas et al. 2005). The RNA component of RNase MRP is structurally related to the RNA component of RNase P (Chamberlain et al. 1998). Though the RNase P RNA has been shown to be catalytically active, this has never been demonstrated for the RNase MRP RNA (Guerrier-Takada et al. 1983; Stohl and Clayton 1992; Pannucci et al. 1999; Kikovska et al. 2007).
Several in vitro assays have been established to test RNase MRP cleavage activity in S. cerevisiae which include characterized in vitro substrates for all three compartments (Stohl and Clayton 1992; Venema and Tollervey 1999; Cai and Schmitt 2001; Gill et al. 2004). With the availability of highly purified NuMRP and partially purified mitochondrial RNase MRP (MtMRP) we examined the biochemical characteristics and enzymatic behavior of these two enzymes. The results strongly support the hypothesis that NuMRP and MtMRP are different identities that possess a common enzymatic RNA but distinct protein components and enzymatic activities.
RESULTS
Protein composition analysis of MtMRP and NuMRP preparations
The S. cerevisiae NuMRP was purified to homogeneity using a tagged version of Pop4 and a modified tandem affinity purification (TAP) technique (see Materials and Methods). The NuMRP preparation contained a single RNA of 340 nucleotides (nt) encoded by the yeast NME1 gene, and 10 proteins (Salinas et al. 2005). The MtMRP was partially purified by conventional ion exchange chromatography and glycerol density-gradient centrifugation. Throughout the purification the enzyme was followed using cleavage of the mitochondrial substrate as an assay (Stohl and Clayton 1992; Cai and Schmitt 2001). The final preparation contained a single RNA of 340 nt that was the same RNA in the NuMRP preparation. Since both NuMRP and MtMRP preparations contained an identical RNA component, RNA levels were used to quantitate amounts of the two enzymes and allow direct comparison.
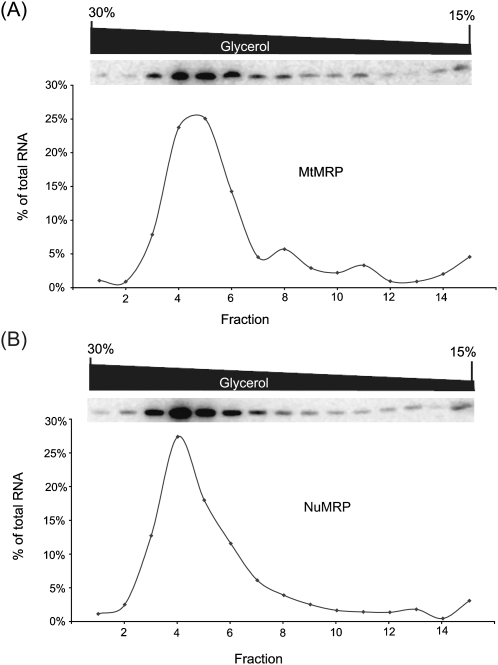
Differences in protein and RNA composition can lead to differences in the density of a ribonucleoprotein complex and a difference in its behavior by density gradient centrifugation. The purified RNase MRP preparations, each containing the same amount of the MRP RNA, were analyzed in parallel in a 15%–30% glycerol density-gradient. Gradients were centrifuged together and then divided into 15 fractions. The RNA levels in each fraction were analyzed by Northern analysis (Fig. 1). The RNA profiles were similar for both of the RNase MRP enzymes. The RNA peak was broader and consistently one fraction later in the MtMRP, but the difference was not significant. This indicates that the density of the two complexes is slightly different but comparable (Fig. 1).
FIGURE 1.
Glycerol gradient centrifugation of NuMRP and MtMRP. Purified NuMRP or MtMRP containing 50–80 ng of the MRP RNA were centrifuged in parallel on a 15%–30% glycerol density-gradient, and fractioned as described in the Material and Methods. RNA isolated from the gradient fractions and the MRP RNA was detected by Northern analysis. (A) MtMRP. (B) NuMRP.
