Abstract
Telomerase adds simple-sequence repeats to chromosome 3′ ends to compensate for the loss of repeats with each round of genome replication. To accomplish this de novo DNA synthesis, telomerase uses a template within its integral RNA component. In addition to providing the template, the telomerase RNA subunit (TER) also harbors nontemplate motifs that contribute to the specialized telomerase catalytic cycle of reiterative repeat synthesis. Most nontemplate TER motifs function through linkage with the template, but in ciliate and vertebrate telomerases, a stem–loop motif binds telomerase reverse transcriptase (TERT) and reconstitutes full activity of the minimal recombinant TERT+TER RNP, even when physically separated from the template. Here, we resolve the functional requirements for this motif of ciliate TER in physiological RNP context using the Tetrahymena thermophila p65-TER-TERT core RNP reconstituted in vitro and the holoenzyme reconstituted in vivo. Contrary to expectation based on assays of the minimal recombinant RNP, we find that none of a panel of individual loop IV nucleotide substitutions impacts the profile of telomerase product synthesis when reconstituted as physiological core RNP or holoenzyme RNP. However, loop IV nucleotide substitutions do variably reduce assembly of TERT with the p65-TER complex in vitro and reduce the accumulation and stability of telomerase RNP in endogenous holoenzyme context. Our results point to a unifying model of a conformational activation role for this TER motif in the telomerase RNP enzyme.
Keywords: ribonucleoprotein biogenesis, protein–RNA interaction, telomerase reverse transcriptase, telomerase holoenzyme, telomere maintenance
INTRODUCTION
Telomeres are chromatin domains that cap the ends of eukaryotic chromosomes, insulating authentic ends from inappropriate recombination, fusion, and degradation (Palm and de Lange 2008). Telomere assembly is nucleated on tandem copies of a DNA repeat, most often a simple sequence such as the TTGGGG repeats first discovered in the ciliated protozoan Tetrahymena thermophila or the TTAGGG repeats later characterized in vertebrates. The DNA that nucleates telomeric chromatin assembly exists in a precarious balance of sequence loss and gain (Gilson and Geli 2007). Terminal sequence erosion is inherent in the process of DNA-templated DNA replication and must be compensated by new DNA synthesis.
The telomerase ribonucleoprotein complex (RNP) accomplishes de novo synthesis of telomeric repeat DNA using an active site formed by telomerase reverse transcriptase (TERT) and a template within the integral telomerase RNA (TER). TERT shares the residues conserved among all reverse transcriptases (RTs) that position metal ions in the active site for catalysis (Autexier and Lue 2006). TERT also shares conserved RT-domain side chains that govern selectivity for dNTPs and contact the 3′ end of primer or product base paired with the template. The N-terminal half of TERT includes two TERT-specific domains, an extreme N-terminal domain involved in DNA binding (the TEN domain) and a unique high-affinity RNA-binding domain (TERT RBD or TRBD), followed by the RT domain and a C-terminal extension (CTE) that is also TERT specific (Blackburn and Collins 2010). A high resolution structure of TERT from an insect lacking the TEN domain suggests that the TRBD and CTE fold away from the RT domain toward each other, forming a large contact surface that could lock the TRBD in a conformation inaccessible for RNA binding (Gillis et al. 2008). This intramolecular association may account for why full-length TERT requires lysate chaperone activities for assembly with TER, while the isolated TRBD does not (Holt et al. 1999; Licht and Collins 1999; O'Connor et al. 2005).
Unlike TERTs, TERs show rapid evolutionary divergence in domain structure. Cloning of TERs from ciliates, vertebrates, and budding yeasts has allowed secondary structure modeling by phylogenetic comparison, with distinct models derived for the TERs from each phylogenetic group (Legassie and Jarstfer 2006; Theimer and Feigon 2006). Overall, TERs share three elements of structure required for telomerase function in vivo: a template, a template-adjacent pseudoknot, and a motif that is either a minimal stem–loop in ciliate TERs (stem–loop IV), a bulged three-way stem junction in budding yeast TERs, or the combination of both in vertebrate TERs (conserved region [CR] 4/5; see Discussion). TERs also have motifs that are specific for a phylogenetic group; these motifs recruit the additional proteins required for cellular function and regulation of an endogenously assembled telomerase holoenzyme (Collins 2006; Blackburn and Collins 2010).
Telomerase-mediated synthesis of single-stranded DNA repeats is mechanistically distinct from the synthesis of a nucleic acid duplex by other RTs (Autexier and Lue 2006; Collins 2009). Telomerase must specify a precise string of residues within TER that can be copied as template, synthesize complementary product DNA, and then release single-stranded product from the product-template hybrid. Remarkably, much of this catalytic cycle specialization appears to involve gains of function by nontemplate motifs of TER (Blackburn and Collins 2010). TER motifs 5′ of the template contribute to template definition by restricting active-site access to residues beyond the template 5′ boundary. Also, the region flanking the template 3′ end serves a substrate alignment function, allowing product released from the template 5′ end to be repositioned at the template 3′ end for processive repeat synthesis. These TER-motif contributions to enabling the use of an internal template require physical connection of the template, flanking motifs, and a TER motif that provides high affinity for TERT.
