Abstract
The combination of ligation-based RNA capture methods and high-throughput sequencing has facilitated the characterization of transcriptomes and the identification of novel noncoding RNAs. However, current ligation-based RNA capture methods require RNA substrates with terminal 3′-hydroxyl groups, limiting their utility for identifying RNAs with modified termini like 2′,3′-cyclic phosphates. Cyclic phosphate-terminated RNAs are generated by endonucleolytic cleavages and self-cleaving ribozymes and are found as stable modifications on cellular RNAs such as the U6 spliceosomal RNA. We developed a method that uses the Arabidopsis thaliana tRNA ligase to add an adaptor oligonucleotide to RNAs that terminate in 2′,3′-cyclic phosphates. The adaptor allows specific priming by reverse transcriptase, which is followed by additional steps for PCR amplification and high-throughput DNA sequencing. Applying the method to total human RNA, we found 2836 sequencing reads corresponding to the 3′ terminus of U6 snRNA, validating the method. In addition to a large background of reads that map throughout abundantly transcribed RNAs, we also found 42,324 reads of specific fragments from several tRNA isoacceptor families, suggesting that this method may identify processing events previously undetected by other RNA cloning techniques.
Keywords: 2′,3′-cyclic phosphate; ligation; high-throughput sequencing; RNA processing
INTRODUCTION
RNAs have been found to participate in an ever-increasing array of cellular processes, affecting gene expression, development, and differentiation, as well as a number of medical disorders (Mattick and Makunin 2006). The widespread nature of RNA activities makes a compelling case for the need to identify and characterize the complete set of RNA molecules present in a cell. The feasibility of completing such a catalog is greatly enhanced by the development of new sequencing technologies (Shendure and Ji 2008) that enable the acquisition of tens of millions of sequence reads at relatively low cost. These technologies provide single base resolution and a large, quantitative dynamic range, yielding data on molecules present at only a few copies per cell up to those present at high abundance.
Various approaches have been devised to convert RNA populations into libraries of cDNAs for sequence analysis, including those that prime reverse transcription with oligo-dT primers or random hexamers (Ying 2004). In some approaches, an oligonucleotide adaptor of known sequence is ligated by T4 RNA ligase to the 3′ ends of a population of RNA molecules. This adaptor serves as a priming site for reverse transcriptase to generate a cDNA copy of the ligated RNA. Because most current high-throughput sequencing technologies are based on short reads, fragmentation of RNA is typically carried out prior to reverse transcription to obtain small fragments of a few hundred base pairs. Following reverse transcription, a second adaptor is ligated to the 3′ end of the cDNA, and PCR is subsequently used to amplify the fragments prior to sequencing.
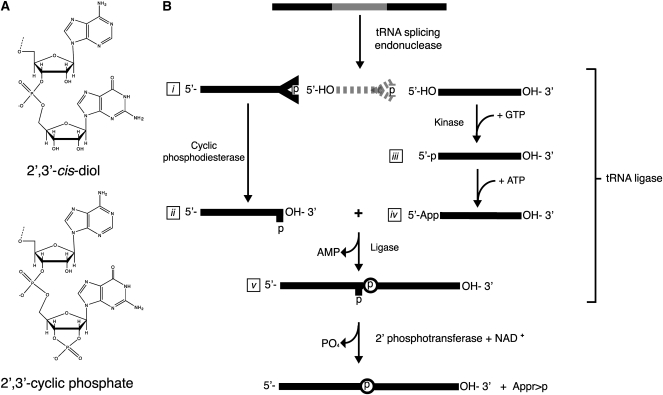
T4 RNA ligases 1 and 2 are the enzymes most commonly used for the initial ligation of RNAs, and both require RNA substrates with terminal 3′-hydroxyl groups (Fig. 1A, top), which represent the majority of cellular RNAs. However, several RNA cleavage events generate termini with 2′,3′-cyclic phosphates (Fig. 1A, bottom), which are unable to be used as substrates for adaptor ligation by T4 RNA ligases. For example, RNAs with terminal 2′,3′-cyclic phosphates are generated during tRNA splicing (Abelson et al. 1998), in endonucleolytic cleavage by several ribonucleases (Deshpande and Shankar 2002), in self-cleavage by RNA ribozymes (Salehi-Ashtiani et al. 2006), and in response to various cellular stresses including the accumulation of unfolded proteins in the endoplasmic reticulum (Ron and Walter 2007; Han et al. 2009) and oxidative stress (Thompson and Parker 2009b). Furthermore, at least one noncoding RNA, the U6 spliceosomal RNA (Lund and Dahlberg 1992), possesses a 2′,3′-cyclic phosphate that regulates its association with spliceosomal proteins (Licht et al. 2008).
FIGURE 1.
RNA termini and tRNA splicing. (A) RNA termini with a 2′,3′-cis diol (top; T4 RNA ligase 1 substrate) or 2′,3′-cylic phosphate (bottom; A. thaliana tRNA ligase substrate). (B) The tRNA splicing pathway generates intermediates with 2′,3′-cyclic phosphate termini. tRNA exons (represented as solid lines) are joined by tRNA ligase. The tRNA intron (broken line) is degraded. The 2′-phosphate at the splice junction is removed by a 2′-phosphotransferase.