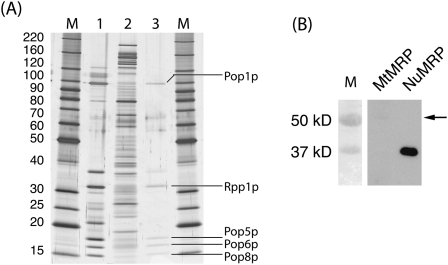
Proteins precipitated from both MtMRP and NuMRP samples containing the same amount of the MRP RNA were separated by SDS-PAGE, and visualized by MALDI compatible silver stain (Fig. 2A). The TAP-tag purified NuMRP prior to the glycerol gradient served as a control. Individual protein bands were isolated, digested in-gel by trypsin, and analyzed by mass spectrometry. All 10 of the previously identified NuMRP proteins were visualized and identified successfully in the control (Fig. 2A, lane 1). In the NuMRP fraction isolated by glycerol gradient centrifugation, the protein concentration was much lower, and only six of the 10 proteins could be visualized and five of these proteins were in a concentration high enough to be identified by mass spectrometry as Pop1p, Rpp1p, Pop5p, Pop6p, and Pop8p (Fig. 2A, lane 3). In the MtMRP fraction most of the major proteins have been successfully identified by mass spectrometry after in-gel trypsin digestion (Table 1). None of the protein components of the NuMRP were found in the MtMRP preparations (Fig. 2A, lane 2). Hence, none of the 10 NuMRP protein components appear to be associated with the MtMRP preparation. Table 1 provides a summary of potential MtMRP proteins identified in MtMRP fractions. Confirmation of any of these as genuine MtMRP proteins still requires further experimentation.
FIGURE 2.
Protein analysis of NuMRP and MtMRP. (A) Protein samples were separated on a 7.5%–17.5% SDS-PAGE gel and visualized using MALDI compatible silver staining. Protein components were excised, digested by trypsin, and analyzed by MALDI-TOF mass spectrometry (see Materials and Methods). M, molecular-weight markers. (Lane 1) 10 μL of TAP-tag purified NuMRP prior to glycerol gradient. All 10 NuMRP protein components were confirmed in this preparation (not shown). (Lane 2) Precipitated proteins from MtMRP containing 1000 picograms (pg) of the MRP RNA. (Lane 3) Precipitated proteins from NuMRP containing 1000 pg of the MRP RNA. Five of the 10 proteins successfully identified by mass spectrometry of this sample are indicated. (B) Proteins precipitated from both MtMRP and NuMRP containing 1000 pg of the MRP RNA were separated on a 12.5% SDS-PAGE gel, transferred to nylon membrane, and Pop4 was detected using anti-TAP antibody. Both preparations were made from the same yeast strain expressing a Pop4:TAP fusion as its only source of Pop4 protein. Part of the protein is specifically cleaved during the NuMRP purification giving a smaller protein, but leaving the antibody epitope. The arrow indicates the size of the full-length uncleaved protein in the MtMRP preparation.
TABLE 1.
Potential RNase MRP proteins that were identified in the MtMRP preparation
Proteins precipitated by TCA from both MtMRP and NuMRP samples with the same amounts of MRP RNA were analyzed by Western blot with anti-TAP-tag antibody (Fig. 2B); while Pop4 is clearly visible in NuMRP preparation, only a trace amount of Pop4 was detected in MtMRP (arrow in Fig. 2B). This result indicates that Pop4 is clearly not a protein component of MtMRP. A similar result was seen with an anti-peptide antibody against Rpp1 (data not shown).
Comparison of enzymatic cleavage on different substrates
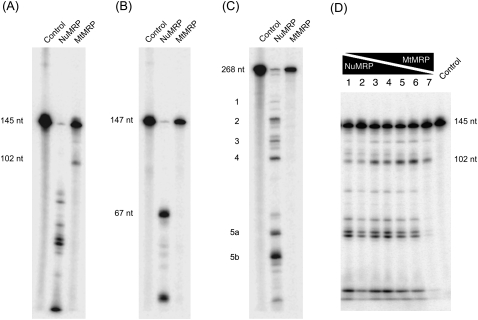
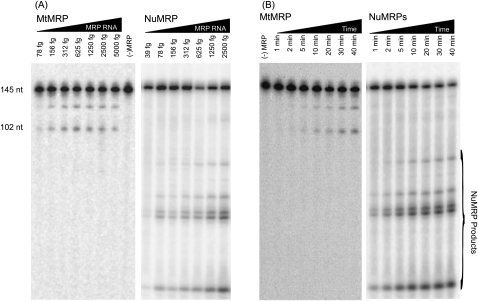
Since the two enzymes appeared to have very different protein compositions, we examined the differences of both nucleolar and MtMRP in substrate recognition and cleavage. The catalytic activities of the two enzymes were compared in vitro on three different substrates: the rRNA substrate, the CLB2 substrate, and the mitochondrial ORI5 substrate. The MtMRP cleaves the mitochondrial ORI5 substrate at the 5′-end of the GC cluster C/CSB II region, leaving a 3′-fragment of ∼102 nt; this is consistent with the reported cleavage pattern (Fig. 3A; Stohl and Clayton 1992; Cai and Schmitt 2001). However, MtMRP was able to cleave neither the rRNA substrate nor the CLB2 substrate when tested under a variety of conditions (Fig. 3B,C). NuMRP cleaved the 147-nt rRNA substrate at the A3 site, leaving a 67-nt 3′-fragment. When NuMRP was incubated with the 268-nt CLB2 substrate, multiple cleavage sites were seen, consistent with previous reports (Gill et al. 2004). NuMRP recognizes mitochondrial ORI5 substrate; however, it cleaves the mitochondrial substrate at multiple positions different than those cleaved by MtMRP, creating five major fragments on the gel. There is no corresponding 102-nt fragment generated by NuMRP even at lower enzyme concentrations (Fig. 4A). When MtMRP is mixed with NuMRP at various ratios, the mixed enzymes cleaved the mitochondrial substrate at both the MtMRP cleavage site and NuMRP cleavage sites. Indeed, cleavage at the mitochondrial site seems to preclude further cleavage by the NuMRP (Fig. 3D).
