Abstract
We present a simple and fast method for large-scale purification of RNA oligonucleotides suitable for biochemical and structural studies. RNAs are transcribed in vitro with T7 RNA polymerase using linearized plasmid DNA templates. After addition of EDTA, the crude transcription reaction is subjected directly to weak anion-exchange chromatography using DEAE-sepharose to separate the T7 RNA polymerase, unincorporated rNTPs, small abortive transcripts, and the plasmid DNA template from the desired RNA product. The novel method does neither require tedious phenol/chloroform extraction of the T7 RNA polymerase nor denaturation of the RNA, which is desirable especially for larger RNAs. In addition, isotopically labeled rNTPs can be easily recycled from the column flow-through and oligomeric RNA aggregates can be separated from the natively folded monomeric RNA product.
Keywords: native RNA purification, in vitro transcription, fast performance liquid chromatography (FPLC), diethylaminoethyl (DEAE) sepharose
INTRODUCTION
Over the past decade, new roles for RNA in biology have emerged and established a central role for RNA in the post-transcriptional regulation of gene expression through RNA interference (RNAi), nonprotein coding regulatory RNAs or cis-acting transport, and translational control elements in mRNAs (Sharp 2009). Many of these regulatory RNA elements conserve the potential to fold into complex structures suggesting that their function is at least partly encoded in their tertiary fold. Thus, a thorough biochemical characterization combined with structural analysis by X-ray crystallography and NMR spectroscopy is imminent if we aim to understand how these RNAs perform their biological functions. Milligram quantities of RNA required for biochemical and structural studies can be easily prepared by in vitro transcription from DNA templates using T7 RNA polymerase, but purification of the final RNA product still remains rather time-consuming (Milligan et al. 1987; Price et al. 1995; Doudna 1997). Preparative denaturing polyacrylamide gel electrophoresis (PAGE) is still most commonly used to purify transcribed RNAs. Although this method provides nucleotide resolution up to 100 nucleotides (nt) RNAs, it is both time-consuming and denatures the RNA, which can lead to misfolded or aggregated RNA species after elution from the denaturing gel (Uhlenbeck 1995; Lukavsky and Puglisi 2004). Alternative methods using anion-exchange HPLC to purify chemically synthesized RNA oligonucleotides or to separate homogeneous-length RNAs from trans-cleavage reactions with a hammerhead ribozyme are typically limited to small RNAs (<40 nt) (Anderson et al. 1996; Shields et al. 1999).
Therefore, several native RNA purification schemes have been developed over the past years (Kieft and Batey 2004; Lukavsky and Puglisi 2004; Batey and Kieft 2007; Kim et al. 2007; McKenna et al. 2007). These methods use linearized plasmid DNA templates for large-scale in vitro transcription and do separate the desired RNA oligonucleotide from the transcription mixture using size-exclusion chromatography or affinity chromatography via ribozyme-cleavable RNA affinity purification tags. Large scale affinity-purification developed by Batey and Kieft uses tandem MS2 coat protein-binding stem–loops (Lim and Peabody 1994; LeCuyer et al. 1995) as affinity tags at the 3′ end of the RNA transcript, which bind to the Ni2+-affinity column matrix via interaction with a hexahistidine-tagged MBP-MS2 coat protein fusion (Zhou et al. 2002). This RNA affinity tag is preceded by the cis-acting glmS ribozyme sequence (Winkler et al. 2004), which can be activated by glucosamine-6-phosphate to elute the desired RNA product (Batey and Kieft 2007). This robust affinity-purification protocol allows the production of milligram quantities of native RNAs with homogeneous 3′ ends, which can be essential for crystallographic applications. Oligomeric RNA species, which can form during transcription, are not separated from the monomeric species by this technique and would require additional purification by size-exclusion chromatography. Furthermore, over 200 nt of the RNA transcript comprise the ribozyme and affinity tag, which in turn significantly lowers the final yield of the desired RNA product per milliliter of transcription reaction. This is obviously not desirable for NMR spectroscopic applications, which require isotope labeling with expensive 13C,15N-labeled or deuterated rNTPs and, therefore, demand high transcription yields per milliliter of reaction to minimize the cost of RNA production.