Splicing of tRNA introns occurs through a nonsplicesomal pathway (Fig. 1B; Abelson et al. 1998)). Cleavage of the precursor tRNA at the exon–intron borders generates a 5′ exon and an intron that have termini consisting of a 5′ hydroxyl and a 2′,3′-cyclic phosphate (Fig. 1B, i). The 5′ exon is further processed by tRNA ligase, a multifunctional enzyme that catalyzes the hydrolysis of the 2′,3′-cyclic phosphate group to a 2′-phosphate (Fig. 1B, ii), the phosphorylation of the 5′ hydroxyl of the 3′ exon (Fig. 1B, iii), the adenylation of the resulting 5′ phosphate on the 3′ exon (Fig. 1B, iv), and finally the ligation of the two exons. A spliced tRNA possesses a 2′-phosphate at the exon–exon ligation junction (Fig. 1B, v), which is removed by a 2′-phosphate-specific phosphotransferase in an NAD+-dependent reaction (Culver et al. 1997).
tRNA ligases from diverse organisms differ in their substrate specificities (Englert and Beier 2005). For example, the tRNA ligase from the yeast Saccharomyces cerevisiae is only partially active on non-tRNA substrates (Apostol and Greer 1991; Nandakumar et al. 2008), whereas the enzymes derived from several plants have relaxed specificities (Schwartz et al. 1983; Pick et al. 1986). We exploited the relaxed specificity of the Arabidopsis thaliana tRNA ligase along with high-throughput DNA sequencing to develop a method to identify RNAs with 2′,3′-cyclic phosphates and demonstrate that when applied to total human cellular RNA, the method identifies the U6 terminus. Other RNA fragments are found that map to the anticodons and 3′ regions of particular isoaccepting tRNA families. Additionally, a large background of sequence reads corresponds to RNA fragments due to hydrolysis or nucleolytic cleavage at positions throughout transcribed RNAs.
RESULTS
Specificity of the A. thaliana tRNA ligase and T4 RNA ligase
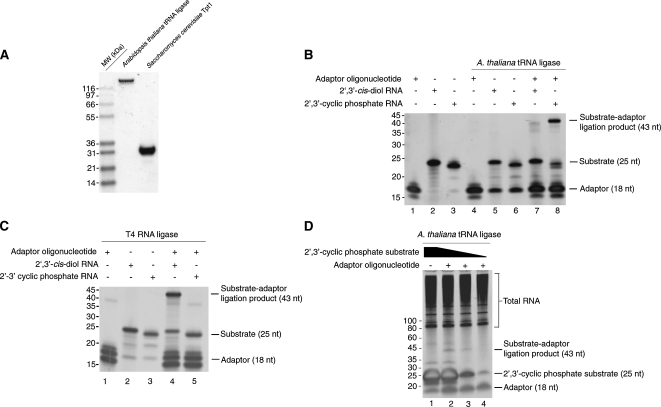
A. thaliana tRNA ligase (Englert and Beier 2005) was expressed and purified from E. coli (Fig. 2A). We sought to examine the substrate specificity of A. thaliana tRNA ligase and T4 RNA ligase 1 for the ligation of an adaptor DNA oligonucleotide to the 3′ terminus of an RNA substrate. The adaptor DNA oligonucleotide (18 nucleotides) (Fig. 2B, lane 1) was preadenylated and blocked at its 3′ end with a dideoxycytidine. The chemical modifications of the adaptor allow the ligation reaction to be performed in the absence of ATP to limit circularization of substrate RNAs in favor of ligation to the adenylated adaptor (Lau et al. 2001).
FIGURE 2.
A. thaliana tRNA ligase is specific for RNAs terminated by 2′,3′-cyclic phosphate groups. (A) Purified A. thaliana tRNA ligase (lane 1) and S. cerevisiae 2′-phosphotransferase Tpt1 (lane 2) were resolved by SDS-PAGE and stained with Coomassie Blue. (B) Substrates bearing 2′,3′-cis diols (lanes 2,5,7) or 2′,3′-cyclic phosphates (lanes 3,6,8) were incubated with purified A. thaliana tRNA ligase (lanes 4–8) and a preadenylated DNA adaptor (lanes 1,4,7,8). A 43-nt ligation product between the 2′,3′-phosphate substrate and adaptor is produced upon incubation with tRNA ligase. (C) Substrates bearing 2′,3′-cis diols (lanes 2,4) or 2′,3′-cyclic phosphates (lanes 3,5) were incubated with T4 RNA ligase 1 (lanes 4,5) and a preadenylated DNA adaptor (lanes 1,4,5). A 43-nt ligation product between the 2′,3′-cis diol substrate and adaptor is produced upon incubation with T4 RNA ligase 1. (D) Tenfold dilutions of a 2′,3′-cyclic phosphate substrate were diluted into S. cerevisiae total RNA and incubated with preadenylated oligonucleotide (lanes 2–4) and A. thaliana tRNA ligase (lanes 1–4). A 43-nt ligation product is observed in the presence of A. thaliana tRNA ligase.
We used a deoxyribozyme of the 10–23 family (Santoro and Joyce 1997) to cleave a 32-nucleotide (nt) target RNA, generating a 25-nt product with a terminal 2′,3′-cyclic phosphate (Fig. 2B, lane 3). In addition, we used an RNA of identical sequence with a 2′,3′-cis diol as a control (Fig. 2B, lane 2). We tested whether either the RNA with the terminal 2′,3′-cyclic phosphate or the RNA with the terminal 2′,3′-cis diol could serve as a substrate in reactions containing the adaptor DNA oligonucleotide and A. thaliana tRNA ligase (Fig. 2B) or T4 RNA ligase 1 (Fig. 2C). In reactions containing tRNA ligase, we observed the formation of a ligation product between the substrate with a 2′,3′-cyclic phosphate and the adaptor oligonucleotide (Fig. 2B, lane 8), but not a specific ligation product between the 2′,3′-cis diol substrate and adaptor (Fig. 2B, lane 7). The ligation products observed due to treatment of the 2′,3′-cis diol substrate with tRNA ligase (Fig. 2B, lane 7) may represent ligation of the preadenylated adaptor to substrates that have undergone hydrolytic events that generate 2′,3′-cyclic phosphate termini. It is also possible that the bacterially purified tRNA ligase preparation contains a contaminating ribonuclease that generates cyclic phosphate termini upon cleavage. The observation of lower molecular species from reactions in which the adaptor has been omitted (Fig. 2B, lanes 5,6) suggests that some degradation of the RNA substrate has occurred.