FIGURE 3.
In vitro cleavage assays of RNase MRP on three different substrates. RNA substrates 3′-end labeled with 32P were incubated with either NuMRP or MtMRP for 30 min at 37°C. The cleavage reactions were analyzed on a 6% polyacrylamide, 7 M urea gel as described in Materials and Methods. (A) Mitochondrial Ori5 substrate. (B) rRNA A3 substrate. (C) CLB2 mRNA substrate, (D) MtMRP and NuMRP were mixed together on the mitochondrial substrate at varying ratios.
FIGURE 4.
Enzyme titration and time course of RNase MRP cleavage by NuMRP and MtMRP. (A) The 32P-labeled mitochondrial substrate was incubated with increasing amounts of MtMRP or NuMRP for 30 min. Enzyme levels were quantitated by the MRP RNA levels, which are indicated. (B) The 32P-labeled mitochondrial substrate was incubated with the MtMRP or NuMRP for increasing amounts of time. The cleavage reactions were analyzed on a 6% polyacrylamide, 7 M urea gel as described in Materials and Methods.
Titration and time course on substrates
We also compared the two enzymes to see if their rate of catalysis was the same and to see if they would demonstrate comparable cleavages under higher enzyme concentrations and longer reaction times. To compare the cleavage abilities of MtMRP and NuMRP, both enzymes were serially diluted and incubated with substrate. Minimal cleavage activity (some detectable cleavage product) was seen in preparations containing 78 femtograms (fg) of RNase MRP RNA for MtMRP, and 39 fg of RNase MRP RNA for NuMRP. Quantitation of the amount of total products generated per fg of RNase MRP RNA (see Materials and Methods) indicated that NuMRP was about four times more active than the MtMRP enzyme (Fig. 4A). This is possibly the result of a partial inactivation during the purification process or the innate ability of the NuMRP to process more efficiently. The MtMRP purification is more complicated, but not considerably harsher than the NuMRP purification.
The RNA cleavage activity of both enzymes was time-dependent. The cleavage products could be seen starting from 1 min of incubation for NuMRP and 5 min for MtMRP. The concentration of cleavage products increased with the incubation time, and saturated after 10–30 min for NuMRP and MtMRP (Fig. 4B, and data not shown). With both enzymes, even at the highest enzyme concentrations and the longer reaction times, different cleavage products were observed. This result indicates that the NuMRP and MtMRP are well separated and that they are distinct enzymes.
Salt sensitivity
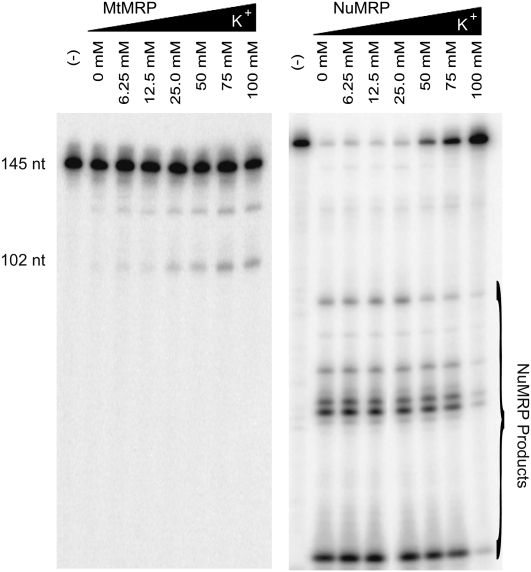
Differences in protein composition between the two enzymes may also lead to differences in optimal cleavage conditions. To examine this possibility we assayed MtMRP on mitochondrial ORI5 substrate and NuMRP on all three substrates at a variety of potassium and magnesium concentrations. The MtMRP activity on the mitochondrial substrate was both K+ and Mg2+ dependent, with the best cleavage activity observed at 75 mM K+ and 10 mM Mg2+ (Fig. 5). In contrast, NuMRP activity preferred no or low salt concentrations. Although Mg2+ is necessary, NuMRP catalytic activity is relatively stable in a broad range of Mg2+ concentrations, and K+ is not essential to NuMRP. This was found to be true for cleavage on all three substrates, indicating an enzyme preference as opposed to changes in substrate structure.
