Abstract
The Escherichia coli endoribonuclease RNase E is an essential enzyme having key roles in mRNA turnover and the processing of several structured RNA precursors, and it provides the scaffold to assemble the multienzyme RNA degradosome. The activity of RNase E is inhibited by the protein RraA, which can interact with the ribonuclease's degradosome-scaffolding domain. Here, we report that RraA can bind to the RNA helicase component of the degradosome (RhlB) and the two RNA-binding sites in the degradosome-scaffolding domain of RNase E. In the presence of ATP, the helicase can facilitate the exchange of RraA for RNA stably bound to the degradosome. Our data suggest that RraA can affect multiple components of the RNA degradosome in a dynamic, energy-dependent equilibrium. The multidentate interactions of RraA impede the RNA-binding and ribonuclease activities of the degradosome and may result in complex modulation and rerouting of degradosome activity.
Keywords: RNA degradosome, DEAD-box helicase, RNA degradation, RraA, natively unstructured proteins
INTRODUCTION
The degradation of messenger RNA affects the abundance of transcripts available for translation and, consequently, the level of the protein products. Accordingly, processes that impact on the rates of RNA turnover contribute to the regulation of gene expression (Grunberg-Manago 1999; Arraiano and Maquat 2003; Wilusz and Wilusz 2004). The capacity to adjust the stability of individual transcripts rapidly enables efficacious responses to a changing environment or the timely progression of developmental pathways. Amongst the bacteria, there are many enzymes that affect RNA stability with impact on genetic regulation, and these include endo- and exoribonucleases, RNA helicases, poly(A) polymerase and RNA pyrophosphohydrolase (RppH), and other RNA-associated proteins, such as the RNA chaperone Hfq (Morita et al. 2008; Vogel 2009).
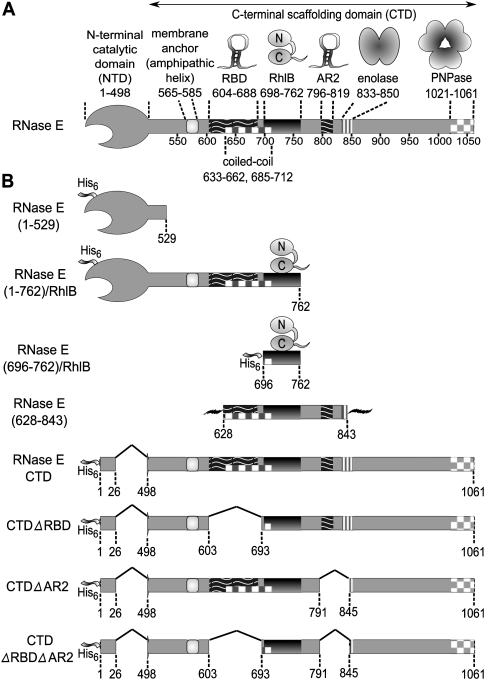
In Escherichia coli, the ribonuclease E (RNase E) is the major endonuclease of mRNA turnover, and it also serves a second key function as a processor of precursors of structured RNA species, such as ribosomal RNA, tRNA, 6S, and tmRNA (Carpousis et al. 2009). RNase E of E. coli and other γ-proteobacteria is a comparatively large protein, consisting of a little more than 1000 amino acids (Fig. 1A). The highly conserved N-terminal half of RNase E encompasses its hydrolytic ribonuclease activity, while the much more variable and mostly unstructured C-terminal half mediates the protein–protein interactions that underpin the assembly of the multienzyme RNA degradosome (Callaghan et al. 2004). The principal components of the degradosome are the phosphorolytic 3′exoribonuclease, polynucleotide phosphorylase (PNPase), the ATP-dependent DEAD-box RNA helicase RhlB (E.C. 3.6.1), and the glycolytic enzyme, enolase (E.C. 4.2.1.11) (Py et al. 1996; Marcaida et al. 2006; Carpousis 2007). The degradosome scaffolding region also contains two RNA-binding sites, RBD and AR2 (Fig. 1A). Variable components are recruited to the degradosome in substoichiometric quantities, such as polyphosphate kinase (Carpousis 2007). The RNA chaperone Hfq also interacts with RNase E and recruits small regulatory RNAs to direct cleavage of specific transcripts (Morita et al. 2005, 2008). The physical association of the components of the degradosome permits their activities to cooperate and to be potentially coordinated. For instance, PNPase and helicase cooperate with RNase E to degrade structured transcripts (Coburn et al. 1999). In this regard, the role of enolase in degradosome function is less clear, but may be related to control mechanisms involving small regulatory RNAs in phosphosugar stress (Morita et al. 2004). Because it impacts globally on transcripts encoding diverse enzymes (Bernstein et al. 2004), the degradosome may be considered as a regulatory hub in E. coli and other γ-proteobacteria, such as Salmonella sp. Given this central role, it may seem very surprising that the degradosome is not widely conserved amongst the eubacteria, and it is not essential for survival in E. coli. Nonetheless, loss of degradosome is inferred to compromise fitness (Kido et al. 1996; Vanzo et al. 1998; Briegel et al. 2006). Taken together, the apparently contradictory aspects of the degradosome suggest that it is a comparatively recent and specialized adaptation that has become assimilated in important regulatory networks (Marcaida et al. 2006).
FIGURE 1.
The organization of protein and RNA-binding sites in RNase E that define the canonical RNA degradosome, and a description of the derived constructs used in this study. (A) Schematic representation of the RNase E scaffolding domain, which brings together degradosome components. (B) The expression constructs used to prepare recombinant RNase E and RNase E/RhlB complexes. The His6 ribbon represents a hexa-histidine affinity tag. The jagged lines flanking RNase E(628–843) construct represent non-RNase E peptide sequences derived from its expression vector (a 13-residue leader and a 20-residue tail).
In contrast to the nonessentiality of the degradosome, the catalytic domain of RNase E is absolutely required for survival of E. coli. In accordance with its vital role, the activity of the RNase E is regulated by a number of mechanisms, including autoregulation through interaction with the 5′ untranslated region of its own transcript (Jiang et al. 2000; Schuck et al. 2009). Another regulatory mechanism potentially involves repression of RNase E catalytic activity by the regulators, RraA and RraB (Lee et al. 2003; Gao et al. 2006; Yeom et al. 2008a,b). The interactions of RraA with RNase E affect the composition of the degradosome and therefore modulate its activity. One surprising finding is that the inhibitory effect of RraA on the hydrolytic activity of RNase E requires the noncatalytic C-terminal portion of RNase E, and it has been proposed that RraA binds a discontinuous region of the C-terminal domain (Lee et al. 2003; Gao et al. 2006). However, it is unclear how this might communicate to the catalytic domain. Furthermore, RraA does not itself bind RNA, so it is not obvious how it might impede the activity of RNase E.