In reactions containing T4 RNA ligase, we observed a ligation product between the adaptor and the substrate with a terminal 2′,3′-cis diol (Fig. 2C, lane 4), whereas in reactions containing A. thaliana tRNA ligase these two could not be ligated (Fig. 2C, lane 5). We also observed a ligation product of ∼36 nt in T4 RNA ligase-treated reactions that included the adaptor (Fig. 2C, lanes 1,4,5), consistent with an adaptor–adaptor ligation product. These data indicate that some portion of the chemically synthesized adaptor lacks the 3′ dideoxy modification and are consistent with the observation of a lower molecular weight species in the control reaction with only T4 RNA ligase and adaptor (Fig. 2C, lane 1). We suggest that these species are adaptor molecules that have undergone circularization. This conclusion is supported by the fact that these species accumulate in the corresponding tRNA ligase reaction (Fig. 2B, lane 4), as the tRNA ligase enzyme is not active on substrates with 2′,3′-cis diol termini.
Overall, these data indicate that the two ligases have mutually exclusive activities. In addition, they confirm that the A. thaliana tRNA ligase will act on an RNA substrate that does not resemble a tRNA in sequence.
We next determined whether A. thaliana tRNA ligase could perform a similar ligation in the presence of a complex population of RNAs. We mixed total RNA from the yeast S. cerevisiae and dilutions of the 25-nt 2′,3′-cyclic phosphate RNA substrate with A. thaliana tRNA ligase and the preadenylated adaptor DNA oligonucleotide (Fig. 2D). The substrate was in ∼40-fold molar excess over total RNA at its most concentrated (Fig. 2D, lanes 1,2) and was serially diluted by 10-fold (Fig. 2D, lanes 3,4). The adaptor oligonucleotide was in approximately twofold molar excess over total RNA in each reaction. We observed a product of the expected length (43 nt) only in the case of coincubation of substrate, adaptor, and A. thaliana tRNA ligase (Fig. 2D, lanes 2–4), and the intensity of this product decreased with decreasing amounts of the 2′,3′-cyclic phosphate substrate. In a similar experiment, we tested the sensitivity of this reaction using PCR and were able to amplify a product corresponding to ligated 2′,3′-cylic phosphate substrate in the presence of excess total RNA (Supplemental Fig. 1). Thus, A. thaliana tRNA ligase is active on its substrates in an in vitro reaction containing a complex mixture of nonsubstrate RNAs.
Removal of the 2′-phosphate is necessary for efficient reverse transcription
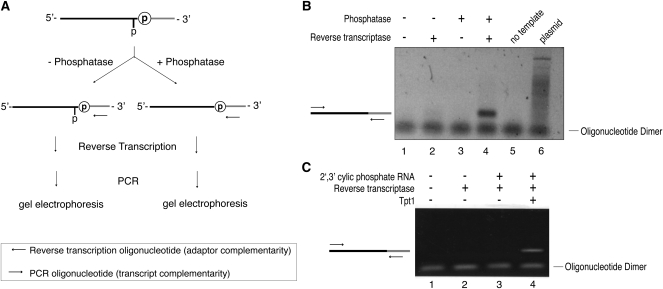
The ligation of tRNA exons by tRNA ligase results in a product with a 2′-phosphate at the ligation junction (Abelson et al. 1998). Because conversion to cDNA requires reverse transcription across the ligation junction, we tested whether the 2′-phosphate at the substrate–adaptor junction blocks reverse transcriptase-mediated conversion of ligated RNA into cDNA.
We purified a 199-nt RNA with a terminal 2′,3′-cyclic phosphate, generated by deoxyribozyme-mediated cleavage of an 802-nt RNA precursor (Santoro and Joyce 1997). This substrate was ligated to a preadenylated adaptor with A. thaliana tRNA ligase (Fig. 3A), and the ligation product was purified from excess adaptor. We treated the ligated product with calf intestinal phosphatase (CIP) to remove the 2′-phosphate present at the ligation junction (Steiger et al. 2001) or left it untreated (Fig. 3A). The samples were reverse transcribed using an oligonucleotide complementary to the ligated adaptor, and the cDNA products were used as a template for PCR (Fig. 3B, lanes 1–4). A PCR product of the expected size (161 base pairs [bp]) required both reverse transcriptase and phosphatase treatment (Fig. 3B, lanes 3,4), indicating that removal of the 2′-phosphate is necessary for reverse transcription across the ligation junction. A PCR product of the expected size was not observed in the absence of template (Fig. 3B, lane 5) or when the plasmid used for the in vitro transcription reaction was used as template (Fig. 3B, lane 6), indicating that production of the PCR product required ligation of the adaptor oligonucleotide to the substrate RNA by A. thaliana tRNA ligase. As the removal of 2′-phosphates by CIP is inefficient (McCraith and Phizicky 1990), we also evaluated whether S. cerevisiae 2′-phosphotransferase Tpt1 (Fig. 2A, lane 2) could remove the 2′-phosphate and found it equally effective at supporting cDNA conversion (Fig. 3C). Because of the unique specificity of Tpt1 for 2′-phosphates (Steiger et al. 2001, 2005), we employed this enzyme in subsequent cloning protocols. These results demonstrate that a 2′-phosphate at the ligation junction inhibits cDNA conversion by reverse transcriptase and that this inhibition is relieved by removal of the 2′-phosphate by treatment with CIP or Tpt1.
FIGURE 3.
A 2′-phosphate at the tRNA ligase ligation junction blocks reverse transcriptase processivity. (A) Schematic depicting the removal of a 2′-phosphate by CIP and detection of subsequent reverse transcription products by PCR. (B) Agarose gel analysis of PCR reactions in which primers specific to the adaptor and the RNA substrate with a terminal 2′,3′-cyclic phosphate were used to test the requirement for phosphatase treatment in the generation of a reverse transcription product. A PCR product of the expected size (161 bp) indicates that a cDNA copy was generated and amplified. (C) Agarose gel analysis of PCR reactions in which primers specific to the adaptor and the RNA substrate with a terminal 2′,3′-cyclic phosphate were used to test 2′ monophosphate removal by purified Tpt1. A PCR product of the expected size (161 bp) indicates 2′ monophosphate removal by Tpt1 and subsequent reverse transcription.