FIGURE 5.
Effect of ionic strength on NuMRP and MtMRP activity. The 32P-labeled mitochondrial ORI5 substrate was incubated with nuMRP and mtMRP for 30 min in buffers with different concentrations of K+ or Mg2+. Except for the indicated ion, standard reaction conditions were used. The cleavage reactions were analyzed on a 6% polyacrylamide, 7 M urea gel as described in Materials and Methods.
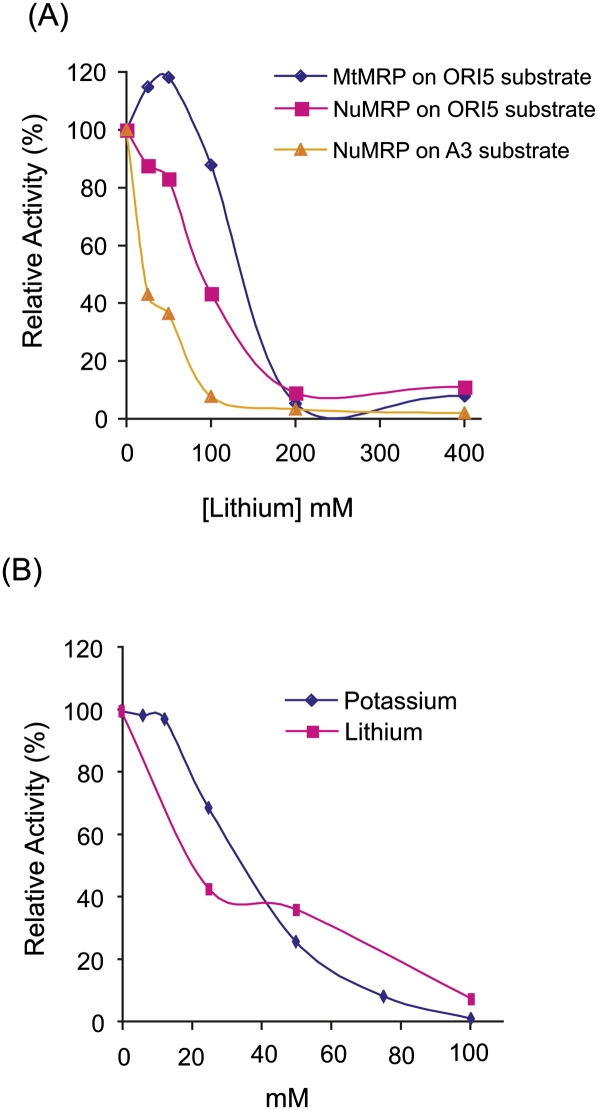
It has been reported that lithium directly inhibits RNase MRP cleavage at the A3 site of pre-rRNA in vivo (Dichtl et al. 1997), though it has never been shown directly in an in vitro assay. We examined whether lithium would inhibit our in vitro RNase MRP assays and whether or not the different enzymes might display different sensitivities. In our assays, Li+ does not significantly inhibit MtMRP cleavage of ORI5 substrate at 100 mM or less concentration, and does not significantly inhibit NuMRP cleavage of ORI5 substrate at 50 mM or less concentration (Fig. 6). This inhibition pattern is similar to the inhibition of RNase MRP activity in high K+ buffer, which indicates that at least in vitro inhibition by Li+ is caused by changes in the ionic strength of the assay buffer as opposed to specific inhibition of the enzyme (Fig. 6).
FIGURE 6.
Effect of lithium on RNase MRP catalytic activities. (A) The 32P-labeled substrate was incubated for 30 min with NuMRP or MtMRP under standard reaction conditions with increasing concentrations of lithium and either the ORI5 or A3 substrates. (B) Effect of lithium or potassium on cleavage of the A3 substrate by NuMRP. The cleavage activity without adding lithium or potassium is set as 100%.
Inhibitors of RNase MRP
Adenosine 3′, 5′-diphosphate (pAp) has been shown to be an inhibitor of a number of exonucleases including the 5′→3′ exonucleases Xrn1p and Rat1p (Dichtl et al. 1997). Xrn1 degrades the CLB2 mRNA after RNase MRP specifically cleaves its 5′-untranslated region (UTR). Our in vitro experiments showed that pAp did not inhibit RNase MRP cleavage activities on any of the three substrates (data not show).
