Abstract
In situ hybridization is widely used to visualize transcribed sequences in embryos, tissues, and cells. For whole mount detection of mRNAs in embryos, hybridization with an antisense RNA probe is followed by visual or fluorescence detection of target mRNAs. A limitation of this approach is that a cDNA template of the target RNA must be obtained in order to generate the antisense RNA probe. Here we investigate the use of short (12–24 nucleotides) locked nucleic acid (LNA) containing DNA probes for whole mount in situ hybridization detection of mRNAs. Following extensive protocol optimization, we show that LNA probes can be used to localize several mRNAs of varying abundances in chicken embryos. LNA probes also detected alternatively spliced exons that are processed in a tissue specific manner. The use of LNA probes for whole mount in situ detection of mRNAs will enable in silico design and chemical synthesis and will expand the general use of in situ hybridization for studies of transcriptional regulation and alternative splicing.
Keywords: chicken embryo; in situ hybridization; mRNA detection; locked nucleic acids, LNA
INTRODUCTION
Whole mount in situ hybridization (ISH) detection of mRNAs in embryos is commonly performed using antisense RNA probes that are transcribed from a cDNA copy of the target mRNA (Nieto et al. 1996). Incorporation of digoxygenin (DIG)-labeled nucleotides (nt) into the antisense RNA every 10–15 nt provides an immunological substrate for probe detection. Since probes are generally at least several hundred nucleotides, many DIG molecules are available for detection per target mRNA. Although this approach is highly sensitive, a significant limitation is that a cDNA for the target mRNA must be obtained through cloning, from cDNA repositories, or from colleagues. Obtaining cDNAs is time consuming, and because certain sequences are underrepresented in cDNA libraries, some cDNAs are exceedingly difficult to acquire. RNA probes are also easily degraded by ubiquitous RNases and are not generally effective if shorter than 300 nt in length, often making them unsuitable for discriminating between splice variants of the same gene.
The ability to use short (12–24) oligonucleotide sequences for ISH detection of mRNAs would eliminate the need for a cDNA template and permit in silico design and chemical synthesis of probes. Oligonucleotide probes would also allow detection of short individual exons in alternatively spliced transcripts. Some studies have tested using short DNA oligonucleotides for microRNA and mRNA detection (Valoczi 2004; Thomsen 2005; Kloosterman et al. 2006). However, hybridization signal is low due to relatively poor affinity of short DNA probes to target mRNA sequences and the inability to incorporate more than one or two DIG molecules per probe. As a consequence, short DNA oligonucleotides have not proven generally useful for whole mount ISH analysis.
Locked nucleic acids (LNAs) are modified nucleotides that contain a methylene bridge between the 2′O and the 4′C on the ribose ring that “locks” the structure into a high binding-affinity, RNA-mimicking conformation (Koshkin et al. 1998; Wengel et al. 2003). DNA oligonucleotide probes containing LNAs at every third position (LNA probes) show dramatically enhanced hybridization specificity and duplex stability (Mctigue et al. 2004). LNA probes are 10-fold more sensitive than DNA probes for Northern analysis and give significantly increased signal for FISH at the subcellular level (Valoczi 2004; Thomsen 2005). LNA probes have also been used to detect 21–22-nt microRNAs by whole mount ISH and in tissue sections (Weinholds et al. 2005; Ason et al. 2006; Darnell et al. 2006; Kloosterman et al. 2006; Nelson et al. 2006; Obernosterer et al. 2007; Silahtaroglu et al. 2007; Nuovo 2008; Pena et al. 2009) and RNAs in yeast cells (Valoczi 2004; Thomsen 2005). For ISH applications LNAs typically show single nucleotide specificity (Kloosterman et al. 2006). The unprecedented specificity of LNA probes for their target sequences have made them useful for SNP analyses (Jacobsen et al. 2002; Latorra et al. 2003; Johnson et al. 2004).
Despite these attributes, the use of LNAs to detect mRNAs by whole mount ISH in vertebrate embryos has not been reported. mRNAs are generally much less abundant than microRNAs, presenting technical challenges related to sensitivity of detection with short probes that contain one or at most a few DIG molecules. Here we explore the use of short (12–24 nt) LNA probes to detect mRNAs by whole mount ISH in chicken embryos. Following extensive protocol optimization, we find that LNA probes designed in silico can readily detect mRNAs of varying abundances in developing chicken embryos. LNA probes can also be used to detect individual exons in alternatively spliced transcripts.