Size-exclusion chromatography-based purification using high-resolution fast performance liquid chromatography (FPLC) systems efficiently removes unincorporated ribonucleotide triphosphates (rNTPs), small abortive transcripts, and the high molecular weight plasmid DNA template from the RNA product under native conditions (Kim et al. 2007; McKenna et al. 2007). While this method is limited in its ability to separate transcribed RNAs with heterogeneous 3′ ends, it permits separation of oligomeric species from the desired monomeric RNA required for most biochemical and structural characterizations. Despite these advantages, the use of size-exclusion chromatography still requires additional, time-consuming steps prior to its application. Multiple phenol/chloroform extractions are needed to remove the T7 RNA polymerase and optimal resolution during size-exclusion chromatography requires desalting of the transcription reaction and concentration of the sample to reduce the loading volume.
Here, we present a method, routinely used in our lab, for rapid, large-scale purification of native, structurally homogeneous RNA suitable for biochemical and structural studies. RNAs are prepared by in vitro transcription using T7 RNA polymerase and linearized plasmid DNA templates, and after addition of EDTA, the crude transcription reaction is applied directly to weak anion-exchange chromatography using DEAE-sepharose without further manipulation. T7 RNA polymerase and unincorporated rNTPs, which do not bind to the column matrix, are found in the flow-through. Small abortive transcripts, the desired RNA product and the plasmid DNA template are separated on the DEAE column over a shallow salt gradient. This, easy to implement, native RNA purification method not only eliminates the need for tedious phenol/chlorofom extraction of the T7 RNA polymerase, but also allows the isotopically labeled rNTPs to be easily recycled, and in addition, oligomeric RNA species can be separated from monomeric RNA, based on their overall difference in charge.
RESULTS AND DISCUSSION
Plasmid DNA template design and in vitro transcription
Large-scale in vitro transcription requires milligram amounts of plasmid DNA template coding for the RNA oligonucleotide of interest and therefore high copy number vectors, such as pUC18 should be used as described previously (Lukavsky and Puglisi 2004). Use of plasmid DNA as a template for in vitro transcription rather than synthetic DNA oligonucleotide, improves the yield for RNA transcripts, larger than 30 nt and is crucial for size-based and charge-based separation of the DNA template from the desired RNA transcript (McKenna et al. 2007). Maximum yield of the large-scale transcription reaction is achieved at optimal magnesium chloride concentration (typically, 20–25 mM). Inclusion of as little as 1 U of pyrophosphatase per milliliter of transcription reaction (Fig. 1A, cf. lanes 3 and 5) also increases the yield by inhibiting buildup of inorganic pyrophosphate, which traps magnesium ions at a molar ratio of 2:1 of Mg:PPi and, in turn, significantly slows down the transcription rate (Kern and Davis, 1997; Lukavsky and Puglisi 2004). Finally, prolonged incubation times of the in vitro RNA transcription reactions (up to 4 h) can also help to improve the yield, however, for certain RNAs the likelihood of hydrolysis will also increase.
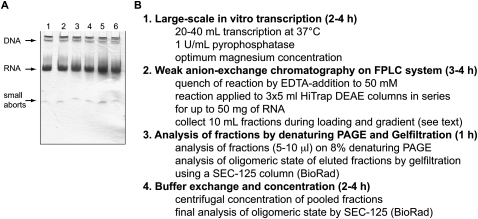
FIGURE 1.
Outline of RNA sample preparation. (A) Addition of pyrophosphatase improves RNA transcription yield. Denaturing PAGE analysis of large-scale transcriptions in the absence (lanes 1–3) or presence (lanes 4–6) of pyrophosphatase (1 U/mL transcription reaction). The yield is analyzed by loading 2 μL of the transcription reaction after 45 (lanes 1,4) and 90 min (lanes 2,3,5,6); in lanes 3,6 (relative to lanes 2,5) the magnesium concentration was increased from 24 to 48 mM by adding 48 uL of a 4.9 M MgCl2 solution to the 10 mL transcription reaction. RNA bands are visualized by staining with 0.1% toluidine blue. The best yield is obtained in the presence of pyrophosphatase without further addition of magnesium (lane 5). (B) Individual steps of RNA sample preparation from in vitro transcription to final concentration of the purified RNA. Estimated time for each step is shown in parentheses.