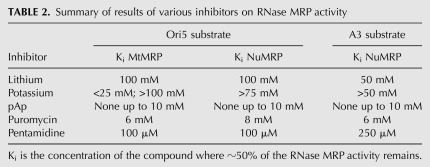
Puromycin, an antibiotic that inhibits protein synthesis, has been previously reported to inhibit mouse RNase P and mouse MtMRP (Vioque 1989; Potuschak et al. 1993). In our in vitro experiment, 6 mM puromycin inhibited approximately half of ORI5 substrate cleavage by MtMRP, and half of the ORI5 substrate cleavage activities by NuMRP were inhibited at 8 mM puromycin (Table 2). This is a concentration 1000-fold higher than that found to inhibit the ribosome (Starck and Roberts 2002), and equivalent to what was seen for the mouse RNase MRP and P enzymes (Potuschak et al. 1993). This confirms puromycin as an inhibitor of RNase MRP but revealed no significant enzymatic differences between the two enzymes. However, similar levels of inhibition of both enzymes by puromycin indicate that it binds the RNA component in order to inhibit activity, exemplifying the role of the RNA in catalytic activity.
TABLE 2.
Summary of results of various inhibitors on RNase MRP activity
Pentamidine, an RNA binding inhibitor of some ribonucleoproteins, was also tested (Sands et al. 1985; Liu et al. 1994; Sun and Zhang 2008). Depending on whether pentamidine binds to the RNA component of RNase MRP or the RNA substrates, it may have a different inhibitory effect on the mitochondrial and NuMRP. Our results showed that pentamidine inhibits substrates cleavage activities of both MtMRP and NuMRP. Pentamidine inhibits about half of ORI5 cleavage activities of both enzymes at the concentration around 100 μM; however, more than twice that amount is needed to inhibit the NuMRP from cleaving the rRNA substrate (Table 2). These results suggest that pentamidine binds to the substrate RNA to inhibit RNase MRP function.
DISCUSSION
RNase MRP is a site-specific ribonucleoprotein endoribonuclease with three distinct functions. These include cleaving RNA transcripts to generate primers for DNA replication in mitochondria (Chang and Clayton 1987; Chang et al. 1987), processing the A3 site of the rRNA precursor to form the 5.8S(S) ribosomal RNA in the nucleolus (Chu et al. 1994; Lygerou et al. 1996), and cleaving the 5′-UTR of CLB2 mRNA, which promotes its degradation at the end of mitosis (Gill et al. 2006). In S. cerevisiae, NuMRP has been well studied and it contains a single RNA and 10 protein components. However, little is known about the composition of the MtMRP, except that it has the same RNA component as the NuMRP. In this study, MtMRP was purified to a single RNA component using conventional biochemical techniques.
When analyzed in parallel by glycerol density gradient centrifugation, MtMRP consistently peaked at a slightly lower density fraction, suggesting its density is less than that of NuMRP. Interestingly, the MtMRP fractions did not contain any of the protein components of NuMRP. Indeed, we identified >50 proteins in these samples. More than 60% of the identified proteins were known to be localized to mitochondria or have a role in mitochondrial metabolism (Table 1). This was expected since the purification starts with a mitochondrial fractionation. Some of these proteins are known to be RNA binding proteins and may be genuine components of the MtMRP complex, though none of these have been directly confirmed yet as genuine components.
Western blot analysis on both NuMRP and MtMRP preparation with the same amount of RNase MRP RNA showed only a trace amount of Pop4 protein in MtMRP, <1% of the amount in NuMRP. Pop4 is a core component of the NuMRP and the nuclear RNase P enzymes. Indeed, addition of this protein is critical for reconstitution of the human RNase P activity (Mann et al. 2003). These results indicate that the protein compositions of NuMRP and MtMRP are different despite sharing the same RNA component. There are no reports of a mitochondrial RNA processing defect associated with any of the known NuMRP protein components. Indeed, our laboratory has directly looked with both NuMRP-specific proteins, Snm1 and Rmp1, and has found nothing (Cai et al. 1999; K Salinas and ME Schmitt, unpubl.). Though it is a negative result, it is still consistent with our findings in this paper. Interestingly, the RNase MRP RNA from a number of eukaryotes without mitochondria, such as the microsporidia, Encephalitozoon cuniculi, are missing a number of the conserved helices in what is believed to be the substrate specificity domain (Woodhams et al. 2007). This suggests that these may be binding platforms for mitochondrial specific proteins.
So far, three in vitro substrates of RNase MRP have been established in S. cerevisiae: the ORI5 substrate for the MtMRP (Cai and Schmitt 2001) and the A3 and CLB2 substrates for the NuMRP (Gill et al. 2004). In our results, MtMRP enzyme cleaves only the mitochondrial substrate while the NuMRP enzyme cleaves all three substrates. However, NuMRP cleaves the ORI5 substrate at multiple sites, different from MtMRP cleavage, generating five major fragments on the gel. In vitro cleavage study with mouse NuMRP on the mitochondrial substrate was reported previously (Karwan et al. 1991). In this particular case, the nucleolar enzyme was found to produce two additional cleavages to the one that was performed by the mitochondrial enzyme. Indeed, the mouse MtMRP may also have a very different protein composition than the NuMRP.