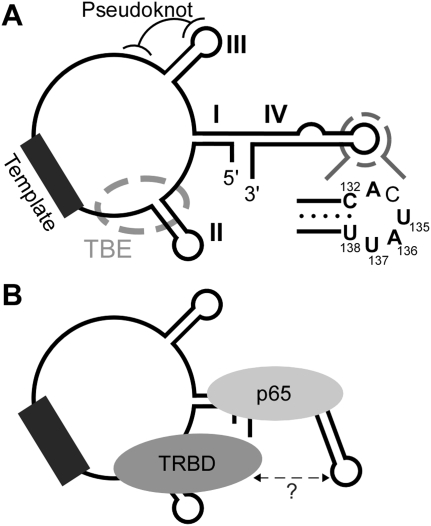
We have used T. thermophila telomerase as a model system to understand the significance of nontemplate motifs of TER. In T. thermophila TER, 5′ template boundary definition and high-affinity interaction with TERT are interdependent functions of the one nontemplate motif conserved among all ciliate TERs, termed the TBE (Fig. 1A). Beyond the template and its flanking regions, all TERs from Tetrahymena species share conserved primary sequence motifs in stem–loop IV, including the loop itself (Fig. 1A). Studies in several laboratories have demonstrated that loop deletion or conserved loop nucleotide substitutions greatly inhibit the activity of the minimal recombinant telomerase RNP reconstituted in rabbit reticulocyte lysate (RRL) by TERT and TER (Licht and Collins 1999; Sperger and Cech 2001; Lai et al. 2003; Mason et al. 2003; Cunningham and Collins 2005; O'Connor et al. 2005). In contrast to other ciliate TER motifs, the full extent of loop IV function can be provided by complementation using an RNA oligonucleotide physically unlinked from the template region (Lai et al. 2003).
FIGURE 1.
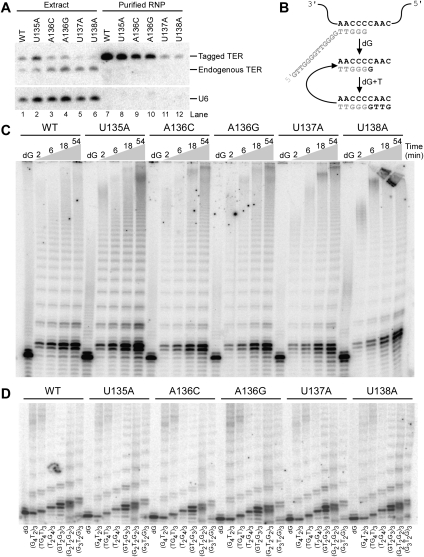
TER structure, motif function, and protein interactions. (A) T. thermophila TER secondary structure elements are indicated by Roman numerals, and functional motifs described in this report are labeled. TBE indicates the template 5′ boundary/high-affinity TERT binding element. Loop IV nucleotides conserved among TERs of Tetrahymena species are in bold. (B) Protein interactions of TER in the p65-TER-TERT RNP are schematized. TRBD indicates the TERT high-affinity RNA-binding domain.
Recent studies indicate that loop IV is not properly positioned by TERT alone. Instead, stable positioning of loop IV in the active RNP requires a hierarchical p65-TER-TERT RNP assembly pathway initiated by p65. The p65-TER-TERT ternary complex reconstitutes the physiological RNP core required for biological stability of TER and TERT (Witkin and Collins 2004; Prathapam et al. 2005). When p65 binds TER to initiate ternary complex assembly, a TER conformational change occurs within the p65 binding site in central stem IV, which greatly stimulates the subsequent recruitment of TERT (O'Connor and Collins 2006; Stone et al. 2007). Single-molecule FRET studies revealed that p65 binding brings the TBE and loop IV in closer proximity (Fig. 1B), and that these motifs are brought closer yet by formation of the p65-TER-TERT ternary complex (Stone et al. 2007). The p65-mediated enhancement of TERT RNP assembly can be reconstituted with the TRBD alone, suggesting that the TRBD could contact both the TBE and loop IV (Fig. 1B). Cross-linking analysis supports a direct contact of loop IV with TERT (Lai et al. 2003), but the TERT domain involved and the functional significance of the contact were not resolved.
Here, we have addressed whether loop IV residues are involved in ternary complex RNP assembly in vitro and in vivo. We find that two conserved loop IV residues, U137 and U138, are particularly important for p65-dependent RNP assembly both in vitro and in vivo. We also addressed whether loop IV residues are important for catalytic activity of the physiological core RNP reconstituted in RRL or the larger holoenzyme complex reconstituted in vivo. Surprisingly, these residues have increasingly less-critical roles in the telomerase catalytic cycle when the RNP context is increasingly physiological. Together, our findings suggest a new hypothesis for function of this nontemplate TER motif. We suggest that loop IV favors a biologically stable and biochemically active conformation of TERT. This mechanism can unify the functional requirements for ciliate TER loop IV observed in vitro and in vivo and also account for the results of functional studies of the likely motif equivalents in vertebrate and yeast telomerases.