We have investigated the interaction of RraA with RNase E in the context of the degradosome and its subassemblies. We present evidence that RraA binds to and masks the RNA-binding domains within the C-terminal domain of RNase E. RraA also binds to the basic C-terminal extension of RhlB, which is required for RNA binding by the helicase (Chandran et al. 2007). By occluding these binding sites in the degradosome, RraA represses the activity of the helicase and, indirectly, PNPase. The RNA-binding sites of the degradosome are natively unstructured and positively charged, and we explain how these may occupy negatively charged surface grooves of RraA. We discuss the implications of these findings for a model in which RraA can modulate and reroute the RNA degradosome, with global as well as specific impact on gene expression.
RESULTS
RraA interacts with the RNA-binding sites in the degradosome-organizing domain of RNase E
To identify interaction sites of RraA within the degradosome, we have used various recombinant constructs of portions of RNase E, some of which were coexpressed and copurified with the RhlB (Fig. 1B). Using surface plasmon resonance, we observed a very weak interaction between RraA and the catalytic domain of RNase E (1–529) (data not shown), in agreement with earlier findings (Lee et al. 2003).
We next explored interactions with the degradosome-organizing domain of RNase E. Highly purified recombinant RraA was found to form a stable complex with RNase E(628–843), corresponding to a fragment of the C-terminal domain that encompasses the RNA-binding domain AR2 and a portion of RBD, and the helicase binding site (see schematic in Fig. 1A). The stable complex could be resolved by electrophoretic mobility shift assays (EMSA) compared with the free components (Fig. 2A). The EMSA conditions used do not provide quantitative binding constants, but the results do suggest that a stable complex can form with stoichiometric subunit composition. The protein–protein interaction was also confirmed by cross-linking using the bifunctional cross-linking agent ANB-NOS (Supplemental Fig. 1A). Using mass spectrometry to analyze the putative intact complex, we observed signals from complexes mostly representing one or two RNase E (628–843) peptides with two RraA trimers (Supplemental Fig. 2A,B). We observe dimers of RraA trimers in crystals of RraA grown under different conditions (data not shown), so the hexamer may be a biologically relevant oligomer of RraA. The observed interaction of RraA with RNase E(628–843) narrows down by half the region previously reported to interact with RraA (Lee et al. 2003).
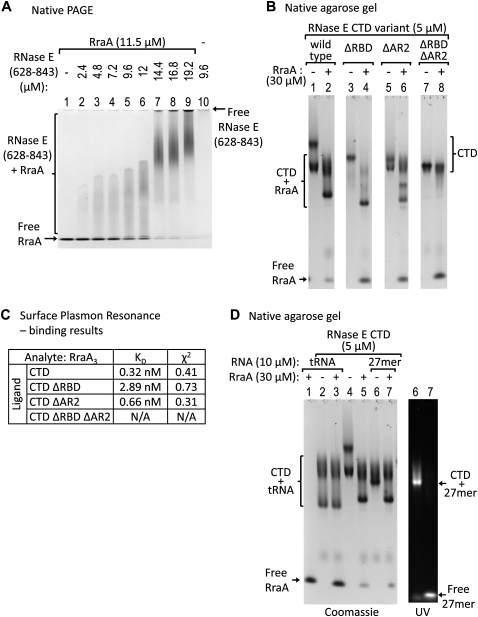
FIGURE 2.
RraA association with RNase E and competition with RNA binding. (A) Increasing amounts of RNase E(628–843) were mixed with the same amount of RraA and analyzed on a polyacrylamide gel under native conditions. Upon addition of RNase E(628–843), a complex with RraA is formed, while the amounts of free RraA decrease. RNase E(628–843) alone does not migrate into the gel due to its positive charge. (B) Recombinant constructs of RNase E CTD were incubated with RraA and analyzed on a native agarose gel. CTD constructs containing at least one of the RNA-binding sites interact with RraA (lanes 2,4,6), but not the construct lacking both the RNA-binding sites: CTDΔRBDΔAR2 (lane 8). The multiple bands are probably due to oligomerization of RNase E CTD, possibly through the RBD region. (C) Analysis of RraA binding to CTD and its deletion constructs from surface plasmon resonance. The kinetic profiles of RraA and CTD, RraA and CTDΔRBD, RraA and CTDΔAR2 interactions fit a 1:1 binding model (see Supplementary material). The affinities were calculated for a RraA trimer. (D) RNase E CTD was first incubated with fluorescein-tagged oligonucleotide (27 mer) or with tRNA, then mixed with RraA and analyzed on a native agarose gel. (Right) UV-fluorescence signal from the 27-mer RNA. CTD binds both tRNA (lane 2) and the 27 mer (lane 6). Upon addition of RraA, the complex of RNase E CTD with tRNA remains unaffected (lane 3). In contrast, in the presence of RraA the 27-mer oligonucleotide is released from RNase E CTD (lane 7) and is replaced by RraA (identical protein band pattern in lanes 5 and 7). B and D are images of the same gel, for clarity shown separately.
To refine the location of the interaction sites of RraA, we tested RNase E C-terminal domain (CTD) proteins having deletions of either one or both RBD and AR2 (Fig. 2B). Using EMSA, RraA was found to change the mobility of the wild-type RNase E CTD (Fig. 2B, cf. lanes 1 and 2) and of the constructs lacking either RBD (Fig. 2B, cf. lanes 3 and 4) or AR2 (Fig. 2B, cf. lanes 5 and 6), but it did not affect the mobility of the CTD containing deletions of both RNA-binding sites (Fig. 2B, cf. lanes 7 and 8). The results from EMSA were corroborated and quantified by surface plasmon resonance binding profiles, which can be modeled as association of one RraA trimer to one RNase E, with estimated KD in the nanomolar concentration range (tabulated in Fig. 2C; experimental profiles are shown in Supplemental Fig. 3). The binding constants are stronger than reported earlier using surface plasmon resonance, but in the earlier studies the RNase E was immobilized by nonspecific amine coupling, which may mask some of the potential binding sites (Lee et al. 2003). The constructs we used are immobilized with an N-terminal affinity tag, which would minimize masking effects.