RESULTS AND DISCUSSION
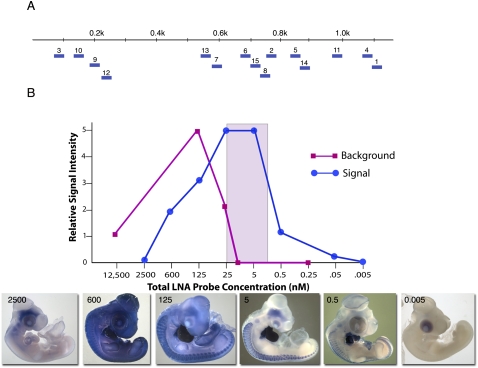
To evaluate the feasibility of using short (20–25 nt) LNA-containing oligonucleotides as hybridization probes for detection of mRNAs by in whole mount ISH, 15 LNA probes were designed against the chicken cardiac troponin T (cTNT) mRNA (Fig. 1A; Supplemental Fig. S1) using the Primer3 software (Untergasser et al. 2007). cTNT is highly expressed in developing heart and skeletal muscle cells of chicken embryos (Cooper and Ordahl 1984; Antin et al. 2002) and so provides an abundant and highly localized target for protocol optimization. Each LNA probe was labeled on the 5′ and 3′ end with one DIG molecule, and an equimolar mixture of all 15 LNA probes was tested across a 2.5 million–fold concentration range for the ability to detect cTNT mRNAs in day 4–5 (d4–5) chicken embryos.
FIGURE 1.
(A) Location of l5 LNA probes along the cTNT mRNA. (B) Graph depicting relative signal and background intensities as a function of LNA probe concentration. An equimolar mixture of the 15 LNA probes shown in A was used for hybridizations. The colored rectangle identifies the probe concentration range over which high signal and low background was obtained. Embryo images show staining patterns obtained at the indicated probe concentrations. cTNT expression is visible in the heart and in the skeletal muscle cells of the somites along the dorsal sides of embryos.
ISH expression patterns with high signal in the heart and skeletal muscle cells of somites and low background were obtained over a relatively narrow probe concentration range of 0.5–25 nM (Fig. 1B). Lower probe concentrations showed reduced signal, while higher concentrations gave increased background. At the highest probe concentrations tested, both signal and background were lost due to competitive binding of anti-DIG antibody by LNA probes that continued to diffuse from the embryo into the buffer even after extended washing (data not shown).
Sensitivity of probe detection is a critical parameter for visualizing mRNAs using LNA probes. For standard antisense RNA probes, DIG-labeled nucleotides are incorporated on average every 10–15 nt. A 1-kb antisense RNA probe may therefore contain 75 or more DIG molecules, which are visualized using a DIG antibody conjugated to alkaline phosphatase. For LNA probes, only one DIG molecule can be added to each end of the probe, resulting in just two DIG molecules hybridizing to each target mRNA per probe. Tailing reactions in which a long string of nucleotides is added to the end of an LNA have been used to detect microRNAs in sections (Johnson et al. 2007); however, tailed probes produce high nonspecific signal in some tissues when used for whole mount ISH (data not shown).
To maximize hybridization signal, we next optimized several aspects of the ISH protocol. A hybridization time course using the mixture of 15 cTNT LNA probes showed that faint signal was observed after just 30 min and that significant signal was observed after 16 h of hybridization (Supplemental Fig. S2A). Longer hybridization times up to 120 h yielded a modest graded increase in hybridization signal. For all following studies, we chose 48 h as a reasonable intermediate time for hybridization.
Other parameters were also optimized. Since background limits the upper concentration of probe that can be used, washing conditions were examined. Extensive analyses revealed that addition of Tween-20 and CHAPS to the Triton X-100 containing wash solution significantly reduced background, presumably by enhancing the rate that unbound LNA probes are removed.