RESULTS
Loop IV nucleotide substitutions affect the overall activity of p65-TER-TERT RNP
Loop IV deletion in the RRL-reconstituted minimal TERT+TER RNP nearly abolished product synthesis, with only a few products of low nucleotide addition processivity (NAP) from incomplete copying to the template 5′ end (Licht and Collins 1999). Substitutions of nucleotides U137 and/or U138 in the conserved 135-UAUU-138 motif of loop IV (Fig. 1A) also reduced NAP, while substitution of other loop positions reduced repeat addition processivity (RAP) or activity overall (Sperger and Cech 2001; Cunningham and Collins 2005; O'Connor et al. 2005). NMR structures of stem–loop IV revealed that loop nucleotide U138 forms a noncanonical base pair with C132, which organizes stacking of all other bases in the loop except the flipped-out and relatively unconstrained U137 (Chen et al. 2006; Richards et al. 2006). This structure suggests a rationale for the functional significance of U137 and for the importance of the U138 in ejecting U137 from the constraints of base stacking.
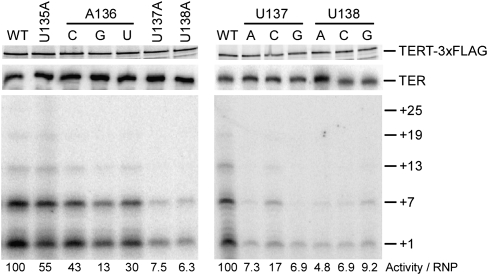
To determine whether substitutions in the conserved UAUU motif of loop IV conferred the same activity defects in the context of the physiological core RNP as in the TERT+TER minimal RNP, we used RRL to reconstitute p65-TER-TERT ternary complexes with TER variants containing individual UAUU-motif nucleotide substitutions. Ternary complex RNPs assembled in the presence of excess p65 were purified from RRL using C-terminally 3xFLAG tagged TERT and assayed by direct telomeric primer extension with radiolabeled dGTP and TTP (Fig. 2). Although loop IV TER variants are not compromised in ability to bind p65, and assembly of the p65-TER complex with TERT is predominantly dependent on TERT binding to the TER TBE motif 5′ of the template (Fig. 1B), RRL-chaperoned ternary complex assembly could have been affected by the loop IV substitutions. Therefore, we normalized product intensity by the recovery of TER to quantify the overall defect in catalytic activity (Fig. 2, bottom). The activity of p65-TER-TERT RNPs with loop IV substitutions was reduced to varying extent, with increasing activity loss 5′ to 3′ across the conserved residues U135, A136, U137, and U138. This rank order parallels previous findings for the minimal TERT+TER RNP (Sperger and Cech 2001). However, to our surprise, none of the individual loop IV substitutions reconstituted as p65-TER-TERT RNP induced the accumulation of low-NAP products from incomplete template copying, as observed for the minimal TERT+TER RNP with substitutions U137A and/or U138A. Consistent with uncompromised NAP, all p65-TER-TERT enzymes with a loop IV nucleotide substitution appeared capable of RAP, with products resulting from addition of a second repeat and several additional repeats as well (Fig. 2; longer exposure and additional data not shown). To gain insight about possible structural requirements for loop nucleotide function, we examined the activity of TERs with all possible A136, U137, or U138 base substitutions assembled as p65-TER-TERT RNP (Fig. 2). None of the A136 substitutions was as deleterious as U137A or U138A, but among them the bulky A136G substitution was the most severe. Likewise, among the U137 substitutions, U137A and U137G were the most severe. At position U138, U138G appeared marginally less severe than the other substitutions, which may be due to partially compensatory formation of a canonical rather than noncanonical base pair with C132. Notably, independent of the introduced nucleotide identity, all substitutions of U137 and U138 led to substantial loss of catalytic activity. In contrast, none of any of the possible substitutions of U135 led to substantial loss of activity (data not shown).
FIGURE 2.
Reduced overall activity of p65-TER-TERT RNPs with loop IV nucleotide substitutions. Input and bound TERT were detected by imaging of the radiolabel after resolution by SDS-PAGE; the TERT polypeptides of less than full-length are created by translation initiation at internal positions. Bound TER was detected by Northern blot. The ratio shown is a normalization of product intensity from second-repeat synthesis to TER.
Loop IV nucleotides are required for p65-enhanced TERT RNP assembly in vitro
T. thermophila TERT-TER interaction requires the TBE (Lai et al. 2001), but motifs beyond the TBE including stem–loop IV contribute to the interaction under some conditions (Sperger and Cech 2001; Lai et al. 2003). In the physiological pathway of p65-dependent RNP assembly, p65 induces a change in TER conformation that increases TERT RNP assembly. The p65-mediated stimulation of TERT RNP assembly can be observed by gel mobility shift assays using the bacterially produced N-terminal half of TERT or the TRBD alone or by single-molecule FRET with these polypeptides or full-length TERT produced in RRL (O'Connor and Collins 2006; Stone et al. 2007). Real-time single-molecule FRET observation of p65-TER-TERT RNP assembly demonstrated that there is an obligate hierarchy: independent of subunit concentrations, the ternary complex forms by initial binding of p65 to TER and subsequent recruitment of TERT (Stone et al. 2007). In gel mobility shift assays, hierarchical assembly is evident in the higher affinity of TERT for p65-TER relative to TER alone, while the p65-TER interaction affinity is independent of TERT (O'Connor and Collins 2006). The p65 enhancement of TERT RNP assembly requires p65-induced remodeling of central stem IV, but whether loop IV residues are also critical for p65 stimulation of TERT RNP assembly has not been previously addressed.