NuMRP and MtMRP were found to have different salt sensitivities. Potassium is essential to MtMRP cleavage activity with the optimum being at 75 mM. However, NuMRP cleaves ORI5, A3, and CLB2 substrate at highest efficiency without potassium in the reaction buffer and even shows partial inhibition at 75 mM KCl. These differences indicate distinct enzymatic difference between the enzymes and not changes in substrate folding.
Lithium has been shown to directly inhibit cleavage at the A3 site of the rRNA precursor in vivo (Dichtl et al. 1997). Since RNase MRP is responsible for that cleavage event it has been proposed that the enzyme is sensitive to lithium. Our in vitro experiments showed both NuMRP and MtMRP cleavage activity were decreased in high Li+ concentrations. NuMRP cleavage activity on the rRNA substrate was inhibited >60% in 50 mM Li+, and catalytic activity was completely inhibited when the Li+ concentration was >100 mM. However, variations in K+ concentration resulted in a similar pattern of inhibition. We conclude that Li+ inhibition of nuclear RNase MRP activity in vitro is linked to high monovalent ion concentration and not a specific inhibition. It is known that high Li+ concentrations directly result in inhibition of the MET22 gene product which is responsible for breaking down pAp, a known inhibitor both in vitro and in vivo of the Xrn1 and Rat1 exonucleases (Dichtl et al. 1997). We found that pAp has no direct inhibition on RNase MRP activity in our in vitro assay. However the Xrn1 and Rat1 exonucleases degrade RNase MRP cleavage products in vivo, inhibition of RNase MRP may be the indirect result of product inhibition. Indeed, we have seen strong product inhibition in our in vitro assays (data not shown). In vivo, this product inhibition must also be strong since reduction of cellular pAp by including methionine in the yeast media still leads to the accumulation of the RNase MRP rRNA substrate in the presence of Li+ (Dichtl et al. 1997).
Several lines of evidence from this study indicate that the MtMRP and NuMRP are distinct entities that contain the same RNA component. First the two enzymes have different protein components in their preparation. Second, the two enzymes have distinct substrate specificities and distinct activities on common substrates. Third the two enzymes have different ionic strength optima for catalysis, even on the same substrate. Despite these differences, both enzymes contain the same RNA component and are endoribonucleases. Both enzymes require magnesium and are inhibited by similar concentrations of puromycin. The differences and similarities indicate that the RNA component is the catalytic center that is shared by both enzymes. Roles of the protein components may be to promote the active structure of the RNA component, ensure its proper localization, and help provide substrate specificity.
MATERIALS AND METHODS
Strains and media
Yeast media and genetic manipulations have been described previously (Gill et al. 2004; Salinas et al. 2005). The YSW1 strain which contains a TAP fusion cassette inserted into the carboxy-terminal of the POP4 gene has the genotype MATa POP4∷TAPTAG∷TRP1ks pep4:LEU2 nuc1∷LEU2 sep1∷URA3 trp-1his3–11,15 can-100 ura3–1 leu2–3,112 and was used for both NuMRP and MtMRP purification.
RNase MRP purification
NuMRP was purified from strain YSW1 using a modified TAP protocol. Briefly, YSW1 cells were resuspended in buffer Z-150 (20 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsufonyl fluoride) and were disrupted by passing through a Microfluidics M-110L microfluidizer (Microfluidics) eight to 10 times. Broken cells were removed at 10,000g for 15 min, and the cell extract was clarified by centrifugation at 100,000g for 100 min. The clarified extract was loaded onto a UNOspere Q (Bio-Rad) column equilibrated with buffer Z-150. The column was washed in four column volumes of buffer Z-150 and proteins were eluted with buffer Z-350 (20 mM Tris-HCl [pH 8.0], 350 mM KCl, 1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsufonyl fluoride). The eluted fraction was then used for NuMRP purification using a TAP protocol previously described in detail (Gill et al. 2004). Addition of the UNOspere Q column removed any residual RNase P that may contaminate the purification (Lygerou et al. 1996).