Using an optimized protocol, we then tested whether we could detect cTNT transcripts in embryos using single LNA probes. Hybridizations were carried out at 22°C below the predicted Tm for each probe (Supplemental Fig. S1). Washing times and conditions were identical for all probes, and the staining reaction was performed for 1 h so that results could be directly compared. Each of the 15 LNA probes gave specific hybridization signal (Fig. 2A), although signal and background levels varied significantly between probes. Since in all cases background could be eliminated with extended washing, the specific probe sequence appeared to affect diffusion of unbound LNA probes from the embryo. Obvious correlations were not identified between background or signal intensity and probe sequences, melting temperatures, or the location of probes in relation to RNA secondary structure.
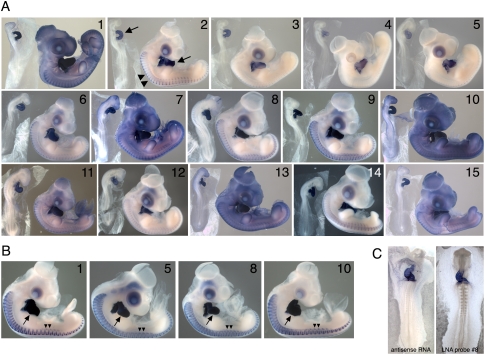
FIGURE 2.
(A) ISH patterns for each of the 15 LNA probes shown in Figure 1A. Hybridization was carried out at 22°C below the predicted Tm for each probe. Washing and staining times were the same for all probes. For each probe, a stage 14 and stage 22–24 embryo is shown. (B) Expression patterns obtained using the indicated LNA probes when washing and staining was optimized for each probe. Arrows indicate the heart; double arrowheads point to staining within the skeletal muscle cells of the somites. (C) Comparison of hybridization signal obtained with a 1.17-kb cTNT antisense probe versus cTNT LNA probe 8.
Hybridizations were repeated using four individual LNA probes that showed different signal and background levels in the experiment above (Fig. 2A, probes 1,5,8,10). Hybridizations were carried out for 48 h at 22°C below the Tm for each probe. Staining and washing times were varied to optimize signal for each probe. Initial staining reactions were performed at room temperature and were terminated when background appeared. The embryos were then washed overnight at 4°C, and the staining–washing cycle was repeated until a high level of signal was evident. Despite the differences observed when all probes were washed and stained identically (Fig. 2A), when handled individually high level signal and low background was obtained for each probe (Fig. 2B). The hybridization signals obtained with LNA probe 8 was similar to the signal obtained using a 1.17-kb cTNT antisense RNA probe (Fig. 2C).
Using the LNA probe 8 as a starting sequence, the effect of probe length on hybridization signal was assessed. Signal obtained with single-DIG labeled LNA probes of 12, 14, 16, 18, 20, and 22 nt were compared. Although the 12-nt probe failed to produce hybridization signal, signal intensity obtained with probes 14–22 nt in length was indistinguishable (Supplemental Fig. S2B). The 24-nt double DIG labeled probe produced significantly higher signal than the 22-nt LNA probe containing one DIG molecule.
We also evaluated probe specificity by comparing the hybridization signal obtained with cTNT probe 8 versus probes containing one to four mutations (Supplemental Fig. S1). Results showed that a single internal substitution of an LNA nucleotide significantly reduced hybridization signal, while two or more LNA nucleotide substitutions abolished signal (Supplemental Fig. S3).