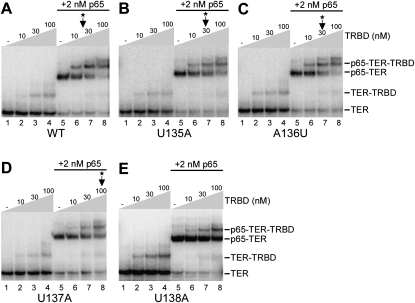
We used gel mobility shift assays to examine the impact of loop IV substitutions on TER-TERT interaction in the absence or presence of p65 (Fig. 3). Radiolabeled wild-type TER or TER variants with individual substitutions in the loop IV UAUU motif were combined with purified recombinant p65 and TERT RBD. Each TER was almost entirely shifted to the p65-TER complex by addition of 2 nM p65 (Fig. 3A–E, lanes 5). Under the assay conditions used here, between 10 and 30 nM of TRBD was sufficient to shift half of the p65-TER to p65-TER-TRBD ternary complex with wild-type TER (Fig. 3A, lanes 5–8; the TRBD concentration required for shift of half of the p65-TER to slower mobility is indicated by an arrow and asterisk). TER with U135A or A136U substitution demonstrated slightly reduced or comparable ability to recruit the TRBD to p65-TER, clearly retaining a preferential assembly of the TRBD with p65-TER (Fig. 3B,C; cf. the formation of p65-TER-TRBD complex in lanes 5–8 and formation of TER-TRBD complex in lanes 1–8). In contrast, TER with U137A or U138A substitution showed greatly reduced p65-TER recruitment of TRBD (Fig. 3D,E). These changes in p65-dependent TRBD assembly were highly reproducible over several assay repetitions (data not shown). Also, assays using the N-terminal half of TERT encompassing both the TRBD and TEN domain gave similar results (data not shown). Thus, as was predicted from previous findings, p65 enables loop IV to have a greater role in recruiting TERT. Also consistent with previous findings, none of the loop IV nucleotide substitutions compromised p65-TER interaction or precluded formation of a discrete p65-TER-TERT ternary complex.
FIGURE 3.
Reduced TERT association with p65-TER complexes formed by TERs with loop IV nucleotide substitutions. Mobility shifts were done in parallel with the same protein preparations and different radiolabeled TERs: wild-type (A), U135A (B), A136U (C), U137A (D), and U138A (E). Lanes 1 contain TER alone, lanes 5–8 contain 2 nM p65, and lanes 2–4 and 6–8 contain a range in concentration of TRBD as indicated. The migration of TER and RNP complexes is annotated at right. The TRBD concentration required for shift of half of the p65-TER to slower mobility is indicated by an arrow and asterisk; for TER U138A in E, this concentration was above the range of the titration shown.
Loop IV nucleotide substitutions reduce RNP accumulation in vivo
In vivo accumulation of T. thermophila TER and TERT is interdependent and also requires p65 (Witkin and Collins 2004). Furthermore, if p65-induced conformational change in TER is impeded by deletion or substitution of the TER central stem IV bulge, RNP accumulation is drastically reduced (Stone et al. 2007). Whether p65-dependent protection of TER and TERT derives simply from stable interaction of the three subunits in the physiological RNP core or whether only specific conformation(s) of this RNP are resistant to turnover is not known.
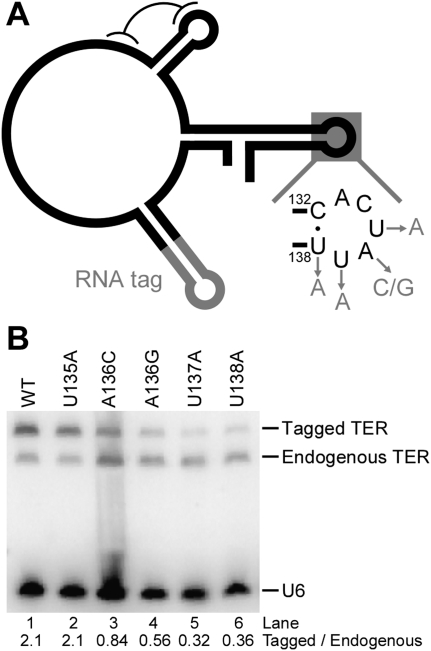
Loop IV substitutions in the UAUU motif can reduce p65 enhancement of TRBD assembly in vitro (Fig. 3). We therefore examined whether loop IV nucleotide substitutions affect RNP accumulation in vivo. As in a previous study (Cunningham and Collins 2005), we expressed variant TERs with a functionally neutral hairpin tag inserted in stem II to distinguish recombinant from endogenous TER by length and to provide a binding site for recombinant TER affinity purification using phage MS2 coat protein (Fig. 4A). TER transgenes were integrated at the nonessential BTU1 locus, with complete replacement verified by Southern blot (data not shown). Increasing TER expression does not increase steady-state TER accumulation, due to limiting levels of p65 and/or TERT. Instead, recombinant TER competes with endogenous TER for assembly into telomerase holoenzyme (Cunningham and Collins 2005). Hairpin-tagged, but otherwise wild-type TER expressed from the transgene locus accumulated in modest excess of endogenous TER, at an ∼2:1 ratio (Fig. 4B, lane 1). Hairpin-tagged U135A TER accumulated at a similar ratio, while substitutions in other positions of the UAUU motif compromised tagged TER accumulation relative to endogenous TER and a U6 small RNA loading control (Fig. 4B). Hairpin-tagged TER with the A136U substitution was undetectable by Northern blot, likely due to the accidental creation of a polyuridine termination signal for RNA polymerase III (data not shown). We therefore tested the two other TER variants at this position, A136C and A136G, which were similarly (A136C) or more severely (A136G) compromised in the reconstitution of core RNP catalytic activity in RRL (Fig. 2). In vivo, substitutions of A136 reduced tagged TER accumulation relative to endogenous TER to ∼40% (A136C) or ∼25% (A136G) of the ratio attained by tagged wild-type TER (Fig. 4B, lanes 3,4). Substitutions U137A and U138A reduced in vivo accumulation of tagged TER to an even greater extent, with only ∼15% of the ratio attained by tagged wild-type TER (Fig. 4B, lanes 5,6). This graded inhibition of in vivo accumulation parallels the loss of catalytic activity of the core RNP reconstituted in RRL (Fig. 2). None of the individual loop IV nucleotide substitutions was as deleterious for RNP accumulation as the previously assayed deletion or substitution of the central stem IV dinucleotide bulge (Stone et al. 2007), suggesting that individual loop IV nucleotide substitutions have less impact than loss of correct loop positioning in TER tertiary structure.