Since RraA can associate with the RNA-binding sites within the RNase E C-terminal domain, we next explored whether RraA might interfere with RNA interaction. The CTD of RNase E was able to bind a fluorescein-labeled 27-mer single-stranded RNA by EMSA, but upon addition of stoichiometric quantities of RraA, this RNA was displaced (Fig. 2D). We also observe that RraA can displace 27-mer RNA bound to the CTDΔAR2 and CTDΔRBD proteins (data not shown). These results suggest that RraA can interfere with RNA binding by either or both of the RNA-binding sites. However, under the same conditions, a complex of CTD and tRNA was mostly unaffected by RraA (Fig. 2D). The interaction with tRNA could be disrupted using a much greater excess of RraA (data not shown). We suggest that the tRNA is less effectively displaced by RraA compared with the 27-mer RNA because it is larger and more structured and thus likely to make more extensive interactions with the RNase E RNA-binding domains. Similarly, Lee et al. (2003) observed that RraA is not effective at disrupting RNase E binding to the much larger pM1 RNA substrate.
RraA forms a stable ternary complex with RNase E and helicase RhlB, in which all components can interact
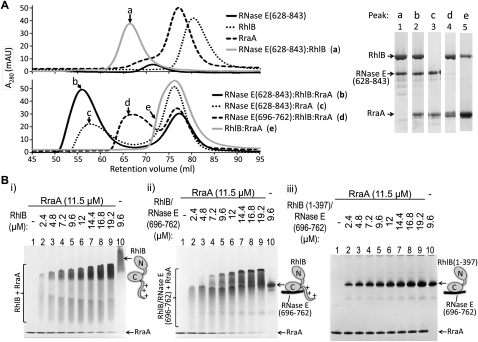
RhlB helicase binds strongly to the isolated 69-residue segment of RNase E that lies between the RBD and AR2 sites (KD of ∼10 nM) (Worrall et al. 2008b). We tested whether RraA would bind to the stable binary complex of RhlB and RNase E(628–843), which encompasses the 69-residue helicase binding site and the flanking RNA-binding regions that were shown to be required for RraA binding in the previous section (Fig. 2B). A stable ternary assembly comprising RNase E(628–843), RhlB, and RraA could be isolated by size-exclusion chromatography (Fig. 3A) and was also seen by EMSA (Supplemental Fig. 4A,B). Thus, RraA does not appear to displace the helicase from the RNase E segment, so it seems likely that the RraA interactions with RNase E(628–843) are mediated predominantly through the two RNA-binding sites and not the helicase-binding site. Using surface plasmon resonance, RraA was found to bind readily to larger complexes such as RNase E(1–762)/RhlB, RNase E CTD/RhlB complexes, as well as the complete recombinant degradosome (data not shown). A competitive effect of RraA for RNA binding was observed for the complex of RNase E(628–843) with RhlB and for recombinant degradosome (Supplemental Fig. 5).
FIGURE 3.
Interaction of RraA with RhlB and its complexes with RNase E. (A) Protein complexes containing RhlB, RNase E peptides, and RraA were analyzed by size-exclusion chromatography. (Left) Overlaid elution profiles from S200 column (the void volume of the column is 45 mL). (Right) SDS-PAGE of selected peak fractions (a–e) showing the composition of the complexes. A 4%–12% gradient PAA gel did not allow for separation of RNase E(696–762) from RraA band (lane 4), but the same samples analyzed subsequently on 4%–20% gradient PAA gel show that both components are present in complex with RhlB (data not shown). (B) Binding of RraA to RhlB was tested in titration electrophoretic mobility shift assays. The same amount of RraA was mixed with increasing amounts of partner protein and analyzed on a polyacrylamide gel under native conditions. RraA was able to change mobility of RhlB (i) or RhlB in complex with its recognition peptide from RNase E, RNase E(696–762) (ii), but no change was observed for RNase E(696–762)/RhlB(1–397), which lacks the C-terminal extension of the helicase (iii).
In the course of evaluating RraA binding to the binary RNase E/RhlB complexes, we discovered that RraA also directly binds RhlB. Evidence for the complex was obtained from size-exclusion chromatography (Fig. 3A), from EMSA (Fig. 3B, “i,” “ii”) and from chemical cross-linking (Supplementary materials Fig. 1B). Using surface plasmon resonance, the preliminary KD for the RhlB/RraA interaction is estimated to be 80 μM (data not shown), so it represents a weak macromolecular interaction. The KD for the RraA/RhlB interaction was estimated using RhlB immobilized to the sensor chip surface by amine coupling, which may mask binding interactions and could therefore potentially underestimate the binding strength. The physical interaction of RhlB with RraA is also suggested by low-resolution molecular envelopes obtained using Small Angle X-ray Scattering (Supplemental Fig. 6). Deletion of the highly basic C-terminal tail of RhlB (residues 398–421) abolishes its binding to RraA in the EMSA, suggesting that the tail is important for the interaction (Fig. 3B, “iii”).
The question arises whether the pairwise interactions of RraA with the CTD and with RhlB can also occur in the context of the ternary complex of RhlB–RraA–RNase E. Using surface plasmon resonance, we observe similar binding affinities of RraA for the RhlB–RNase E complex in comparison with the binding to the isolated CTD (data not shown), suggesting that binding of RraA at the three potential contact sites are not mutually exclusive or negatively cooperative. Whether they are simultaneous and multidentate remains to be established. Nonetheless, our data suggest that RraA can interact with three possible binding sites in the subassembly of the degradosome comprising the helicase and two flanking RNA-binding sites from RNase E.
SrmB and RhlE are two other E. coli RNA helicases from the DEAD-box family that are known to associate with RNase E and to functionally replace RhlB under certain growth conditions (Khemici et al. 2004; Prud'homme-Généreux et al. 2004). We observe that both SrmB and RhlE can form a stable complex with RraA (Supplemental Fig. 4C–E). However, these helicases may interact with RraA in a different way from RhlB. For example, while the C-terminal tail of RhlB is required for binding RraA, the RhlE C-terminus (residues 392–453), which has a similar overall basic charge, is not required (Supplemental Fig. 4E). It is possible that RraA interacts with the body of the three helicases to different extents.