Application of 2′,3′-cyclic phosphate capture to human total RNA
There are multiple situations in which RNAs that terminate in 2′,3′-cylic phosphates are present. These include stable modifications (e.g., the 3′ terminus of U6), cleavage by endonucleases, for example, Ire1 (Gonzalez et al. 1999), RNase L (Dong et al. 2001) and tRNA endonucleases (Thompson and Parker 2009a), and self-cleaving ribozymes (Salehi-Ashtiani et al. 2006). We, therefore, performed a cloning experiment with the A. thaliana tRNA ligase in an effort to identify known and novel cellular RNAs with 2′,3′-cylic phosphates.
We applied the cloning protocol to a total RNA sample isolated from human brain (Supplemental Fig. 3). Due to the cost and yield constraints of synthesizing preadenylated adaptors of sufficient length, we elected to perform the initial ligation with a 5′ phosphorylated adaptor in the presence of ATP and A. thaliana tRNA ligase. Ligated products were purified from excess adaptor oligonucleotide, and recovered RNAs were treated with Tpt1 to remove 2′-phosphates. The RNA pool was chemically fragmented (Cloonan et al. 2008) and reverse transcribed using a primer complementary to the adaptor. Following reverse transcription, the cDNA population was incubated with T4 RNA ligase and a preadenylated adaptor to ligate a unique DNA sequence to the 3′ end of the cDNAs (Troutt et al. 1992). The sample was PCR amplified and sequenced on an Illumina Genome Analyzer to collect 36-base reads, using a sequencing primer complementary to the tRNA ligase adaptor. The sequencing data were aligned to the human genome (Mar. 2006 build, hg18) and visualized using the University of California Santa Cruz (UCSC) genome browser (Karolchik et al. 2008).
We mapped a total of 831,012 sequence reads to the human genome (build hg18 and its associated annotations). Of these, 48% (n = 398,514) map to regions in the genome encoding well-characterized noncoding RNAs, including ribosomal RNAs (n = 284,376), tRNAs (n = 42,324), spliceosomal RNAs (n = 19,999), and Ro-associated Y-RNAs (n = 6272).
Another 15% (n = 122,931) of the reads map to the exons of protein-coding genes and putative noncoding transcripts (Hsu et al. 2006). Some of these correspond to brain-specific mRNAs (e.g., 1613 reads map to CDR1, which is abundantly expressed in brain tissue [Chen et al. 1990]) or long noncoding RNAs (e.g., 1277 reads map to the MALAT1 locus [Wilusz et al. 2008]).
Of the remaining 37% (n = 309,567), 170,239 map to repeats annotated by RepeatMasker (Smit et al. 2004), including fragments of ribosomal RNA (n = 114,968) and L2 LINE elements (n = 5328). Finally, the rest of the reads (n = 139,328) distribute among 36,346 unique genomic positions, with a mean of 3.8 reads per position.
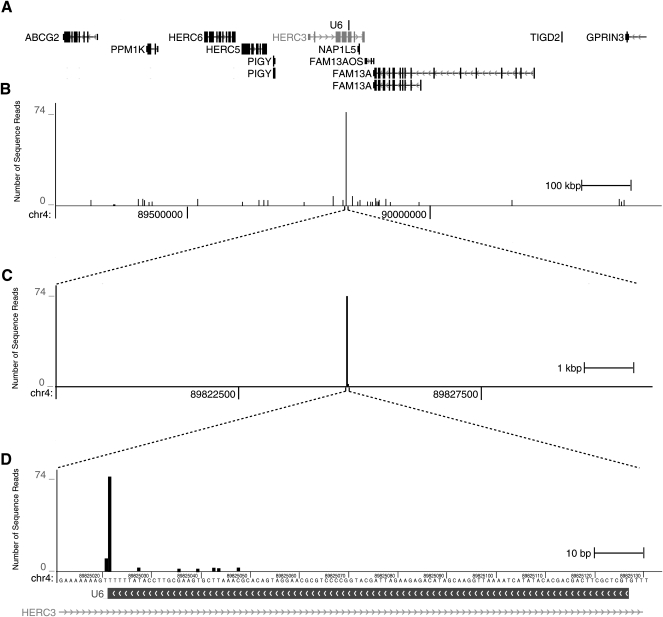
We examined the distribution of reads mapping to U6 spliceosomal RNA (Fig. 4) to determine whether we could capture an RNA known to possess a stable 2′,3′-cyclic phosphate (Lund and Dahlberg 1992). Unlike the case for the bulk of our data for which sequence reads distribute along the encoded RNAs, of the 2836 reads that map to U6, the great majority (n = 2549; 90%) correspond to capture at a single position: the 3′-terminus. The U6 snRNA gene is present in multiple copies (at least 49 in hg18) in the human genome, and the locus shown in Figure 4 is representative of the distribution of data across all U6 loci. In addition to those that map to the 3′ terminus of U6, we found another 287 reads that map to 25 other positions within U6. The accumulation of sequence reads mapping to the terminus of U6 indicates that this method is capable of capturing biologically relevant RNAs from complex RNA mixtures.
FIGURE 4.
The 2′,3′-cylic phosphate terminus of U6 snRNA is identified by the cloning scheme. (A) A 1.2-Mb region (chr4: 89,225,071–90,425,071) from the human genome (hg18) is shown, which contains 12 gene models and a U6 snRNA gene (center). (B) Sequencing reads mapping to the 12-kb interval (chr4: 89,819,071–89,831,071) are plotted, with a maximum of 74 reads. The highest signal in the region corresponds to the U6 snRNA gene, whereas several other positions have one or two mapped reads. (C) Magnification of the region surrounding the U6 snRNA gene (120 kb). (D) Magnification of the region surrounding the U6 gene to base pair resolution (chr4: 89,825,011–89,825,131) showing reads mapping to the 3′ terminal base.