FIGURE 4.
Reduced RNP accumulation due to TER loop IV nucleotide substitutions. (A) The stem II hairpin tag of transgene-encoded TERs is depicted along with a summary of the individual loop IV nucleotide substitutions tested for impact on TER accumulation in vivo. (B) Northern blot detection of TER and U6 loading control RNA in whole-cell samples of total RNA. The ratio shown is an internal ratio of transgene-encoded to endogenous TER in each lane.
Holoenzymes harboring loop IV substitutions suffer enhanced degradation in extract
When a hairpin-tagged TER with the double substitution U137A, U138A was overexpressed in replacement of endogenous TER, the variant accumulated to normal TER level (because there was no competition from endogenous TER) and allowed maintenance of very short telomeres (Cunningham and Collins 2005). However, RNP harboring the tagged TER proved impossible to affinity purify from extract, precluding studies of holoenzyme catalytic activity. Because the single-nucleotide loop IV substitutions studied here were graded in their inhibition of RNP assembly in vitro and RNP accumulation in vivo, we used a panel of single-nucleotide loop IV substitution variants to investigate loop IV influence on telomerase holoenzyme properties.
Telomerase holoenzymes containing hairpin-tagged TER were purified using MS2 coat protein fused to tandem Protein A domains as described previously by adding the coat protein to cell extract and then recovering associated RNPs on IgG agarose (Cunningham and Collins 2005). This method is highly specific for enrichment of tagged TER, without any contamination by endogenous TER (Fig. 5A). We noticed that even for affinity purifications performed largely at 4°C, the disparity in final yield of eluted RNP comparing wild-type tagged TER and the loop IV TER variants (Fig. 5A, lanes 7–12) was exaggerated compared with their relative levels in input cell extracts (Fig. 5A, lanes 1–6) or whole cells (Fig. 4B). The severity of RNP loss during purification parallels the disadvantage to RNP accumulation in vivo, suggesting that loop IV substitutions increase the percentage of RNP in a degradable conformation. Our holoenzyme purification results do not resolve whether the degradable RNP pool is in dynamic equilibrium between conformations more or less resistant to degradation or whether the initial process of holoenzyme assembly creates RNP subpopulations with distinct conformations, with loop IV substitution favoring the conformation that is more readily degraded.
FIGURE 5.
Normal catalytic activity of holoenzyme RNPs with loop IV single-nucleotide substitutions. (A) Input extracts and purified holoenzyme RNP samples were compared for levels of TER and U6 small nuclear RNA. Equal fractions of the input cell extracts or eluted RNPs were used for RNA isolation. (B) The primer used for assays shown in C aligns to the template such that a single nucleotide should be added in reactions with dGTP only (dG); in the presence of both dGTP and TTP (dG+T), major products will result from copying to the template 5′ end in each repeat addition. Because primer is in excess of enzyme or product in the activity assay, long products result from RAP. (C) Reactions with dGTP only were performed for 54 min. (D) Reactions were performed for 54 min. Reactions with dGTP only contained primer (G4T2)3.
Purified holoenzymes with loop IV substitutions retain a normal profile of catalytic activity
For the tagged TERs with single-nucleotide loop IV substitutions, we were able to enrich enough holoenzyme for assays of catalytic activity. As demonstrated previously (Cunningham and Collins 2005), no telomerase activity was recovered in parallel control purifications from wild-type cell extract (data not shown). We used primer (GT2G3)3 to monitor two types of activity (Fig. 5B): addition of a single dGTP, an activity readout insensitive to differences in NAP or RAP, and the processive repeat synthesis reaction in dGTP and TTP. Holoenzymes with hairpin-tagged wild-type or variant TER were cross-compared in reactions performed using a normalized content of TER, determined by Northern blot hybridization of purified RNP preparations (Fig. 5A, lanes 7–12).