Native RNA purification scheme using weak anion-exchange chromatography
A typical transcription reaction contains protein and nucleic acid components, which differ greatly in their overall charge. Unincorporated rNTPs and small abortive transcripts contain a small number of negatively charged phosphate groups, larger RNA transcripts (>30 nt) expose an increased, size-dependent negative charge, and, finally, the large plasmid DNA template (>3 kb) contains the highest overall phosphate charge per molecule. In addition, the T7 RNA polymerase with its near-neutral pI also differs in overall charge from the nucleic acid components and therefore only binds below 100 mM NaCl to a weak anion-exchange DEAE-sepharose column matrix (Grodberg and Dunn 1988). This advantageous charge difference of all the transcription reaction components should therefore allow separation of the T7 RNA polymerase, rNTPs, and small abortive transcripts from the desired RNA transcript and the DNA template using anion-exchange chromatography. DEAE-sepharose, a weak anion-exchange matrix, was chosen, since it would require lower salt concentrations to elute the RNA product as compared to a strong anion-exchange matrix, such as Mono Q resin, which can retain RNA products even at high NaCl concentration (Pikovskaya et al. 2009). The experimental setup and the purification protocol are described below (Fig. 1B).
Our weak anion-exchange chromatographic purification of RNA oligonucleotides from large-scale transcription reactions (10–40 mL), uses an AKTA prime FPLC system equipped with a 50 mL superloop and three 5 mL HiTrap DEAE-sepharose FastFlow (FF) columns connected in series corresponding to a total binding capacity of at least 50 mg of RNA. Alternatively, a single HiPrep 20 mL FF DEAE column can be used with very similar performance (Fig. 3A, below). Ideally, this FPLC system should be dedicated to RNA purifications only and not be used for protein purifications from cell lysates to avoid RNase contamination of the RNA samples. The crude transcription reaction is stopped upon addition of EDTA to a final concentration of 50 mM and loaded directly into the superloop. The DEAE columns are equilibrated with three column volumes of the low salt buffer A (50 mM sodium phosphate at pH 6.5, 150 mM sodium chloride, and 0.2 mM EDTA) and the crude transcription is then loaded onto the DEAE column at a flow rate of 1 mL/min, while collecting 10 mL fractions. A typical elution profile during sample application shows a large peak containing both the T7 RNA polymerase and the unincorporated rNTPs, which do not efficiently bind the DEAE matrix at this salt concentration (Fig. 2A). After loading, a short salt gradient up to 10% buffer B (buffer A with 2 M sodium chloride) is applied to ensure complete removal of unincorporated rNTPs and the T7 RNA polymerase, followed by a shallow salt gradient up to 30% buffer B. At the beginning of this gradient, the small abortive transcripts are eluted around 330 mM NaCl (Fig. 2A). RNA oligonucleotides elute depending on their overall phosphate charge per molecule starting with fraction 15 and 16 for 30 nt RNAs (400 mM NaCl) up to fraction 28 for 500 nt RNAs (570 mM NaCl), while the plasmid DNA template elutes as a broad peak over several fractions toward the end of the shallow gradient at 630–700 mM NaCl (Fig. 2A). Therefore, this very shallow gradient is especially required for purification of larger RNAs. Denaturing PAGE analysis of eluted fractions shows that the plasmid DNA template is well separated from the desired RNA product (Fig. 2B). Following the elution of the plasmid DNA, a short gradient to 100% and back to 0% buffer B is employed in order to wash and equilibrate the column for the next RNA purification (Fig. 2A). The combined RNA fractions are then equilibrated into appropriate buffers using Centriprep centrifugal devices and concentrated for subsequent experiments (Fig. 1B).