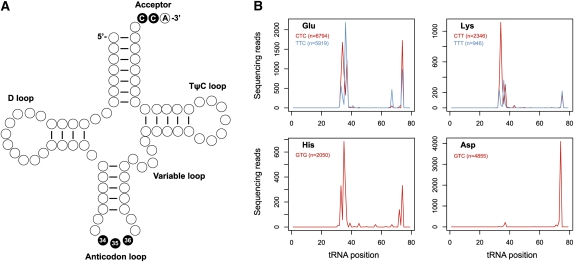
We adopted the pattern of cleavage observed for U6 snRNA as a heuristic to analyze the rest of the data, because this pattern of a majority of sequence reads mapping to an individual position suggests a functional enrichment in vivo (as opposed to nonspecific breakdown in vitro, e.g., during sample preparation). Applying a criterion of ≥100 reads at a single position accounting for ≥80% of the total for a given transcript, we found that tRNAs also exhibit patterns of focal cleavage. Thousands of fragments map to the 3′ ends of tRNA molecules (Fig. 5, Supplemental Fig. 2), with five (Arg-ACG, Arg-TCG, Asn-GTT, Asp-GTC, and Ile-AAT) meeting the criterion. These all exhibit focal cleavage in their CCA-acceptors, with the predominant site of cleavage between the CC dinucleotide, and lower levels of cleavage between the CA dinucleotide. Using a more relaxed threshold of ≥100 reads among 2–4 positions accounting for ≥80% of the total, we found an additional 25 tRNAs (e.g., Glu-CTC, Glu-TTC, Gln-TTG, His-GTG, Lys-CTT, and Lys-TTT) that exhibit more complex patterns of cleavage within both the anticodon loop and CCA-acceptor stem (Fig. 5B; Supplemental Fig. 2). Because tRNA splicing intermediates contain 2′,3′-cyclic phosphates, we also analyzed tRNA exon and intron junctions to determine whether the 3′ end of tRNA 5′ exons (corresponding to position 37 in most tRNAs) might be captured by the protocol. However, among all tRNA subtypes, we found very few reads mapping to position 37, indicating that these are not abundant in the library. This absence of reads is likely due to the efficiency of the splicing reaction in vivo. It is also possible that these molecules may have been lost during gel purification as they would migrate near the limit chosen for size selection (Supplemental Fig. 3).
FIGURE 5.
Fragments of specific tRNA subtypes are identified by the cloning scheme. (A) Diagram of a canonical tRNA. The majority of 2′,3′-cyclic phosphate RNAs correspond to cleavage at the positions in black. (B) Compiled sequence reads mapping to tRNAs of the Glu, His, Lys, and Asp families. Positions along the x-axis are relative to tRNA start site.
Finally, we searched for other transcribed regions meeting the criterion of ≥100 reads at a single position accounting for >80% of the total reads. We found no other protein-coding mRNA transcripts able to satisfy the criterion. Thousands of captured RNA fragments distribute throughout transcribed regions (e.g., Fig. 4, top). Of these, the majority derives from ribosomal RNA loci and protein-coding exons, and their prevalence approximately correlates with the abundance of the RNA molecules. However, the pattern of captured fragments for any single RNA transcript is not uniform across the sequence, suggesting that many fragments may derive from more specific cleavage events. For example, within the 18S ribosomal RNA, we observed 119,408 fragments distributed across 1269 positions. However, only 31 of these positions account for 46% (n = 55,092) of the total, suggesting that these positions may be targeted for cleavage by an endonuclease or may correspond to sites that have undergone geometry-influenced in-line hydrolysis (Soukup and Breaker 1999), either of which could leave cyclic-phosphate termini. Similar patterns of cleavage within 18S rRNA resulted from independent cloning experiments (data not shown); however, we cannot rule out patterns attributable to cloning bias (Ingolia et al. 2009) or a contaminating ribonuclease in the tRNA ligase preparation.
DISCUSSION
We provide a protocol for capturing RNAs with 2′,3′-cyclic phosphates by using A. thaliana tRNA ligase to specifically ligate the RNAs to an adaptor oligonucleotide, facilitating cDNA conversion and sequencing. The activity of A. thaliana tRNA ligase contrasts with that of T4 RNA ligase 1, which requires a 3′ hydroxyl for its catalytic activity. The product of ligation by tRNA ligase contains a 2′-phosphate at the adaptor-substrate junction, which can be removed by either calf intestinal phosphatase or the S. cerevisiae 2′-phosphotransferase Tpt1 to facilitate the conversion of ligated RNA to cDNA. By combining the ligation and phosphatase treatments with steps to prepare a library of DNAs for sequencing by the Illumina Genome Analyzer, we demonstrate that this protocol can be applied to the complex RNA population derived from human brain to identify RNAs with 2′,3′-cyclic phosphates, such as the U6 snRNA.
Using the features of the U6 fragments that were captured as a heuristic, we identified several tRNA families whose cleavage products map to anticodon loops and CCA acceptors. Cleavage at these positions within tRNAs has been previously observed. For example, glutamyl- and lysyl-tRNAs of S. cerevisiae are targets of Kluyveromyces lactis γ-toxin, a tRNA endonuclease that cleaves these tRNAs 3′ of the wobble position (i.e., between positions 34 and 35) (Lu et al. 2005), producing fragments with cyclic-phosphate termini. In addition, a conserved eukaryotic response to stress targets specific tRNAs for destruction via endonucleolytic cleavage (Thompson and Parker 2009a). Thus, the captured fragments within tRNA anticodon loops may represent specific cleavage events that are related to cellular stress (Yamasaki et al. 2009) or might correspond to anticodon cleavages in pre-tRNAs by an unknown mechanism.