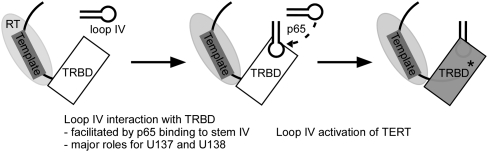
Holoenzymes with tagged wild-type or loop IV variant TERs supported comparable levels of single dGTP addition (Fig. 5C, lanes labeled dG). We were surprised to observe that holoenzymes harboring wild-type and loop IV variant TERs also achieved comparable synthesis of long products with no difference in overall activity (Fig. 5C). We attempted to tease out potential defects in NAP or RAP using a variety of assay conditions, including time courses of product synthesis (Fig. 5C) and panels of staggered primer permutations that oblige different 3′ end positioning relative to the template (Fig. 5D). In these and other assays, we detected no reproducible difference in the amount or profile of products generated by purified holoenzymes with wild-type and variant TERs. This rescue of catalytic activity by holoenzyme assembly is even more pronounced than the improvement obtained by p65-directed assembly of TERT and TER in RRL (Fig. 2). These unexpected results suggest that no individual nucleotide of the conserved UAUU motif plays a critical role in the catalytic cycle. Rather, loop nucleotides make their primary contribution to active-site function indirectly, perhaps by favoring a conformational change in TERT (Fig. 6). For the percentage of holoenzyme that becomes resistant to degradation by forming a loop IV–TERT interaction despite the single-nucleotide substitution, or by stabilizing an active TERT conformation without TERT–loop IV interaction, the catalytic cycle progresses in an unperturbed manner.
FIGURE 6.
Model for loop IV function. The template is indicated by a thick black box over a thinner line, representing the template-flanking regions of TER. Loop IV is also schematized, with the p65-dependent conformational change illustrated. Other TER regions have been omitted for clarity. The TRDB and RT domains of TERT are shown, but the TEN and CTE domains are omitted for clarity. TRBD* indicates conversion of the TRBD to a state required for RNP catalytic activation; this could be a conformational change within the TRBD and/or a change in TRBD interaction with other TERT domains.
DISCUSSION
Stem–loop IV harbors the most conserved primary sequence elements of ciliate TER aside from the template and immediately template-flanking motifs. TERs from Tetrahymena species show absolute conservation of both the central stem IV bulge region and the loop IV UAUU motif (McCormick-Graham and Romero 1995). Highly divergent ciliate TERs appear to share a stem IV bulge and uridine-rich loop (Lingner et al. 1994), although for most ciliate TERs the precise base-pairing register of stem IV has not been experimentally established.
Previous studies implicated loop IV as a site of direct TERT interaction (Lai et al. 2003). However, loop IV was not found to be a major determinant of TER-TERT interaction in minimal RNP context, even though it was a major determinant of catalytic activity (Licht and Collins 1999; Lai et al. 2001, 2003; Sperger and Cech 2001). Here, we show that individual loop IV nucleotide substitutions reduce TERT-TER interaction in the presence of p65 (Fig. 3). Combined with previous studies, we conclude that p65-dependent positioning of loop IV closer to the TBE favors loop IV interaction with TERT and increases TERT affinity for the p65-TER complex relative to TER alone. In the presence of p65, loss of TERT interaction affinity from loop IV nucleotide substitutions may be partially compensated by the enforced bending of central stem IV, allowing the physiological core RNPs with loop IV nucleotide substitutions to support both NAP and RAP.
Activity assay results obtained for the RRL-reconstituted physiological RNP core (Fig. 2) and the holoenzyme reconstituted in vivo (Fig. 5) suggest that loop IV nucleotides are not required for progression through the biochemically distinct steps of a telomerase catalytic cycle. Although loop IV nucleotides are not essential mediators of the catalytic cycle, it remains possible that they contribute to active-site function by playing a role that is more evident in minimal RNP context than in the context of the larger holoenzyme. In RRL reconstitutions where RNP degradation is minimal, p65-TER-TERT RNPs have different specific activities, depending on the individual loop IV nucleotide substitution (Fig. 2). In endogenous cell extracts where inactive RNP is subject to turnover, there is differential yield of holoenzyme, depending on the individual loop IV nucleotide substitution (Fig. 5A). We suggest that loop IV functions by triggering a change in TERT conformation (Fig. 6), either influencing the structure of the TRBD or ordering other TERT domains relative to the TRBD. This conformational activation step could specifically involve A136, which seems to be less important for binding to the TRBD in vitro (Fig. 3) than for catalytic activity (Fig. 2) or holoenzyme RNP accumulation in vivo (Fig. 4).