RraA affects the ATPase activities of RhlB and other E. coli DEAD-box helicases
To explore whether the physical interaction with RraA affects the ATPase activity of RhlB, we monitored phosphate release accompanying ATP turnover. As RhlB has poor ATPase activity in the absence of RNase E and RNA (Worrall et al. 2008b), we used for the assay the avid complex of RhlB with its 69-residue recognition segment of RNase E that lacks the RNA-binding segments [RNase E(696–762)]. The ATPase activity of the RhlB/RNase E(696–762) complex was stimulated slightly by RraA in the absence of RNA substrate (see the table in Fig. 4A). It is possible that the interaction of RraA with the C-terminal tail of RhlB mimics the binding of RNA and exerts a similar, but lesser stimulating effect. However, in the presence of yeast bulk RNA, the activity was inhibited in a dose-dependent manner (Fig. 4A, left). Unlike RhlB, the DEAD-box helicases SrmB and RhlE are inhibited by RraA regardless of the presence of RNA (data not shown), which suggests a different mode of interaction and inhibition of those helicases.
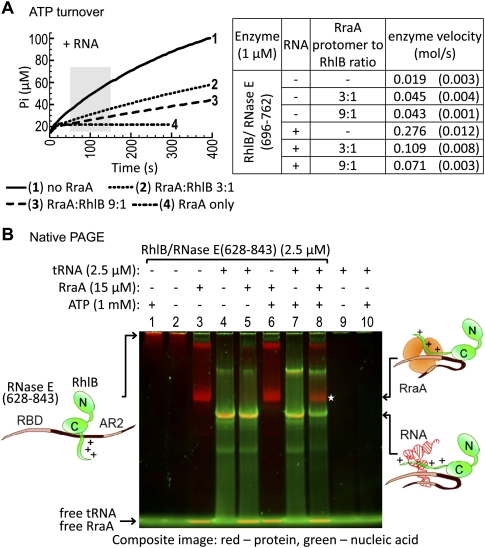
FIGURE 4.
Effect of RraA on the ATPase activity of RhlB and ATP-driven remodeling of RNase E/RhlB complexes by RhlB. (A) Effect of RraA on ATPase activity of RhlB in complex with its activator peptide RNase E(696–762). Phosphate release was monitored in the presence and absence of S. cerevisiae bulk RNA, at various RraA to RhlB molar ratios. The ATPase activity of RhlB was inhibited by RraA in the presence of RNA (left), but stimulated in the absence of RNA (see table). Enzyme velocity was calculated from the 100-sec period between the first 50 and 150 sec of the reaction (shaded zone), as the mole of phosphate produced per second per mole of enzyme. The curves, average velocities, and their standard deviations (in parentheses) were calculated from three reactions each, except the RraA controls. (B) The composite image is an overlay of Coomassie protein staining (red) and SYBR Gold staining of nucleic acid (green). RNase E(628–843)/RhlB complex was premixed with tRNA, incubated with RraA, and supplemented with ATP where indicated. After a short incubation period, the reactions were analyzed by native PAGE. The RNase E(628–843)/RhlB/tRNA complex is stable in the presence of RraA (lane 5). However, upon addition of ATP, tRNA is released from the protein complex and replaced by RraA [*, lane 8, compare with identical complex of RNase E(628–843)/RhlB with RraA in lane 3]. Free RraA and tRNA comigrate with the buffer/dye front and, hence, RraA appears yellow, though there is no evidence for any interaction of RraA with RNA.
RraA binding to the degradosome is facilitated by protein–RNA remodeling through RhlB
The ATPase activity of RhlB can remodel structured RNA or protein–RNA complexes to present the nucleic acid as substrate for the ribonucleases of the degradosome (Coburn et al. 1999). We explored whether the ATPase activity of RhlB affects the recruitment of RraA in the presence of RNA using EMSA. The experimental results are shown in Figure 4B as a composite image of a polyacrylamide gel that was sequentially stained for RNA (green) and protein (red); in this overlay, putative protein–nucleic acid complexes are yellow. The RhlB/RNase E (628–843) binary complex will form a ternary complex with E. coli bulk tRNA (Fig. 4B, lane 4, yellow band) and this complex is not apparently displaced in the presence of ATP (Fig. 4B, lane 7). The RNase E (628–843)/RhlB/tRNA ternary protein–RNA complex is only slightly affected by the presence of RraA (Fig. 4B, lane 5). However, the addition of ATP triggers the release of the bound RNA and formation of an RNase E(628–843)/RhlB/RraA ternary complex (Fig. 4B, lane 8). It is interesting to note that the ATP-driven activity of RhlB cannot be as easily detected by EMSA in the absence of RraA, perhaps because the protein complex rapidly rebinds the released RNA in a dynamic equilibrium. Thus, the presence of ATP catalyzes the remodeling of the protein–RNA complex, so that RNA-binding sites become available for interaction and the RNase E/RhlB assembly can be trapped in complex with RraA.
The same nucleotide- and RraA-dependent effect can be observed for RNase E(628–843)/RhlB(1–397), which lacks the basic C-terminal extension of the helicase, but has the two RNA-binding sites in RNase E, and again for the RNase E(696–762)/RhlB complex, which lacks the two RNA-binding sites from RNase E, but has the RNA-binding basic-tail of the helicase. These results suggest that either of the RNA-binding regions of RNase E or the C-terminal extension of RhlB are sufficient for remodeling (data not shown).
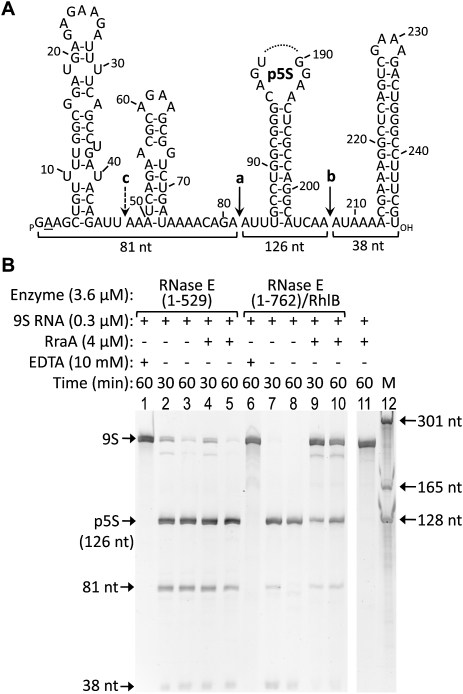
RraA inhibits processing of 9S RNA by RNase E
RNase E catalytic domain, corresponding to residues 1–529, and RNase E(1–762)/RhlB, which encompasses the RBD, were used to test the effect of RraA on processing of the 9S precursor of 5S rRNA by RNase E (Fig. 5A). Stoichiometric amounts of RraA significantly inhibited RNase E(1–762)/RhlB (Fig. 5B, cf. lanes 9,10 and control lanes 7,8), while the activity of RNase E(1–529) was not detectably affected (Fig. 5B, cf. compare lanes 2,3 and 4,5). As shown previously for the processing of the M1 precursor RNA by RNase E (Lee et al. 2003), inhibition of the activity of the isolated catalytic domain could be achieved only with 30-fold molar excess of RraA (data not shown). The results in Figure 5B show that the presence of RhlB and RNase E RBD, which both provide interactions with RraA, were sufficient to promote the inhibitory effect on the catalytic domain of RNase E. The recombinant degradosome containing RNase E, RhlB, enolase, and PNPase was similarly inhibited by RraA, though the result is less clear due to the copurifying contaminant RNA (data not shown).