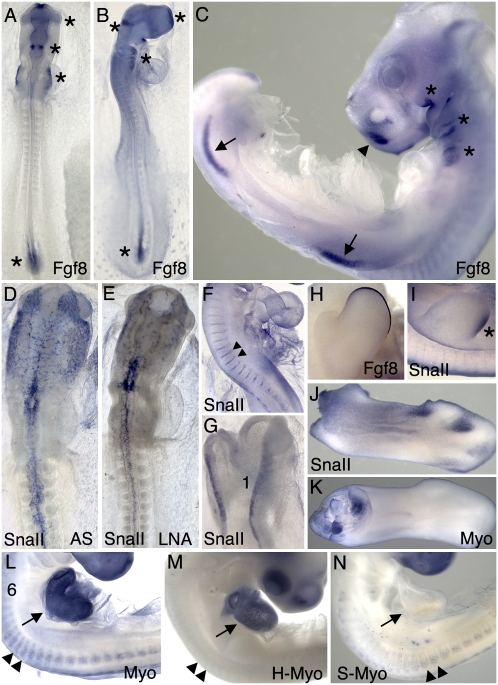
We next designed LNA probes for two less abundant mRNAs that code for proteins having important functions during embryonic development (Supplemental Fig. S1). FGF8 is a member of the fibroblast growth factor family involved in cell cycle regulation, differentiation, and patterning, and also in the pathology of some cancers (Dorkin et al. 1999; Alsan and Schultheiss 2002; Karabagli et al. 2002a,b; Gimeno and Martinez 2007). Snail II (SNAII) is a zinc finger transcription factor that acts as a transcriptional repressor (Nieto 2002). SNAII is expressed in numerous cell types and structures during embryonic development, including premigratory neural crest cells and intersomitic arteries (Ladher et al. 2000; Marin and Nieto 2004). As shown in Figure 3, FGF8 LNA probes produced highly specific staining in the rhombomeres, hindbrain, and tail bud, as well as in the pharyngeal arches and limb bud apical ectodermal ridges. These expression patterns are identical to the published FGF8 expression patterns obtained with antisense RNA probes. LNA probes for SNAII also revealed the known expression patterns of this gene in the migrating neural crest, intersomitic blood vessels, and neural folds (Fig. 3D,F,G). Hybridization signal obtained with one LNA probe was only slightly lower than obtained when three LNAs were combined (Fig. 3E,I,J). These findings demonstrate that a single LNA probe containing two DIG molecules can be used to detect mRNAs in developing embryos by whole mount ISH.
FIGURE 3.
Visualization of FGF8, SNAII, and MYOM1 expression patterns with LNA probes. (A–C) Two LNA probes for FGF8 revealed expression in the frontonasal prominence, isthmus, future maxillomandibular and pharyngeal arch region, primitive streak, and posterior segmental plate mesoderm at HH stage 11 (A), stage 14 (B), and stage 20 (C). At stage 20 and 23, expression was also observed in the apical ectodermal ridge of the limb buds (C, two LNA probes; H, one LNA probe). Using three LNA probes for SNAII, expression pattern and intensity were similar to that obtained with an 850-nt antisense RNA probe (cf. D and E). A single LNA probe also detected SNAII expression in the intersomitic arteries at stage 15 (F) and in the neural crest at stage 11 (G). SNAII expression was also detected in the caudal wing bud at stage 23 (I) and in the interdigital webbing of the limb at stage 31 (J). Six myomesin LNAs, three from the common region and three from the EH exon, labeled dorsal and ventral muscle masses from a stage 30 wing (K) and the somitic myotome (arrowheads) and heart myocardium (arrow) at stage 19 (L). Three LNAs to the cardiac-specific EH exon labeled only the heart at stage 20 (M), whereas two skeletal-exon specific LNA probes labeled only the somite myotome in a stage 19 embryo (N).
Another potential advantage of using short LNA probes is the ability to detect alternatively spliced, differentially expressed exons. To investigate the feasibility of detecting individual exons with LNA probes, LNA probes were designed against alternatively spliced exons in the myomesin (MYOM1) RNA. MYOMI is a component of the striated myofibril in skeletal and cardiac muscle cells and has cardiac-specific and skeletal-predominant isoforms (H- and S-myomesin, respectively) that are generated by alternative splicing. Several LNA probes were designed against exons found only in the H- or S- MYOMI mRNAs (Supplemental Fig. S1). LNA probes were also designed against the common regions of the mRNA. LNA probes recognizing common MYOMI RNA sequences visualized MYOMI RNAs in both the somites and heart (Fig. 3L). Three LNA probes recognizing the H-specific MYOMI exon detected mRNAs only in the heart, while two LNAs recognizing the S-specific exons detected mRNAs only in skeletal muscle cells of the somites (Fig. 3M,N).
The results presented here show that LNA probes are effective for detecting a variety of mRNAs in chicken embryos. Following careful optimization of probe concentration and hybridization parameters, abundant and moderately abundant mRNAs coding for structural, signaling, and transcriptional regulatory proteins can be detected. The ability to detect mRNA sequences, including individual exons, using short LNA probes allows for the localization of specific splice variants and allows probes to be designed in silico and chemically synthesized, eliminating the need for a cDNA template.