Like stem–loop IV of ciliate TER, the stem–loop-containing CR4/5 of vertebrate TER binds TERT directly and can function when physically unlinked from the template (Mitchell and Collins 2000). Substitution or altered context of the stem–loop motif within CR4/5 eliminates the catalytic activity of minimal RNP reconstituted in RRL (Chen et al. 2002). As we propose for ciliate TER loop IV, vertebrate TER CR4/5 may function as a conformational activator of TERT. The potentially analogous three-way stem junction motif of budding yeast TERs could play the same role as well. S. cerevisiae telomerase assayed in vitro has relatively low NAP and RAP compared with T. thermophila or human telomerase, with product yield limited by dissociation of the template hybrid. Perhaps for this reason, RRL reconstitution of S. cerevisiae telomerase with a minimal TER lacking the three-way stem junction motif yields activity that closely resembles purified holoenzyme (Zappulla et al. 2005). However, in vivo disruption of this motif does compromise telomerase function, as evident from telomere shortening (Livengood et al. 2002; Brown et al. 2007).
MATERIALS AND METHODS
RNA synthesis, purification, and detection
TER sequence substitutions were made by site-specific mutagenesis of a double-stranded DNA plasmid vector and confirmed by DNA sequencing. RNAs were generated by in vitro transcription using T7 RNA polymerase and Fok I digested plasmid templates, followed by purification on a 6% (19:1), 0.6X TBE, 7 M urea gel. RNAs were eluted in 0.5 M NH4OAc, 1 mM EDTA, 0.2% SDS at 37°C overnight, followed by organic extraction and ethanol precipitation. The concentration of purified RNA was determined by absorbance. TERs for gel mobility shift assays were internally labeled by incorporation of [α32P]UTP (NEN/PerkinElmer). All transcripts were verified and cross-compared in quality and quantity by SYBR Gold staining of aliquots subject to denaturing PAGE.
TER accumulation in vivo was evaluated using total RNA prepared by TRIzol (Invitrogen) from an aliquot of whole cells. Northern blots were performed using 5 μg of total RNA resolved on a 5% (19:1), 0.6X TBE, 7 M urea gel, probed with a DNA oligonucleotides end-labeled with [γ32P]ATP (NEN/PerkinElmer). The oligonucleotide used to probe for TER is perfectly complementary to endogenous TER and all transgene-encoded stem II-tagged TERs. RNA was isolated from RNP reconstituted in RRL, from cell extract, and from affinity purified holoenzyme samples by TRIzol.
RRL-reconstituted RNP purification and activity assays
Untagged p65 and TERT with a C-terminal triple-FLAG tag were expressed separately by coupled transcription and translation in RRL (Promega). TERT was labeled with 35S-methionine (NEN/PerkinElmer). Immediately after protein synthesis, 12 μL of combined RRL translation reactions was supplemented with 2 pM of TER and 4 pM of nonspecific E. coli tRNA (Sigma) and allowed to assemble at 30°C for 10 min. Ternary complexes were purified by adding 500 μL of T2MG (20 mM Tris-HCl at pH 8.0, 1 mM MgCl2, 10% glycerol) with 5 mM DTT and FLAG M2 monoclonal antibody resin (Sigma), followed by end-over-end incubation at 4°C for 2 h. Beads were washed twice with T2MG with 5 mM DTT at room temperature and then used directly in activity assays as half the reaction volume.
Activity assays on RNPs reconstituted in RRL were performed in buffer containing final concentrations of 50 mM Tris-acetate (pH 8.0), 10 mM spermidine, 5 mM DTT, 2 mM MgCl2, 200 μM dTTP, 5 μM unlabeled dGTP, 20 nM [α32P]dGTP (NEN/PerkinElmer), and 1 μM (G4T2)2 primer. Reactions were incubated at 30°C for 18 min and stopped by adding 80 μL of TE (10 mM Tris-HCl at pH 8.0, 1 mM EDTA). Products were extracted and precipitated with 10 μg of tRNA and 10 μg of linear polyacrylamide as carriers and then resolved on a 10% (19:1), 0.6X TBE, 7 M urea gel.
Electrophoretic mobility shift assays
Bacterial expression, purification, and gel mobility shift assays of six-histidine tagged p65 and TRBD were accomplished as described previously (O'Connor et al. 2005; O'Connor and Collins 2006). Binding was performed in 10 μL reactions containing T2MG, 0.1 M NaCl, 5 mM DTT, 5 μg of bovine serum albumin (New England Biolabs), 0.25 μL of RNasin (Promega), 500 ng of nonspecific tRNA, and loading dyes. Binding reactions were incubated at room temperature for 10 min, followed by electrophoresis on a 5% (37.5:1), 0.5X TBE gel at 4°C.
T. thermophila strain construction
Cultures were grown at 30°C in 2% proteose peptone, 0.2% yeast extract, 10 μM FeCl3, 150 μg/mL ampicillin and streptomycin, and 1.25 μg/mL amphotericin B. BTU1 transgene integration cassettes expressing hairpin-tagged TER from a region of the endogenous RNA Polymerase III TER promoter were created as described previously (Cunningham and Collins 2005), except for inclusion of a neo2 cassette downstream of the RNA polymerase III termination signal prior to the 3′ BTU1 flanking sequence. Cells were transformed by particle bombardment. Selection and assortment for transgene integration were performed for the neo2 cassette using paromomycin largely as described previously (Witkin and Collins 2004).