FIGURE 5.
RraA inhibits 9S RNA processing by RNase E. (A) Secondary structure of 9S RNA, a precursor of 5S rRNA. Indicated are RNase E cleavage sites (a-b, and a minor cleavage site c). Part of the processing product, p5S, was omitted. RNA used in this study has a G instead of an A at the +2 position (underlined). (B) 9S RNA was incubated for the indicated time with RNase E constructs in the presence and absence of RraA. Products of the reactions were analyzed by denaturing PAGE and detected using SYBR Gold. RNase E processes the 9S substrate correctly to the p5S precursor of 5S ribosomal RNA. RraA inhibits the reaction significantly in the case of the degradosome sub-assembly RNase E(1–762)/RhlB (lanes 9,10), but not if only the catalytic domain of RNase E is present (lanes 4,5).
DISCUSSION
In this study, we have explored the interactions of RraA with RNase E and its subassemblies from the RNA degradosome. Our results suggest that RraA can make complex, multidentate interactions with discontinuous sites in the RNA degradosome. Using deletion analyses, we have mapped the principal sites to the two RNA-binding elements of the RNase E C-terminal half, namely the RNA-binding domain (RBD) and the arginine-rich region 2 (AR2) (see Fig. 1A). We have mapped a third binding site of RraA to the C-terminal tail of the ATP-dependent RNA helicase of the RNA degradosome, RhlB. Although these sites interact independently with RraA, it is expected that the three sites could, in principle, cooperate and boost each other's apparent affinities due to their spatial colocalization. Our observation of multiple interaction sites for RraA in the degradosome may account for earlier findings that RraA bound to all deletion mutants of the RNase E CTD (Gao et al. 2006). The association of RraA with the RNA-binding site in the CTD of RNase E interferes with its interactions with tRNA or a 27-mer single-stranded RNA. A similar masking effect has been reported for the L4 ribosomal protein on RNase E, which may occlude the RNA-binding site (Singh et al. 2009).
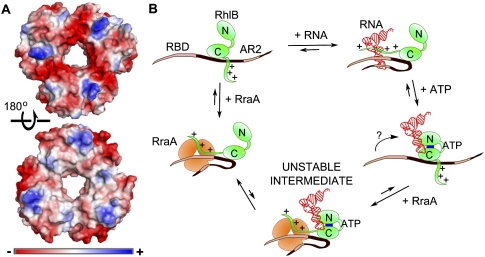
The question arises how RraA can mediate interactions with so many different proteins. RraA is a structurally well-defined trimer, and there is little indication that it might be structurally flexible to accommodate different protein partners through conformational adjustment. Instead, the trimer is rigid conformationally, with well-defined electronegative grooves at the subunit interfaces (Fig. 6A). The grooves are seen in the RraA homolog from Mycobacterium tuberculosis and were hypothesized to bind linear peptide epitopes (Johnston et al. 2003). Sequence comparisons indicate that the electronegative character of these grooves is maintained in RraA homologs among divergent bacterial species, including species completely lacking a potential RNase E partner, such as Thermus thermophilus (Rehse et al. 2004). We suggest that these grooves could bind positively charged peptides in extended conformation, such as the arginine-rich RNA-binding regions of RNase E or the C-terminal tail of RhlB. We envisage that such an interaction would be analogous to the binding of a polyproline-rich segment with the Sm heptameric ring of the eukaryotic spliceosome (Pomeranz Krummel et al. 2009).
FIGURE 6.
Surface charge of RraA and the suggested mechanism for participation of RraA in protein–RNA remodeling by RhlB. (A) Surface character of the Escherichia coli RraA (PDB code 1Q5X, Monzingo et al. 2003). Surface color represents electrostatic potential calculated by Pymol. (Top) The face of the RraA trimer exhibiting electronegative grooves radiating from the central hole along the monomer–monomer interface. The opposite face of the trimer is more neutral (bottom), and interacts with another trimer in the crystal (see Supplementary Material, Fig. 2). (B) Schematic of a possible mechanism of ATP-driven remodeling of RNase E(628–843)/RhlB/RNA complexes by RhlB. Positively charged, unstructured RNA-binding regions, such as RBD or AR2 in RNase E or the C-terminal extension of the helicase (denoted by positive charge marks) are able to bind RNA or RraA. In the case of a structured RNA, such as tRNA, the affinity for the nucleic acid is stronger than that for RraA. Addition of ATP causes remodeling of RNA interactions with the RNA-binding segments so that they can interact with RraA. It is possible that an intermediate containing both RraA and RNA is formed, but it must be comparatively unstable, since no super-shift could be observed in EMSA. There is no evidence whether all RNA-binding sites can simultaneously engage the same RraA trimer, but since any of them is sufficient for the remodeling and can bind to RraA, for simplicity the interactions with RraA or RNA are depicted as concurrent.
While RraA and RNase E are well conserved in the eubacteria (Lee et al. 2003), there is poor conservation of the two RNA-binding sites in RNase E that we have identified as the sites of RraA interaction. It is therefore unclear if the regulatory mechanism is common to those divergent species. However, it is possible that what is held in common is the binding of the RraA through unstructured basic regions, and these are less likely to be conserved. We thus envisage that the multidentate interactions we described here may be general throughout diverse bacterial species, including pathogenic Gram-negative species such as Salmonella or Actinomycetes such as Mycobacterium tuberculosis.