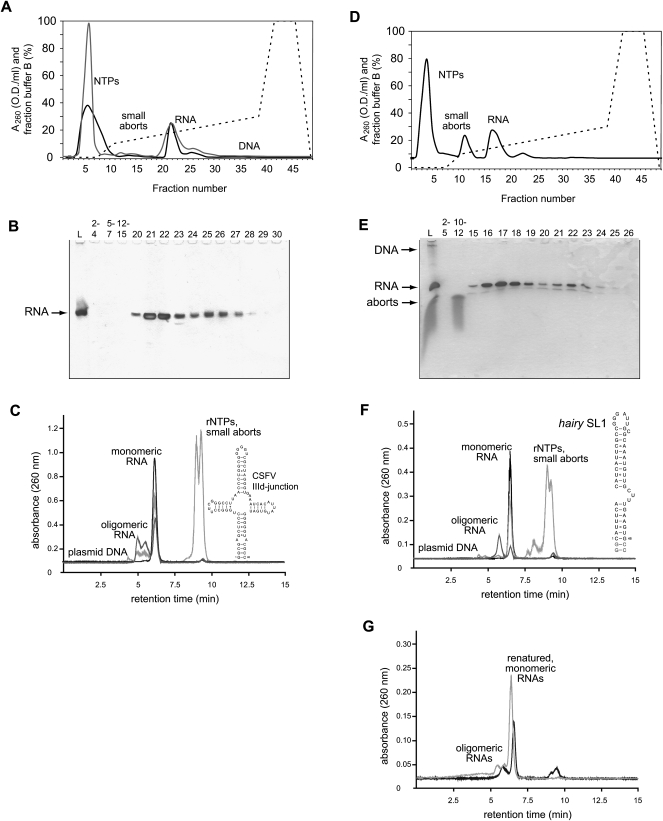
FIGURE 3.
Separation of monomeric from oligomeric RNA species using weak anion-exchange FPLC. (A) Elution profile of the 80 nt CSFV four-way junction RNA purified from a 20 mL in vitro transcription reaction using DEAE-sepharose chromatography. The RNA product elutes in a major peak (fractions 20–23) at lower NaCl concentration followed by a smaller, broad peak (approximately fractions 24–28) at higher NaCl concentration. Three 5 mL HiTrap DEAE-sepharose FF columns (GE Healthcare) connected in series (black UV trace) or one 20 mL HiPrep DEAE-sepharose FF column (gray UV trace) yield very similar elution profiles and resolution. (B) Denaturing PAGE analysis loading 5 μL of the eluted fractions. RNA bands are visualized by staining with 0.1% toluidine blue. The crude transcription reaction is shown in lane L, other lanes are numbered according to the fraction number in A. (C) Gel filtration of the purified CSFV four-way junction RNA. UV trace of the crude in vitro transcription (light gray line) and pooled monomeric RNA fractions (17–21, black line) and the pooled oligomeric RNA fractions (22–28, gray line) are shown. Plasmid DNA, RNA species, rNTPs, and small abortive transcripts are indicated and the secondary structure of the CSFV four-way junction RNA is shown. (D) Elution profile of the 50 nt hairy SL1 RNA purified from a 20 mL in vitro transcription reaction using DEAE-sepharose chromatography. The RNA product elutes in a major peak (fractions 15–18) at lower NaCl concentration followed by a smaller, broad peak (approximately fractions 19–23) at higher NaCl concentration. (E) Denaturing PAGE analysis loading 5 μL of the eluted fractions. RNA bands are visualized by staining with 0.1% toluidine blue. The crude transcription reaction is shown in lane L, other lanes are numbered according to the fraction number in D. (F) Gel filtration of the purified hairy SL1 RNA. UV trace of the crude in vitro transcription (light gray line) and pooled monomeric RNA fractions (15–18, black line) and the pooled dimeric RNA fractions (19–23, gray line) are shown. Plasmid DNA, RNA species, rNTPs, and small abortive transcripts are indicated and the secondary structure of the hairy SL1 RNA is shown. (G) Renaturation of the oligomeric RNA species by heating and rapid cooling on ice analyzed by gel filtration. UV trace of the renatured CSFV four-way junction RNA (gray line) and hairy SL1 RNA (black line) are shown. The corresponding oligomeric species are shown in C and F.
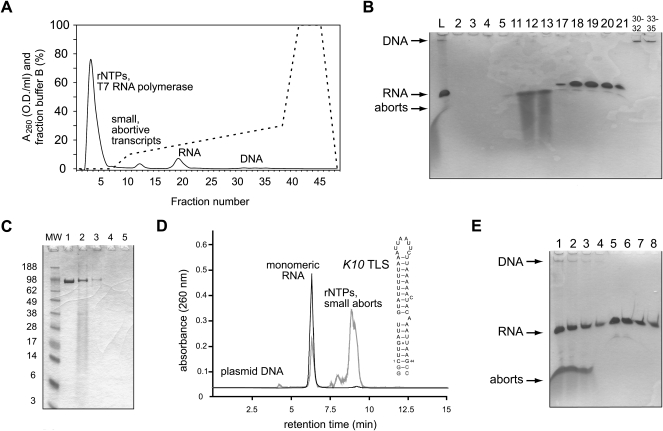
FIGURE 2.