Holoenzyme purification and activity assays
Cells were grown to a density of 4 × 105/mL at 30°C, washed with 10 mM Tris-HCl (pH 7.5), and lysed in T2MG, 15 mM β-mercaptoethanol, and 0.2% NP-40 with protease inhibitor cocktail. Extracts were clarified by centrifugation at 100,000g for 1 h at 4°C and frozen in liquid nitrogen. Hairpin-tagged TER holoenzymes were purified using MS2 coat protein as described previously (Cunningham and Collins 2005), in T2MG adjusted to 50 mM NaCl with coat protein incubation in extract at 4°C for 1 h, followed by addition of IgG agarose and additional incubation at 4°C for 1 h. Beads were then washed three times with T2MG with 0.1 M NaCl, 0.1% NP-40, 1 mM DTT, and 0.1 M phenylmethylsulfonyl fluoride, and washed twice with TEV elution buffer (10 mM Tris-HCl at pH 8.0, 0.1 M NaCl, 0.1% NP-40, 1 mM DTT) prior to elution with TEV protease at room temperature. Glycerol was added to 10% final concentration prior to quick freezing in liquid nitrogen.
Holoenzyme activity assays were performed as described above for RRL-reconstituted RNPs, except for a reduced concentration of unlabeled dGTP (300 nM unlabeled dGTP added to 20 nM [α32P]dGTP) and 50 nM of an 18-nucleotide telomeric-repeat primer indicated in Figure 5 or its legend. To normalize for TER, holoenzyme samples were diluted in T2MG with 5 mM DTT to constitute half of the final reaction volume. Reactions were incubated at 30°C for the times indicated in Figure 5 or its legend.
ACKNOWLEDGMENT
This work was funded by N.I.H. GM054198.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1936410.
REFERENCES
- Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown Y, Abraham M, Pearl S, Kabaha MM, Elboher E, Tzfati Y. A critical three-way junction is conserved in budding yeast and vertebrate telomerase RNAs. Nucleic Acids Res. 2007;35:6280–6289. doi: 10.1093/nar/gkm713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Opperman KK, Greider CW. A critical stem–loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fender J, Legassie JD, Jarstfer MB, Bryan TM, Varani G. Structure of stem–loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Forms and functions of telomerase RNA. In: Walter NG, Woodson SA, Batey RT, editors. Non-protein coding RNAs. Vol. 13. Springer; Berlin, Germany: 2009. pp. 285–301. [Google Scholar]
- Cunningham DD, Collins K. Biological and biochemical functions of RNA in the Tetrahymena telomerase holoenzyme. Mol Cell Biol. 2005;25:4442–4454. doi: 10.1128/MCB.25.11.4442-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes & Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol. 2001;21:990–1000. doi: 10.1128/MCB.21.4.990-1000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Miller MC, Collins K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol Cell. 2003;11:1673–1683. doi: 10.1016/s1097-2765(03)00232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legassie JD, Jarstfer MB. The unmasking of telomerase. Structure. 2006;14:1603–1609. doi: 10.1016/j.str.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Licht JD, Collins K. Telomerase RNA function in recombinant Tetrahymena telomerase. Genes & Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Livengood AJ, Zaug AJ, Cech TR. Essential regions of Saccharomyces cerevisiae telomerase RNA: Separate elements for Est1p and Est2p interaction. Mol Cell Biol. 2002;22:2366–2374. doi: 10.1128/MCB.22.7.2366-2374.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason DX, Goneska E, Greider CW. Stem–loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol Cell Biol. 2003;23:5606–5613. doi: 10.1128/MCB.23.16.5606-5613.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell. 2000;6:361–371. doi: 10.1016/s1097-2765(00)00036-8. [DOI] [PubMed] [Google Scholar]
- O'Connor CM, Collins K. A novel RNA binding domain in Tetrahymena telomerase p65 initiates hierarchical assembly of telomerase holoenzyme. Mol Cell Biol. 2006;26:2029–2036. doi: 10.1128/MCB.26.6.2029-2036.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor CM, Lai CK, Collins K. Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem. 2005;280:17533–17539. doi: 10.1074/jbc.M501211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Prathapam R, Witkin KL, O'Connor CM, Collins K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat Struct Mol Biol. 2005;12:252–257. doi: 10.1038/nsmb900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RJ, Wu H, Trantirek L, O'Connor CM, Collins K, Feigon J. Structural study of elements of Tetrahymena telomerase RNA stem–loop IV domain important for function. RNA. 2006;12:1475–1485. doi: 10.1261/rna.112306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperger JM, Cech TR. A stem–loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry. 2001;40:7005–7016. doi: 10.1021/bi0103359. [DOI] [PubMed] [Google Scholar]
- Stone MS, Mihalusova M, O'Connor CM, Prathapam R, Collins K, Zhuang X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–461. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J. Structure and function of telomerase RNA. Curr Opin Struct Biol. 2006;16:307–318. doi: 10.1016/j.sbi.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Witkin KL, Collins K. Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes & Dev. 2004;18:1107–1118. doi: 10.1101/gad.1201704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Goodrich K, Cech TR. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat Struct Mol Biol. 2005;12:1072–1077. doi: 10.1038/nsmb1019. [DOI] [PubMed] [Google Scholar]