We are unaware of specific cleavage activities associated with CCA-acceptors, but the existence of a bacterial tRNA repair enzyme that can hydrolyze 2′,3′-cyclic phosphates (Yakunin et al. 2004) suggests that a CCA-acceptor repair pathway may exist in human cells. These and other data that link tRNA splicing and non-tRNA processing events (Paushkin et al. 2004; Renzi et al. 2006; Han et al. 2009; Hollien et al. 2009) suggest that cyclic phosphate-terminated RNAs are likely to be more prevalent than previously appreciated.
The sequence reads represent not just RNA species known to possess 2′,3′-cyclic phosphates but also a background signal that largely correlates with the abundance of RNA species (e.g., ribosomal RNAs, MALAT1, and CDR1). Based upon the reproducible pattern of captured and sequenced RNA fragments, these signals may be due to geometry-influenced backbone hydrolysis. The susceptibility of any individual phosphodiester bond to geometry-influenced hydrolysis reflects RNA base geometry and therefore provides information about the three-dimensional geometry of surrounding RNA bases. This principle is the basis of the in-line assay (Soukup and Breaker 1999). Therefore, the cyclic-phosphate capture method may also provide information on RNA structure.
We did not observe fragments corresponding to the cleavage sites of the genomic self-cleaving ribozymes previously identified using in vitro selection (Salehi-Ashtiani et al. 2006). It is possible that our inability to detect these intermediates is due to their absence from the total RNA population or it may reflect the low abundance or short half-lives of these intermediates in vivo.
In summary, we present a method for the selective capture and identification of RNAs with terminal 2′,3′-cyclic phosphates that enables the analysis of a branch of cellular RNA biology previously not amenable to high-throughput experimentation.
MATERIALS AND METHODS
Protein expression and purification
The tRNA ligase gene from A. thaliana (Englert and Beier 2005) was subcloned into pET-28a (Novagen), which fuses the tRNA ligase to an N-terminal hexahistidine tag. The open reading frame of TPT1 was cloned from a yeast ORF collection (pBY011-TPT1) (Hu et al. 2007) into pET-53-DEST (Novagen) via Gateway (Invitrogen) to generate pET-53-TPT1, which fuses Tpt1 to an N-terminal hexahistidine tag and C-terminal Strep-tag. These plasmids were transformed into the E. coli expression strain BL21 (DE3) RIPL (Stratagene). Expression and purification were carried out based on a previous protocol (Khalid et al. 2005). Single colonies were inoculated into 50 mL of Luria broth containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) and were grown overnight with shaking. These cultures were diluted (OD600 = 0.2) into 500 mL of terrific broth containing ampicillin (50 μg/mL) and chloramphenicol (34 μg/mL) and were shaken at 37°C for ∼2 h until OD600 reached 0.6, at which point the cultures were chilled on ice for 30 min. Ethanol (final concentration 2%) was added to the cultures, and isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to a final concentration of 0.4 mM. The cultures were shaken at 17°C for 20 h, at which point cells were harvested by centrifugation, frozen, and stored at −80°C.
Cell pellets were resuspended in Buffer A (50 mM Tris-HCl at pH 7.5, 250 mM NaCl, and 10% sucrose) containing 0.2 mg/mL lysozyme and were rotated at 4°C for 30 min. Lysates were briefly sonicated to reduce viscosity, and Triton X-100 was added to 0.1%. Lysates were clarified by centrifugation at 15,000 RPM for 30 min, and the supernatants were incubated with 2 mL of Nickel-NTA sepharose that had been equilibrated in Buffer A. The suspensions were rotated for 1 h at 4°C, and the beads were collected by low-speed centrifugation. Beads were washed twice in batch with 15 mL of Buffer E (50 mM Tris-HCl at pH 7.5, 250 mM NaCl, and 10% glycerol) containing 25 mM imidazole and were then poured into a fritted column. Buffer E containing 300 mM imidazole was added, and five 2-mL fractions were collected. These fractions were checked by SDS-PAGE, and peak fractions containing tRNA ligase or Tpt1 were pooled and incubated with 2 mL of heparin sepharose that had been equilibrated in Buffer D (50 mM Tris-HCl at pH 7.5, 10% glycerol, 5 mM DTT, and 1 mM EDTA) containing 150 mM NaCl. The suspension was rotated for 1 h and washed twice in batch with 15 mL of Buffer D containing 150 mM NaCl. Resin was poured into a fritted column, and Buffer D containing 500 mM NaCl was added. Five 2-mL fractions were collected and checked by SDS-PAGE, and peak fractions were pooled. Protein purity was confirmed by SDS-PAGE, and concentrations were calculated using the Bradford assay by comparing to bovine serum albumin standards.
Oligonucleotides
All oligonucleotides were obtained from Integrated DNA Technologies.
Generation of 2′,3′-cyclic phosphate substrates by 10–23 deoxyribozyme cleavage
DNA–RNA hybrid oligo S33T7 (5′-GCTGATTCTAATCGACTrCrArCrUrArUrArUrGrGrArCrArCC-3′) (1.675 nmol) and the 10–23 enzyme 33.T3–T7b (5′-GGTGTCCAGGCTAGCTACAACGAATAGTGAGTC-3′) (2.575 nmol) were heated to 85°C for 1 min and cooled to 37°C for 3 min, followed by the addition of MgCl2 (final concentration of 10 mM) to initiate cleavage. Reactions were incubated at 37°C for 1 h and were stopped by the addition of loading buffer (42.5% formamide, 25 mM EDTA at pH 8.0, 0.01% [w/v] bromophenol blue, and xylene cyanol) followed by purification on 15% denaturing acrylamide gels (Invitrogen). The band corresponding to the cleaved product was visualized by ultraviolet shadowing and excised with a razor blade. Gel slices were pulverized with pellet pestles (Kimble–Kontes), resuspended in nuclease-free water (Ambion), and heated to 70°C for 10 min. This slurry was applied to a DTR gel filtration column (Edge Biosystems), and column eluate was precipitated with 0.3 M sodium acetate and 70% ethanol in the presence of 15 μg of GlycoBlue (Ambion). Following incubation at −80°C for >20 min, precipitation reactions were centrifuged at 20,000g for 15 min at 4°C. The pellet was washed with 70% ethanol, centrifuged at 20,000g for 5 min at 4°C, and resuspended in nuclease-free water to a final concentration of 10 μM.