MATERIALS AND METHODS
Embryo collection and preparation
Fertile chicken eggs (HyLine; not a commercially available source) were incubated in a forced-draft, humidified incubator at 37.5°C for 2–5 d, depending on the stages desired. Embryos were collected into chilled chick saline (123 mM NaCl), removed from the vitelline membrane, and cleaned of yolk. Extra-embryonic membranes and large body cavities (brain vesicles, atria, allantois, eye) were opened to minimize trapping of the in situ reagents. Embryos were fixed overnight at 4°C in fresh 4% paraformaldehyde.
Embryos were rinsed in PBS, then in PBS plus 1% Tween-20 (PBT), and dehydrated by steps (25%, 50%, 75%, 100%, 100%) into methanol before being cooled to −20°C overnight (or up to 10 d). Rehydration reversed this series. Embryos were rinsed twice in PBS, and older embryos were treated with proteinase K: stages 8–13 and 14–18 at 10 μg/mL of proteinase K for 10 and 20 min, respectively; stages 19 and older at 20 μg/mL of proteinase K for 20 min. Embryos were rinsed repeatedly in PBT to stop the digestion and were then transferred to prehybridization solution (see below). Embryos were stored until use either at the methanol step or in prehybridization at −20°C for fewer than 10 d.
Probe design
LNA modified DNA oligonucleotides probes, containing an LNA nucleotide at every third position, and labeled at the 5′ end only, or at the 5′ and 3′ ends, with DIG, were supplied by Exiqon, Inc. Optimization studies by others have shown that an LNA nucleotide at every third position provides the best balance between increased annealing characteristics, background, and hybridization temperature (Karabagli et al. 2002b; Valoczi 2004). Probes were designed using the Primer3 primer design program (Untergasser et al. 2007) and checked using the LNA Oligo Optimizer tool on the Exiqon website (www.exiqon.com). Each probe sequence was screened against all known chicken sequences using BLAST. LNA probes typically show single nucleotide specificity (Kloosterman et al. 2006), nevertheless probes showing significant homology with other chicken sequences were eliminated from consideration.
In situ hybridization
Prepared embryos were transferred to prehybridization solution containing the following: 50% formamide, 5× SSC, 2% blocking powder, 0.1% Tween-20, 0.1% CHAPS, 50 μg/mL yeast RNA, 5 mM EDTA, 50 μg/mL heparin, and DEPC water. Prehybridizations and hybridizations were carried out 22°C below the predicted melting temperature of the LNAs, or at the indicated temperatures. LNA probes were added to 1 mL fresh prehyb buffer and hybridization carried out for 48 h. Following hybridization, embryos were transferred to wash solution containing 2× SSC, 0.1% Chaps prewarmed to the hybridization temperature and washed three times for 20 min. This was followed by three times for 20-min washes in 0.2× SSC, 0.1% CHAPS. Embryos were rinsed twice in KTBT (50 mM Tris at pH 7.5, 150 mM NaCl, 10 mM KCl, 1% Tween-20) and then incubated in 20% sheep serum in KTBT at 4°C for 2–3 h or longer. Anti-DIG antibody binding (1:2000–1:4000) was carried overnight at 4°C on a nutator shaker. Following antibody incubation, embryos were washed five times for 1 h in KTBT, but often including overnight at 4°C. Embryos were then washed twice 10 min in NTMT (100 mM NaCl, 100 mM Tris at pH 9.5, 50 mM MgCl2, 0.1% Tween-20). Color reactions were carried out in NTMT containing 3 μL of 75 mg/mL NBT in dimethylformamide (DMF) plus 2.3 μL of 50 mg/mL BCIP in DMF per milliliter of NTMT. Staining reactions were carried out for 1–6 h at room temperature on a nutator until signal or background became visible, followed by overnight washing in KTBT. A second or third round of color reaction followed until each probe had yielded strong signal. Labeling reactions were permanently terminated by washing with PBS. Embryos were then dehydrated through a graded methanol series to remove background and enhance signal and then rehydrated and stored in PBS plus 0.1% sodium azide. Embryos were photographed on a Leica PlanApo stereomicroscope using a digital acquisition system and transmitted, lateral and/or direct illumination.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Exiqon, Inc., for providing some of the LNA probes. We also thank Maricela Pier and Terry Sesepasara for technical assistance. This work was supported by NIH grant HD044767 to P.B.A.
Footnotes
Abbreviations: LNA, locked nucleic acids; DIG, digoxygenin.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1775610.