Taken together, data from mass spectrometry in nondenaturing conditions and from surface-binding experiments suggest that either one RraA trimer engages one of the three possible sites, but under conditions where the RraA is more concentrated and forms a hexamer, two sites could be engaged either on the same RNase E CTD or linking CTDs from two neighboring RNase E molecules within the degradosome. The X-ray structure of the Escherichia coli RraA (Monzingo et al. 2003) reveals trimers packing as dimers in the crystal lattice, with the electronegative grooves exposed on the outside (Fig. 6A). We observe dimers of RraA trimers in crystals of RraA grown under different conditions (data not shown), so the hexamer may be the biologically relevant oligomer. Thus, it is possible that the RraA hexamer could form on the degradosome to link RNase E CTDs that are in proximity.
The interaction of RraA with RNase E has been observed to affect the degradosome composition under conditions of RraA overexpression (Gao et al. 2006). Earlier findings suggest that RraA displaces RhlB from the degradosome, but we do not observe loss of RhlB from any of the degradosome subassemblies we have explored. For instance, we did not observe displacement of RhlB upon addition of RraA to any of the RNase E/RhlB complexes. However, we did observe displacement of some PNPase from the complex with RNase E CTD and RhlB (data not shown).
The interaction of RraA with three of the E. coli DEAD-box RNA helicases is likely to impact on their function in vivo. DEAD-box proteins can be considered molecular “motors” that can often function to unwind duplex nucleic acids or remodel protein–nucleic acid interactions using the free energy of NTP binding and hydrolysis (Pyle 2008). In our in vitro experiments, RraA served as an efficient probe of remodeling of RNase E/RhlB/RNA complexes by the DEAD-box helicase RhlB. The presence of ATP leads to remodeling of the degradosome–RNA complex, so that RNA-binding sites become available for interaction and could be trapped in complex with RraA. It was recently shown that DEAD-box proteins can unwind using only ATP-binding energy (Chen et al. 2008; Liu et al. 2008); the hydrolysis of ATP is required to reset the helicase for another round of binding and unwinding. A possible mechanism of the protein/RNA remodeling by RhlB, in which RraA plays a passive role, is shown schematically in Figure 6B.
RraA inhibits RhlB and other DEAD-box helicases that can associate with RNase E under different physiological conditions. This inhibition, in turn, must impact on degradation of structured RNAs by the degradosome-associated PNPase. Similarly, the interaction of RraA with RNase E C-terminal half is also likely to impede helicase and (indirectly) PNPase activities that might require co-operation with the RNA-binding sites in RNase E. We have found that RraA might also directly affect PNPase in polymerization or degradation of RNA (data not shown). Altogether, RraA has emerged as a very complex and versatile regulator of degradosome components with a multitude of interactions and their consequences for RNA metabolism.
MATERIALS AND METHODS
Overexpression and purification of proteins
Recombinant degradosome and RNase E(1–762)/RhlB were overexpressed and purified as described previously (Worrall et al. 2008a). The catalytic domain of RNase E, corresponding to residues 1–529 [RNase E(1–529)] was purified as described by Callaghan et al. (2003), except the size-exclusion chromatography step was omitted. RNase E(628–843) (which has also been previously referred to as R-domain or RneHC2), PNPase, and RNase E CTD constructs were purified as described by Callaghan et al. (2004). RNase E(628–843) construct is a fusion of RNase E sequence with a 13-residue leader and 20-residue C-terminal tail derived from its expression vector, pET11c. RNase E(696–762)/RhlB and RNase E(696–762)/RhlB(1–397) were purified as described previously (Worrall et al. 2008b). During purification of these constructs, fractions from chelating chromatography lacking RNase E(696–762) were used as the source of RhlB or RhlB(1–397) alone. RhlB, SrmB, and RhlE constructs were purified as described by Worrall et al. (2008b).
RraA was purified from BL21(DE3) harboring pET28A-RraA (Lee et al. 2003). Expression was induced with 1 mM IPTG for 3 h, and the harvested cells were resuspended in 50 mM Tris-HCl (pH 7.8), 100 mM NaCl, 50 mM KCl, 5 mM MgCl2, and lysed using EmulsiFlex-05 cell disruptor (Avestin). The lysate was clarified by centrifugation (37,500g, 30 min, 4°C), and the soluble fraction was loaded onto a HiTrap Q column (GE Healthcare). The protein was eluted with 0–0.8 M NaCl gradient in 50 mM Tris-HCl (pH 8.0). Fractions containing RraA were pooled, supplemented with 1.4 M ammonium sulphate, filtered, and loaded on HiTrap Butyl HP column (GE Healthcare) equilibrated with buffer A (50 mM Na/K phosphate at pH 7.6, 2 M ammonium sulphate). Protein was first washed with 10% buffer B, and then eluted with an isocratic gradient of buffer B (50 mM Na/K phosphate at pH 7.6, 50 mM NaCl). The UV absorption profile of the purified protein showed that there was no detectable RNA.
The elution profiles in Figure 3A were obtained from a S200 column (Superdex 200 HiLoad 16/60; GE Healthcare) equilibrated with buffer C (50 mM Tris-Cl at pH 7.9, 100 mM NaCl, 50 mM KCl, 5% v/v glycerol).
ATP turnover assays
The EnzCheck Phosphate Assay kit (Molecular Probes) was used to monitor phosphate release accompanying ATPase activity of helicases. Reactions were assembled in the reaction buffer and supplemented with 40 μg/mL Bakers’ yeast RNA (Sigma) where indicated. Stock solutions of helicases and RraA were prepared in 20 mM MOPS (pH 7.2), 50 mM NaCl, 5 mM DTT, and their total volume constituted 3%–7% v/v of the final reaction. Helicase was added to a final concentration of 1 μM, and concentration of RraA in the control reactions with ATP was 9 μM. Proteins were added to the reaction mix and preincubated for 10 min at room temperature before the reaction was started by the addition of 1 mM ATP. Change in absorbance at 360 nm wavelength was monitored for at least 300 sec using a Shimadzu BioSpec-1601 spectrophotometer thermostatted at 25°C. Absorbance values were converted to the corresponding phosphate concentration using a phosphate standard curve, and the initial rates were calculated from the reaction curve over the 100-sec period between 50 and 150 sec from the reaction start. Reported rates are the amount of phosphate released/sec/mol of helicase and constitute averages of three independent experiments. Data were plotted in graphical format with Plot software.