A. thaliana tRNA ligase reactions
Reactions (10 μL) were assembled containing 10 pmol of substrate and 50 pmol of adenylated oligo (Linker 1: 5′-rAppCTGTAGGCACCATCAATddC-3′), 1× tRNA ligase buffer (10 mM Tris–HCl at pH 7.5, 100 mM KOAc, 0.3 mM spermine, 6 mM Mg(OAc)2, 0.5 mM DTT, 0.5% Triton X-100) (Englert and Beier 2005), 14% polyethylene glycol, and A. thaliana tRNA ligase (1 μM). Reactions were incubated at 30°C for 1 h and then ethanol precipitated in the presence of sodium acetate and GlycoBlue. Pellets were resuspended in formamide loading buffer and resolved on 15% denaturing acrylamide gels prior to staining with SYBR Gold (Invitrogen) and imaging on a Storm 860 Molecular Imager (GE Healthcare Life Sciences).
T4 RNA ligase reactions
Reactions (10 μL) were assembled with 10 pmol of substrate, 50 pmol of adenylated oligonucleotide Linker 1, T4 RNA ligase 1 (20 U) in 1× T4 ligation buffer (NEB). Reactions were incubated for 2 h at 37°C prior to ethanol precipitation in the presence of sodium acetate, resuspension in formamide loading buffer, and purification or visualization (via SYBR gold staining) on 15% denaturing acrylamide gels.
Cyclic phosphate spike-in experiment
Reactions were assembled as described above with the following modifications: 1 μg of S. cerevisiae total RNA, preadenylated Linker 1 at a final concentration of 2.5 μM, and cleaved DNA–RNA hybrid oligonucleotide S33T7 at final concentration of 50, 5, or 0.5 μM were included. Reactions were incubated for 1 h at 30°C prior to analysis on a 15% denaturing acrylamide gels.
In vitro transcription, purification, and cleavage
Plasmid CFP-PGEM-T was linearized by restriction with NotI (NEB) overnight at 37°C and was purified on Qiaquick PCR Cleanup columns (Qiagen). Transcription reactions (100 μL) containing 1 μg of linearized template and 100 units of T7 RNA polymerase (Ambion) were incubated at 37°C for 1 h, followed by treatment with Turbo DNase (Ambion) and extraction with phenol:chloroform:isoamyl alcohol. Products were ethanol precipitated in the presence of sodium acetate. Pellets were resuspended in formamide loading buffer and resolved on 6% denaturing acrylamide gels. Full-length product (802 nt) was excised and isolated in the same manner as the cleaved S33T7.
Full-length transcript was cleaved with a 1.5 molar excess of a 10–23 deoxyribozyme (5′-CAGATGAAGGCTAGCTACAACGATTCAGGGT-3′), and the cleavage product (199 nt) with a 2′,3′-cyclic phosphate was purified on a 6% denaturing acrylamide gel. Following ethanol precipitation, the substrate was ligated to the preadenylated oligonucleotide Linker 1 with A. thaliana tRNA ligase. Reactions were precipitated with sodium acetate and ethanol. Pellets were resuspended in water, and the reactions were split in half and incubated with calf intestinal phosphatase (24 U; NEB) at 37°C for 30 min (McCraith and Phizicky 1990) or 50% glycerol. RNA samples were resuspended in formamide loading buffer and purified on a 6% denaturing acrylamide gel. The gel was stained with SYBR Gold, and ligated RNA was visualized by blue light transillumination (Clare Chemical Research). Ligated RNA was excised from the gel and isolated as described previously. Resuspended RNA templates were reverse transcribed in 20-μL reactions with Superscript III (Invitrogen) and one of two primers (CFP 1023.revII: 5′-CTTGCCGTAGGTGGCATC-3′ or linker1.rev: 5′-GATTGATGGTGCCTACAG-3′). cDNA templates (10%) were PCR amplified with Taq DNA Polymerase (Promega) in 50-μL reactions using the following program: 35 cycles of (94°C for 30 sec, 50°C for 30 sec, 72°C for 20 sec) with CFP 1023.fwd (5′-GTCGACATGGTGAGCAAGG-3′) and the oligonucleotide used as the primer for reverse transcription. PCR reactions were resolved on a 1% agarose gel and visualized by ethidium bromide staining and ultraviolet transillumination.
A. thaliana tRNA ligase reaction library preparation
Total RNA (4.6 μg) from human brain (Ambion) was incubated with A. thaliana tRNA ligase in ligation buffer in the presence of 1 mM ATP and 50 pmol of adaptor (SLXA PE; RNA oligo: 5′-phosphate-AGAUCGGAAGAGCGUCGUGUAGGGAAAGAGUGUAGAUCUCGGUGGUCGCC-3′). Ligation reactions were precipitated, resuspended in formamide loading buffer, and purified on a 6% denaturing acrylamide gel. The acrylamide gel was stained with SYBR Gold and visualized by blue light transillumination. RNAs larger than 86 nt were excised, eluted from the acrylamide gel, and ethanol precipitated. Pellets were resuspended in Tpt1 reaction buffer (20 mM Tris-HCl at pH 7.5, 5 mM MgCl2, 2.5 mM spermidine, 0.1 mM DTT, and 0.4% Triton X-100) (Culver et al. 1997) and incubated with Tpt1 (10 μM) in the presence of NAD+ (1 mM) at 30°C for 1 h (Steiger et al. 2001). RNA was ethanol precipitated and chemically fragmented (RNA fragmentation kit, Ambion) at 70°C for 2 min. Fragmented RNA was ethanol precipitated and reverse transcribed using a primer complementary to the ligated adaptor (SLXA PE RT oligo: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTC-3′). cDNAs were treated with exonuclease I (40 U; USB) and shrimp alkaline phosphatase (8 U; USB) for 15 min at 37°C, followed by phenol:chloroform extraction and ethanol precipitation. RNA was hydrolyzed with 10 mM sodium hydroxide for 5 min, and the reactions were neutralized with an equal volume of 10 mM HCl prior to ethanol precipitation (Troutt et al. 1992).