MtMRP was purified from the same strain that was used for NuMRP preparation. The yeast strain YSW1 was grown in 12 L of YPD at 30°C to 2 × 108 cells/mL with vigorous aeration. Cells were harvested by centrifugation and washed with water. The cells were preincubated in three volumes of zymolyase preincubation buffer (0.1 M Tris-HCl [pH 9.3], 5 mM EDTA, 10 mM DTT) at 30°C for 30 min. Collected cells were washed with three volumes of zymolyase digestion buffer (0.02 M sodium phosphate [pH 7.4], 1.2 M sorbitol, 5 mM EDTA, 1 mM DTT), and then incubated in three volumes of zymolyase buffer with zymolyase 100T at 1 mg per mL of original packed cells at 30°C with gentle agitation. The spheroplasts were washed three times with three volumes of cold 1.2 M sorbitol-1 mM EDTA and resuspended in one volume of cold breaking buffer (10 mM Tris-HCl [pH7.5], 0.6 M Sorbitol, 1 mM EDTA, and 0.5 mM PMSF). Spheroplasts were broken in a Dounce homogenizer using 15 strokes, and cell debris was removed by centrifugation at 3000g for 5 min. Mitochondria were pelleted by centrifugation of the supernatant at 18,000 rpm for 15 min in a Beckman JA-20 rotor. The mitochondrial pellets were resuspended in breaking buffer, and the low-speed/high-speed spin cycle was repeated three more times. Mitochondrial pellets were resuspended at 10–20 mg/mL protein in mitochondria lysate buffer (20 mM Tris-HCl [pH 8.0], 1 mM EDTA, 1% Triton X-100, 0.2 M KCl, 10% glycerol, 1 mM PMSF, 1 mM DTT). The mitochondria were lysed by 15 strokes in a glass Dounce homogenizer. Mitochondrial crude extracts were dialyzed for 2 h against 2 L of buffer A-100 (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 5 mM EDTA, 10% Glycerol, 0.1% [v/v] Tween 20, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol). The cell extract was clarified by centrifugation at 100,000g for 100 min. Ion-exchange chromatography with DEAE-Sephacel (GE Healthcare Bio-Sciences) and Bio-Rex 70 (Bio-Rad) were performed. Eluate of Bio-Rex 70 was concentrated with a Centricon YM-100 ultrafiltration device, and could be stored at −70°C. The concentrated eluate from the Bio-Rex 70 was applied on a glycerol gradient (see below). The peak fractions with RNase MRP RNA were pooled for further analysis (Glick and Pon 1995; Cai and Schmitt 2001; Boldogh and Pon 2007).
Glycerol gradient centrifugation
For glycerol gradient centrifugation, 200 μL of the concentrated Bio-Rex70 eluate were loaded onto 15%–30% glycerol gradients in 20 mM Tris-HCl (pH 8.0), 50 mM KCI, 0.5 mM EDTA, and 1 mM DTT, and centrifuged for 13.5 h at 38,000 rpm in a Beckman SW55Ti rotor. Gradients were fractionated from the bottom using a peristaltic pump.
RNA analysis and quantitation
RNase MRP RNA was analyzed by denaturing polyacrylamide gel electrophoresis followed by ethidium bromide staining or by Northern analysis when the RNA concentration was low as described previously (Cai et al. 1999, 2002). The probe used for Northern analysis was a 440-base pair PCR fragment of NME1 DNA. The fluorescence intensity of ethidium bromide stained RNA or the intensity of the Northern blot hybridization signals were captured on a Typhoon-9410 Imager (GE Healthcare Bio-Sciences). RNA concentrations were calculated using ImageQuant 5.0 software (GE Healthcare). The known concentration of RNA transcripts generated from plasmid pMES206 served as a control and measure for quantitation. Transcription from this plasmid using the SP6 RNA polymerase can generate a nearly exact duplication of the endogenous RNase MRP RNA, except for the addition of a guanosine at the 5′-end and seven fewer nucleotides at the 3′-end.
In vitro RNA cleavage assay
RNA substrate preparation, 3′-end labeling, and in vitro RNA cleavage assays were carried out as previously described (Cai and Schmitt 2001; Gill et al. 2004). The 145-nt transcript from plasmid p64/HS40-ori5/4.4 served as the mitochondrial substrate (Cai and Schmitt 2001), the 147-nt transcript of pJA110 containing the A3 site served as the rRNA substrate, and the 268-nt RNA transcript of pJA108 containing the 5′-UTR of CLB2 served as CLB2 substrate (Gill et al. 2004). The results of all experiments were captured on a Typhoon-9410 Imager (GE Healthcare Bio-Sciences). This allowed for accurate quantitation of cleavage products. When more than one product was observed, the signal corresponding to all of the products was summed together to determine the total amount of product.
Protein analysis and identification
Proteins in RNase MRP samples containing ∼8 ng of RNase MRP RNA were precipitated with the addition of trichloroacetic acid (Brown et al. 1989). The pellets were resuspended in 15 μL of loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 10% glycerol, 0.1 M DTT, and 0.01% bromophenol blue), and proteins were separated on a 7.5%–17.5% gradient SDS-PAGE gel. Proteins were stained using a MALDI compatible silver stain (Mortz et al. 2001). Individual protein bands were excised from the gel using a fresh razor blade, the gel pieces were digested with trypsin and subjected to MADI-TOF mass spectrometry (Shevchenko et al. 1996). Mass spectrometry data were collected at the Proteomics Core Facilities at the State University of New York Upstate Medical University using a TOF Spec 2E mass spectrometer (Waters Corp.) or at the State University of New York at Oswego, Department of Biological Science using an AutoFlex II (Bruker BioSciences).