RNA purification using weak anion-exchange FPLC. (A) Elution profile of the 48 nt K10 TLS purified from a 20 mL in vitro transcription reaction using DEAE-sepharose chromatography. Unincorporated rNTPs, T7 RNA polymerase, small abortive transcripts, and the plasmid DNA are well separated from the desired RNA product. The gradient trace (fraction of buffer B in %) is shown as a dashed line. (B) Denaturing PAGE analysis of the eluted fractions. RNA and DNA bands are visualized by staining with 0.1% toluidine blue. The crude transcription reaction is shown in lane L, other lanes are numbered according to the fraction number in A. The very dilute DNA fractions were pooled (30–32 and 33–35) and the plasmid DNA purified from 1 mL of the pooled fractions using QIAGEN PCR purification kit. (C) The purified RNA is free of T7 RNA polymerase. Denaturing SDS PAGE analysis of the eluted fractions. Resuspended pellets from TCA precipitation of 1 mL of the crude transcription reaction (lane 2), the pooled flow-through (lane 3), abortive transcripts (lane 4), and the pooled RNA fractions (lane 5) are loaded. T7 RNA polymerase (lane 1) and a molecular weight marker are loaded as references and the molecular weight is indicated on the left. T7 RNA polymerase bands are visualized by coomassie staining. (D) Gel filtration of the purified K10 TLS RNA. UV trace of the crude in vitro transcription of K10 TLS RNA (gray line) and pooled RNA fractions (17–21) purified by weak anion-exchange chromatography (black line) are shown. Plasmid DNA, RNA species, rNTPs, and small abortive transcripts are indicated and the secondary structure of K10 TLS RNA is shown. (E) The purification scheme recovers over 90% of the transcribed RNA from the crude in vitro transcription. Denaturing PAGE analysis of serial dilutions of the crude 20 mL transcription (lane 1, 2 μL; lane 2, 1 μL; lane 3, 0.5 μL; lane 4, 0.25 μL) and the 50 mL pooled, purified RNA fractions 17–21 (lane 5, 5 μL; lane 6, 2.5 μL; lane 7, 1.25 μL; lane 8, 0.625 μL). RNA bands are visualized by staining with 0.1% toluidine blue.
In order to verify that the final RNA sample is free of the T7 RNA polymerase, 1 mL aliquots of the pooled flow-through, abortive transcript, and final RNA product fractions were TCA-precipitated and analyzed by SDS PAGE. The T7 RNA polymerase was only detected in the application flow-through and not in any of the following pooled fraction, demonstrating that the desired RNA can be purified from all other components in the transcription reaction using weak anion-exchange chromatography (Fig. 2C). Analysis of the pooled RNA fractions by size-exclusion chromatography further demonstrates the purity of the RNA and shows that the pooled fractions (17–21) of the purified K10 transport and localization signal RNA (K10 TLS) (Serano and Cohen 1995) contain a single and—as judged from the retention time—monomeric RNA species (Fig. 2D). Moreover, this purification scheme typically recovers >90% of the transcribed RNA from the crude transcription as judged by the virtually identical RNA band intensities seen in the denaturing PAGE of serial dilutions of the crude transcription, in comparison to the pooled RNA fractions (Fig. 2E).
Using this method, we routinely purify RNA samples ranging from 30 to 500 nt in length in less than 4 h. When smaller RNAs around 30 nt are purified according to this method, slight contamination with abortive transcripts can be observed, but they are easily removed during the buffer exchanges with Centriprep centrifugal devices. When purifying RNA samples larger than 500 nt, care has to be taken not to pool fractions contaminated with the linearized plasmid DNA and therefore only the utmost peak fractions (3–5 fractions) are pooled avoiding the long tail fractions, which are more likely to contain the linearized plasmid. This method also helps to reduce the preparation cost of isotopically labeled RNA samples for NMR spectroscopic studies, since the expensive, unincorporated rNTPs can be easily recycled by pooling, lyophilizing, and desalting using boronate-affinity chromatography as described (Batey et al. 1992).