cDNA was ligated with preadenylated Linker-1 (5′-rApp-CTGTAGGCACCATCAAT-ddC-3′) and T4 RNA ligase 1 (NEB) in the absence of ATP (Troutt et al. 1992). Following ligation, reactions were phenol:chloroform extracted, ethanol precipitated in the presence of GlycoBlue, resuspended in formamide loading buffer, and purified on a 6% denaturing acrylamide gel. The gel was stained with SYBR Gold and visualized by blue light transillumination. DNAs between 100 and 250 nt were excised, eluted from the acrylamide gel, and ethanol precipitated. Precipitated cDNA was PCR amplified with Phusion DNA polymerase (NEB) using primers specific to the ligated linkers (PE SLXA Linker 1 AMP: 5′-CAAGCAGAAGACGGCATACGAGATGATTGATGGTGCCTACAG-3′ and KS OCA216: 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′). PCR products between 100 and 250 bp were selected on a 6% native TBE acrylamide gel and eluted (Quail et al. 2008).
High-throughput sequencing
DNA libraries were sequenced on a Genome Analyzer II (Illumina), collecting 36-base reads from a single end using a sequencing primer (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′) complementary to the tRNA ligase adaptor. A total of 6,035,594 36-base sequence reads were collected in a single lane and mapped to the March 2006 version of the human genome (hg18) using the short-read aligner bowtie, version 0.10.1 (Langmead et al. 2009). For alignment, we trimmed nine bases from the 3′ end of the reads (option “–3 9”), as many of reads contain linker sequences, and disallowed reads that mapped to more than 100 positions in the genome (option “-m 100”). Data were visualized in the UCSC genome browser (Karolchik et al. 2008), and genomic positions relative to RNA features were determined.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Eric Phizicky and Roy Parker for comments on the manuscript, Hildburg Beier (University of Wurzburg) for providing a plasmid encoding the A. thaliana tRNA ligase gene, and Michael Robertson (Scripps Research Institute) for technical advice. The research was funded in part by the NIH (P41 RR11823). S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1934910.
REFERENCES
- Abelson J, Trotta CR, Li H. tRNA splicing. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- Apostol BL, Greer CL. Preferential binding of yeast tRNA ligase to pre-tRNA substrates. Nucleic Acids Res. 1991;19:1853–1860. doi: 10.1093/nar/19.8.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Rettig WJ, Yenamandra AK, Kozak CA, Chaganti RS, Posner JB, Old LJ. Cerebellar degeneration-related antigen: A highly conserved neuroectodermal marker mapped to chromosomes X in human and mouse. Proc Natl Acad Sci. 1990;87:3077–3081. doi: 10.1073/pnas.87.8.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Culver GM, McCraith SM, Consaul SA, Stanford DR, Phizicky EM. A 2′-phosphotransferase implicated in tRNA splicing is essential in Saccharomyces cerevisiae. J Biol Chem. 1997;272:13203–13210. doi: 10.1074/jbc.272.20.13203. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Shankar V. Ribonucleases from T2 family. Crit Rev Microbiol. 2002;28:79–122. doi: 10.1080/1040-840291046704. [DOI] [PubMed] [Google Scholar]
- Dong B, Niwa M, Walter P, Silverman RH. Basis for regulated RNA cleavage by functional analysis of RNase L and Ire1p. RNA. 2001;7:361–373. doi: 10.1017/s1355838201002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Hu Y, Rolfs A, Bhullar B, Murthy TV, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17:536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res. 2005;33:6349–6360. doi: 10.1093/nar/gki934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Licht K, Medenbach J, Lührmann R, Kambach C, Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA. 2008;14:1532–1538. doi: 10.1261/rna.1129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- McCraith SM, Phizicky EM. A highly specific phosphatase from Saccharomyces cerevisiae implicated in tRNA splicing. Mol Cell Biol. 1990;10:1049–1055. doi: 10.1128/mcb.10.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- Pick L, Furneaux H, Hurwitz J. Purification of wheat germ RNA ligase. II. Mechanism of action of wheat germ RNA ligase. J Biol Chem. 1986;261:6694–6704. [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzi F, Caffarelli E, Laneve P, Bozzoni I, Brunori M, Vallone B. The structure of the endoribonuclease XendoU: From small nucleolar RNA processing to severe acute respiratory syndrome coronavirus replication. Proc Natl Acad Sci. 2006;103:12365–12370. doi: 10.1073/pnas.0602426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz RC, Greer CL, Gegenheimer P, Abelson J. Enzymatic mechanism of an RNA ligase from wheat germ. J Biol Chem. 1983;258:8374–8383. [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. 2004. http://www.repeatmasker.org.
- Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger MA, Kierzek R, Turner DH, Phizicky EM. Substrate recognition by a yeast 2′-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog. Biochemistry. 2001;40:14098–14105. doi: 10.1021/bi011388t. [DOI] [PubMed] [Google Scholar]
- Steiger MA, Jackman JE, Phizicky EM. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: Evidence for a conserved two-step reaction mechanism. RNA. 2005;11:99–106. doi: 10.1261/rna.7194605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009a;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009b;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troutt AB, McHeyzer-Williams MG, Pulendran B, Nossal GJ. Ligation-anchored PCR: A simple amplification technique with single-sided specificity. Proc Natl Acad Sci. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM. The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J Biol Chem. 2004;279:36819–36827. doi: 10.1074/jbc.M405120200. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SY. Complementary DNA libraries: An overview. Mol Biotechnol. 2004;27:245–252. doi: 10.1385/MB:27:3:245. [DOI] [PubMed] [Google Scholar]