ACKNOWLEDGMENTS
We thank K. Salinas for comments on the manuscript and for helpful discussions during this investigation. This work was supported by grants GM063798 and GM064634 from the National Institute of General Medical Sciences to M.E.S., and by grant GM085149 to A.S.K.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1893710.
REFERENCES
- Boldogh IR, Pon LA. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2007;80:45–64. doi: 10.1016/S0091-679X(06)80002-6. [DOI] [PubMed] [Google Scholar]
- Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: Elimination of interfering substances. Anal Biochem. 1989;180:136–139. doi: 10.1016/0003-2697(89)90101-2. [DOI] [PubMed] [Google Scholar]
- Cai T, Schmitt ME. Characterization of ribonuclease MRP function. Methods Enzymol. 2001;342:135–142. doi: 10.1016/s0076-6879(01)42541-9. [DOI] [PubMed] [Google Scholar]
- Cai T, Reilly TR, Cerio M, Schmitt ME. Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol Cell Biol. 1999;19:7857–7869. doi: 10.1128/mcb.19.11.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161:1029–1042. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes & Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang DD, Fisher RP, Clayton DA. Roles for a promoter and RNA processing in the synthesis of mitochondrial displacement-loop strands. Biochim Biophys Acta. 1987;909:85–91. doi: 10.1016/0167-4781(87)90029-7. [DOI] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Stevens A, Tollervey D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997;16:7184–7195. doi: 10.1093/emboj/16.23.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol Cell Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill T, Aulds J, Schmitt ME. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Gold HA, Topper JN, Clayton DA, Craft J. The RNA processing enzyme RNase MRP is identical to the Th RNP and related to RNase P. Science. 1989;245:1377–1380. doi: 10.1126/science.2476849. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene Ontology annotations at SGD: New data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwan R, Bennett JL, Clayton DA. Nuclear RNase MRP processes RNA at multiple discrete sites: Interaction with an upstream G box is required for subsequent downstream cleavages. Genes & Dev. 1991;5:1264–1276. doi: 10.1101/gad.5.7.1264. [DOI] [PubMed] [Google Scholar]
- Kikovska E, Svard SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc Natl Acad Sci. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tidwell RR, Leibowitz MJ. Inhibition of in vitro splicing of a group I intron of Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:31–38. doi: 10.1111/j.1550-7408.1994.tb05931.x. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: Role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Mortz E, Krogh TN, Vorum H, Gorg A. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics. 2001;1:1359–1363. doi: 10.1002/1615-9861(200111)1:11<1359::AID-PROT1359>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Rossmanith W, Karwan R. RNase MRP and RNase P share a common substrate. Nucleic Acids Res. 1993;21:3239–3243. doi: 10.1093/nar/21.14.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Ridanpää M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage–hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- Sands M, Kron MA, Brown RB. Pentamidine: A review. Rev Infect Dis. 1985;7:625–634. doi: 10.1093/clinids/7.5.625. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes & Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Characterization of a unique protein component of yeast RNase MRP: An RNA-binding protein with a zinc-cluster domain. Genes & Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Starck SR, Roberts RW. Puromycin oligonucleotides reveal steric restrictions for ribosome entry and multiple modes of translation inhibition. RNA. 2002;8:890–903. doi: 10.1017/s1355838202022069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl LL, Clayton DA. Saccharomyces cerevisiae contains an RNase MRP that cleaves at a conserved mitochondrial RNA sequence implicated in replication priming. Mol Cell Biol. 1992;12:2561–2569. doi: 10.1128/mcb.12.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Zhang Y. Pentamidine binds to tRNA through nonspecific hydrophobic interactions and inhibits aminoacylation and translation. Nucleic Acids Res. 2008;36:1654–1664. doi: 10.1093/nar/gkm1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Ruschendorf F, Sticht H, Spranger J, Muller D, et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am J Hum Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel CT, Mortier G, Kaitila I, Reis A, Rauch A. Type and level of RMRP functional impairment predicts phenotype in the cartilage hair hypoplasia-anauxetic dysplasia spectrum. Am J Hum Genet. 2007;81:519–529. doi: 10.1086/521034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Vioque A. Protein synthesis inhibitors and catalytic RNA. Effect of puromycin on tRNA precursor processing by the RNA component of Escherichia coli RNase P. FEBS Lett. 1989;246:137–139. doi: 10.1016/0014-5793(89)80269-8. [DOI] [PubMed] [Google Scholar]
- Woodhams MD, Stadler PF, Penny D, Collins LJ. RNase MRP and the RNA processing cascade in the eukaryotic ancestor. BMC Evol Biol. 2007;7(Suppl 1):S1–S13. doi: 10.1186/1471-2148-7-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]