RNA preparation and degradation assays
9S RNA was produced and used for assay of RNA processing by RNase E constructs as described by Worrall et al. (2008a). Wild-type catalytic domain of RNase E, RNase E N305D (1–762)/RhlB, and recombinant degradosome were exchanged into 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 100 mM KCl, 5 mM MgSO4, and 5% (v/v) glycerol. RraA was exchanged into 50 mM NaCl, 50 mM MOPS (pH 7.4) using a PD-10 column (GE Healthcare). A total of 4 μL of 6.3 μM RNase E contruct was incubated with 2 μL of 1 μM 9S RNA in deionized water and 1 μL of either of the following: 28 μM RraA or 50 mM NaCl, 50 mM MOPS (pH 7.4), or 70 mM EDTA. Each reaction was incubated for 30 or 60 min at 37°C. Reactions were quenched by adding 1 vol of 2 × Proteinase K buffer with 0.2 mg/mL Proteinase K and incubating for 30 min at 50°C. After the addition of formamide/urea loading dye, the samples were analyzed on an 8% polyacrylamide gel containing 7 M urea. Nucleic acids were detected using SYBR Gold stain (Invitrogen). TotalLab (Nonlinear Dynamics) was used to estimate the molecular weight of the cleavage products.
Electrophoretic mobility shift assays
For the titration EMSA experiments, 75 μM RraA and 25 μM RhlB and/or RNase E constructs were prepared by exchange into buffer C (100 mM NaCl, 50 mM KCl, 50 mM Tris-HCl at pH 7.5, 5% v/v glycerol) using Micro Bio-Spin 6 columns (Bio-Rad). Reactions (13 μL) were prepared in buffer C and contained the indicated concentration of RraA and partner proteins and 1 μL of loading buffer (250 mM DTT, 50% v/v glycerol, 0.05% w/v Bromophenol blue, 50 mM Tris, 384 mM glycine at pH 8.3). Alternatively, 216 μM of RraA stock in original purification buffer was used in the case of RNase E(696–762)/RhlB(1–397) and RhlE, resulting in additional 60 mM ammonium sulphate in the reaction conditions.
Nontitration EMSA reactions with RhlE and SrmB were prepared similarly as described above, but in 8–10-μL volume and included 1.5–2 μL of loading buffer. Reactions were typically incubated for 10 min at 25°C and separated on 5% native PAA gel (acrylamide:bisacrylamide 37.5:1, 200 mM Tris-HCl at pH 8.5, 10% v/v glycerol) in 1 x Tris-Glycine running buffer at 120 V for 155–160 min at 4°C. Gels were stained with Coomassie Brilliant Blue.
FAM-RNA, 5′-(Fl)-GGAUCGGAGUUUUAAAUUAAUAAUAUA-3′ (Dharmacon) was prepared as 100 μM stock in water. E.coli tRNA (Sigma) was purified on s200 column and exchanged into buffer C. Reactions with RNA were set up as described above, with RraA added last to the reaction.
For the remodeling experiments, 10 mM ATP/MgCl2 stock was prepared in buffer C. Reactions (10 μL) contained, in the order of addition, 2.5 μM RhlB and/or RNase E(628–843) in buffer C, 2 μL of loading buffer, 2.5 μM tRNA, 7.5 μM RraA, and 1 mM ATP-Mg (or the appropriate volume of buffer C in the control reactions). Addition of tRNA and RraA was followed by 5-min incubation steps at 25°C, and after addition of ATP, samples were loaded immediately on 8% native PAA gel. PAA gels with RNA were stained first with SYBR Gold (Invitrogen), followed by Coomassie staining.
For RNA competition assay using native agarose gels, 150 μM RraA stock was prepared in buffer F (50 mM NaCl, 20 mM Tris-HCl at pH 7.5, 0.5 mM MgCl2). Reactions (10 μL) contained 2 μL of loading buffer, 5 μL of RNase E CTD constructs (in 20 mM Na-K-phosphate at pH 7.7) or 4 μL of recombinant degradosome (in 250 mM NaCl, 20 mM Tris-HCl at pH 7.5, 2.5 mM MgCl2, 2.5 mM DTT), RNAs and RraA at the indicated concentrations, and the volume was brought up with buffer C. RraA was added last after short incubation with RNA, and the reactions were analyzed on 0.6% agarose in 1X TG buffer at 4°C, run at 120 V for 140–155 min. Gels were first visualized in UV, and also with SYBR Gold in case of recombinant degradosome, and finally stained with 0.025% (w/v) Coomassie R-250 in 40% (v/v) methanol and 7% (v/v) acetic acid.
Selected bands from native gels were analyzed by SDS-PAGE as follows. Bands were excised, incubated for 15 min at 25°C in 1 M β-mercaptoethanol in 1X TG, and the gel slices were fitted in the wells of 10% Bis-Tris NuPAGE SDS-PAGE gels (Invitrogen). Composite images showing overlayed nucleic acid and protein staining of the same gel were constructed using ImageJ software.
Biacore
Surface plasmon resonance was performed at 25°C using a Biacore T100 instrument (Biacore Inc.-GE Healthcare). A total of 1000–2000 RUs of purified His-tagged RNase E C-terminal domain (CTD) and its deletion constructs CTDΔAR2 and CTDΔRBD were immobilized on nitrilotriacetic acid (NTA) chip in running buffer HBS-P (10 mM HEPES at pH 7.4, 150 mM NaCl, 0.05% v/v Tween 20) supplemented with 50 μM EDTA. Purified RraA at a concentration range of 0–2 μM was injected at a 60 μL/min constant flow rate. Data were evaluated by Biacore T100 evaluation software and best fits to the data were obtained with 1:1 binding models. The equilibrium dissociation constants (KD) were calculated from RraA trimer concentration ranges of 0–39 nM by kinetic analysis.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Leonora Poljak and Agamemnon Carpousis (Centre National de la Recherche Scientifique) for generously providing malEF RNA and plasmids encoding the individual degradosome components and RNAs. We thank G. Georgiou for the gift of RraA expression plasmid and Elaine Stephens for advice for protein–protein cross-linking. We thank Kenny McDowall for helpful comments and stimulating discussions and Helen Vincent for valuable comments on the manuscript. We thank DESY for support for access to the SAXS beamline and to Dmitri Svergun for use of the facilities. M.W.G. is supported by a European Union training fellowship of the European Union Early-Stage Training Site ChemBioCam. Z.P. is supported by Biotechnology and Biological Sciences Research Council. This work is supported by the Wellcome Trust.
Footnotes
Abbreviations: AR2, arginine-rich domain 2 of RNase E, corresponding to residues 796–819; PNPase, polynucleotide phosphorylase, also known as polyribonucleotide nucleotidyltransferase (EC 2.7.7.8); RBD, RNA-binding domain of RNase E, corresponding to residues 604–688.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1858010.