REFERENCES
- Alsan BH, Schultheiss TM. Regulation of avian cardiogenesis by Fgf8 signaling. Development. 2002;129:1935–1943. doi: 10.1242/dev.129.8.1935. [DOI] [PubMed] [Google Scholar]
- Antin PB, Zhang W, Bales MA, Garriock R, Yatskievych TA. Precocious expression of cardiac troponin T during early avian embryogenesis. Dev Dyn. 2002;225:135–141. doi: 10.1002/dvdy.10148. [DOI] [PubMed] [Google Scholar]
- Ason B, Darnell DK, Wittbrodt B, Berezikov E, Kloosterman WP, Wittbrodt J, Antin PB, Plasterk RH. Differences in vertebrate microRNA expression. Proc Natl Acad Sci. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Ordahl CP. A single troponin T gene regulated by different programs in cardiac and skeletal muscle development. Science. 1984;226:979–982. doi: 10.1126/science.6095446. [DOI] [PubMed] [Google Scholar]
- Darnell DK, Kaur S, Stanislaw S, Konieczka JK, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;236:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- Dorkin TJ, Robinson MC, Marsh C, Bjartell A, Neal DE, Leung HY. FGF8 over-expression in prostate cancer is associated with decreased patient survival and persists in androgen independent disease. Oncogene. 1999;18:2755–2761. doi: 10.1038/sj.onc.1202624. [DOI] [PubMed] [Google Scholar]
- Gimeno L, Martinez S. Expression of chick Fgf19 and mouse Fgf15 orthologs is regulated in the developing brain by Fgf8 and Shh. Dev Dyn. 2007;236:2285–2297. doi: 10.1002/dvdy.21237. [DOI] [PubMed] [Google Scholar]
- Jacobsen N, Bentzen J, Meldgaard M, Jakobsen MH, Fenger M, Kauppinen S, Skouv J. LNA-enhanced detection of single nucleotide polymorphisms in the apolipoprotein E. Nucleic Acids Res. 2002;30:e100. doi: 10.1093/nar/gnf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MP, Haupt LM, Griffiths LR. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32:e55. doi: 10.1093/nar/gnh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Karabagli H, Karabagli P, Ladher R, Schoenwolf G. Comparison of the expression patterns of several fibroblast growth factors during chick gastrulation and neurulation. Anat Embryol (Berl) 2002a;205:365–370. doi: 10.1007/s00429-002-0264-7. [DOI] [PubMed] [Google Scholar]
- Karabagli H, Karabagli P, Ladher RK, Schoenwolf GC. Survey of fibroblast growth factor expression during chick organogenesis. Anat Rec. 2002b;268:1–6. doi: 10.1002/ar.10129. [DOI] [PubMed] [Google Scholar]
- Kloosterman W, Wienholds E, De Bruijn E, Kauppinen S, Plasterk R. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine, and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- Ladher RK, Church VL, Allen S, Robson L, Abdelfattah A, Brown NA, Hattersley G, Rosen V, Luyten FP, Dale L, et al. Cloning and expression of the Wnt antagonists Sfrp-2 and Frzb during chick development. Dev Biol. 2000;218:183–198. doi: 10.1006/dbio.1999.9586. [DOI] [PubMed] [Google Scholar]
- Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22:79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- Marin F, Nieto MA. Expression of chicken slug and snail in mesenchymal components of the developing central nervous system. Dev Dyn. 2004;230:144–148. doi: 10.1002/dvdy.20027. [DOI] [PubMed] [Google Scholar]
- Mctigue PM, Peterson RJ, Kahn JD. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)–DNA duplex formation. Biochemistry. 2004;43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Patel K, Wilkinson DG. Methods in cell biology. Vol 51. Academic Press; New York: 1996. In situ hybridization analysis of chick embryos in whole mount and tissue sections. [DOI] [PubMed] [Google Scholar]
- Nuovo GJ. In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods. 2008;44:39–46. doi: 10.1016/j.ymeth.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silahtaroglu AN, Nolting D, Dyrskjot L, Berezikov E, Moller M, Tommerup N, Kauppinen S. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- Thomsen R. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11:1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A. Sensitive and specific detection of microRNAs by Northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wengel J, Petersen M, Frieden M, Troels K. Chemistry of locked nucleic acids (LNA): Design, synthesis, and bio-physical properties. Lett Pept Sci. 2003;10:237–253. [Google Scholar]