REFERENCES
- Arraiano C, Maquat L. Post-transcriptional control of gene expression: Effectors of mRNA decay. Mol Microbiol. 2003;49:267–276. doi: 10.1046/j.1365-2958.2003.03545.x. [DOI] [PubMed] [Google Scholar]
- Bernstein J, Lin P, Cohen S, Lin-Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci. 2004;101:2758–2763. doi: 10.1073/pnas.0308747101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel K, Baker A, Jain C. Identification and analysis of Escherichia coli ribonuclease E dominant-negative mutants. Genetics. 2006;172:7–15. doi: 10.1534/genetics.105.048553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan A, Grossmann J, Redko Y, Ilag L, Moncrieffe M, Symmons M, Robinson C, McDowall K, Luisi B. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry. 2003;42:13848–13855. doi: 10.1021/bi0351099. [DOI] [PubMed] [Google Scholar]
- Callaghan A, Aurikko J, Ilag L, Günter Grossmann J, Chandran V, Kühnel K, Poljak L, Carpousis A, Robinson C, Symmons M, et al. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J Mol Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Carpousis A. The RNA degradosome of Escherichia coli: An mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Carpousis A, Luisi B, McDowall K. Endonucleolytic initiation of mRNA decay in Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:91–135. doi: 10.1016/S0079-6603(08)00803-9. [DOI] [PubMed] [Google Scholar]
- Chandran V, Poljak L, Vanzo N, Leroy A, Miguel R, Fernandez-Recio J, Parkinson J, Burns C, Carpousis A, Luisi B. Recognition and cooperation between the ATP-dependent RNA helicase RhlB and ribonuclease RNase E. J Mol Biol. 2007;367:113–132. doi: 10.1016/j.jmb.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Potratz J, Tijerina P, Del Campo M, Lambowitz A, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn G, Miao X, Briant D, Mackie G. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes & Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, Moore C, Cohen S, Georgiou G. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol. 2006;61:394–406. doi: 10.1111/j.1365-2958.2006.05246.x. [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Jiang X, Diwa A, Belasco J. Regions of RNase E important for 5′-end-dependent RNA cleavage and autoregulated synthesis. J Bacteriol. 2000;182:2468–2475. doi: 10.1128/jb.182.9.2468-2475.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J, Arcus V, Morton C, Parker M, Baker E. Crystal structure of a putative methyltransferase from Mycobacterium tuberculosis: Misannotation of a genome clarified by protein structural analysis. J Bacteriol. 2003;185:4057–4065. doi: 10.1128/JB.185.14.4057-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V, Toesca I, Poljak L, Vanzo N, Carpousis A. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: Functional replacement of RhlB by RhlE. Mol Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- Kido M, Yamanaka K, Mitani T, Niki H, Ogura T, Hiraga S. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J Bacteriol. 1996;178:3917–3925. doi: 10.1128/jb.178.13.3917-3925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhan X, Gao J, Qiu J, Feng Y, Meganathan R, Cohen S, Georgiou G. RraA. a protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli. Cell. 2003;114:623–634. [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcaida M, DePristo M, Chandran V, Carpousis A, Luisi B. The RNA degradosome: Life in the fast lane of adaptive molecular evolution. Trends Biochem Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Monzingo A, Gao J, Qiu J, Georgiou G, Robertus J. The X-ray structure of Escherichia coli RraA (MenG), A protein inhibitor of RNA processing. J Mol Biol. 2003;332:1015–1024. doi: 10.1016/s0022-2836(03)00970-7. [DOI] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol. 2004;54:1063–1075. doi: 10.1111/j.1365-2958.2004.04329.x. [DOI] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Yagi M, Aiba H. Analyses of mRNA destabilization and translational inhibition mediated by Hfq-binding small RNAs. Methods Enzymol. 2008;447:359–378. doi: 10.1016/S0076-6879(08)02218-0. [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel D, Oubridge C, Leung A, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme-Généreux A, Beran R, Iost I, Ramey C, Mackie G, Simons R. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold-shock protein, CsdA: Evidence for a ‘cold shock degradosome’. Mol Microbiol. 2004;54:1409–1421. doi: 10.1111/j.1365-2958.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- Py B, Higgins C, Krisch H, Carpousis A. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Pyle A. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Rehse P, Kuroishi C, Tahirov T. Structure of the RNA-processing inhibitor RraA from Thermus thermophilis. Acta Crystallogr D Biol Crystallogr. 2004;60:1997–2002. doi: 10.1107/S0907444904021146. [DOI] [PubMed] [Google Scholar]
- Schuck A, Diwa A, Belasco J. RNase E autoregulates its synthesis in Escherichia coli by binding directly to a stem–loop in the rne 5′ untranslated region. Mol Microbiol. 2009;72:470–478. doi: 10.1111/j.1365-2958.2009.06662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Chang S, Lin P, Averina O, Kaberdin V, Lin-Chao S. Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc Natl Acad Sci. 2009;106:864–869. doi: 10.1073/pnas.0810205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo N, Li Y, Py B, Blum E, Higgins C, Raynal L, Krisch H, Carpousis A. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes & Dev. 1998;12:2770–2781. doi: 10.1101/gad.12.17.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. A rough guide to the noncoding RNA world of Salmonella. Mol Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- Wilusz C, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Worrall J, Górna M, Crump N, Phillips L, Tuck A, Price A, Bavro V, Luisi B. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008a;382:870–883. doi: 10.1016/j.jmb.2008.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall J, Howe F, McKay A, Robinson C, Luisi B. Allosteric activation of the ATPase activity of the Escherichia coli RhlB RNA helicase. J Biol Chem. 2008b;283:5567–5576. doi: 10.1074/jbc.M708620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom J, Go H, Shin E, Kim H, Han S, Moore C, Bae J, Lee K. Inhibitory effects of RraA and RraB on RNase E-related enzymes imply conserved functions in the regulated enzymatic cleavage of RNA. FEMS Microbiol Lett. 2008a;285:10–15. doi: 10.1111/j.1574-6968.2008.01205.x. [DOI] [PubMed] [Google Scholar]
- Yeom J, Shin E, Go H, Sim S, Seong M, Lee K. Functional implications of the conserved action of regulators of ribonuclease activity. J Microbiol Biotechnol. 2008b;18:1353–1356. [PubMed] [Google Scholar]