An additional benefit of this purification method is that monomeric and multimeric RNA species can be also separated, if their overall charge is significantly different. During purification of an 80-nt RNA four-way junction from the classical swine fever virus (Pestova et al. 1998), we observed that the RNA eluted in two distinct peaks along the shallow salt gradient (Fig. 3A). Denaturing PAGE showed that both peak fractions contained the clean RNA product, but analysis of the individual fractions by gel filtration revealed that the main peak contains the monomeric RNA species, while the fractions eluting at higher salt concentration represent dimeric and tetrameric species, which are efficiently separated from the desired monomer (Fig. 3B,C). Similarly, the RNA hairpin comprising stem–loop 1 of the transport signal of Drosophila hairy mRNA (hairy SL1) eluted as monomeric (90%) and dimeric (10%) species (Fig. 3D–F; Bullock et al. 2003). In both cases, gel filtration of the crude transcription reactions demonstrates that these oligomers already formed during the in vitro transcription and most likely represent stably folded duplex or tetrameric species. Heating and rapid cooling on ice of a tenfold dilution of the oligomeric RNA fractions with water (in order to lower the salt content to 50 mM, which favors formation of the monomer), recovers up to 90% monomer, which could then be repurified by DEAE chromatography (Fig. 3G).
Finally, this protocol is not limited to large-scale purifications. For biochemical applications such as 4-thio-uridine labeling of RNA for UV cross-linking experiments or 5′ or 3′ end and internal fluorophore-labeling of RNA, often require only very small transcription reactions (<200 μL). For such small RNA quantities, purification can be achieved using a 1 mL DEAE-sepharose column and 1/15 buffer volume for each gradient step, yielding pure RNA in less than 1 h (data not shown).
The novel purification protocol presented here yields milligram quantities of pure RNA from crude transcription reactions in less than 4 h. Rapid RNA preparation is especially beneficial for biochemical and structural applications, where usually many different RNA constructs have to be prepared for subsequent analyses. While this method does not purify RNAs with homogeneous 3′ ends often crucial for X-ray crystallographic studies, it yields structurally homogeneous, natively folded RNA, which is essential for many biochemical and biophysical applications. Since the protocol is robust and easy to implement in laboratories equipped with FPLC systems, easily scaled to large and small quantities of RNA samples and since DEAE-sepharose columns are available in different sizes at low cost, it should be accessible and beneficial to most groups investigating RNA structure and function.
MATERIALS AND METHODS
Preparation of the plasmid DNA template
Double-stranded DNA templates are prepared by standard PCR methods using overlapping primers as previously described (Lukavsky and Puglisi 2004). Large-scale plasmid purification from a 2 L of 2xYT-carbenicillin culture follows QIAfilter plasmid MEGA and GIGA protocols (QIAGEN) and typically yields 3–6 mg of plasmid DNA, which is dissolved in water to a final concentration of 700 μg/mL and linearized by incubating with 50 U/mL BbsI or BsaI and the appropriate buffer at 37°C (BbsI) or 50°C (BsaI) overnight.
In vitro transcription of RNA from linearized plasmid DNA templates
All procedures use deionized water (Millipore) and chemicals purchased from Sigma. His6-tagged T7 RNA polymerase is prepared in-house (Grodberg and Dunn 1988). In vitro transcriptions are optimized for each individual DNA template in 25 μL trial reactions by varying the magnesium concentration from 4 to 52 mM as previously described (Lukavsky and Puglisi 2004). Large-scale 10–40 mL transcription reactions contain 4 mM each rNTP, 70 μg/mL linearized plasmid, 1200 U/mL T7 RNA polymerase, 40 mM Tris-HCl (pH 8.1) at 37°C, 1 mM spermidine, 5 mM dithiothreitol, 0.1% Triton-X 100, 1 U/mL inorganic pyrophosphatase, and the optimal magnesium chloride concentration determined by the 25 μL trial reactions. After 2–4 h of incubation at 37°C, the reaction is stopped upon addition of EDTA to a final concentration of 50 mM. The transcription reaction can be stored frozen at −20°C until RNA purification.
Purification of in vitro transcribed RNA using weak anion-exchange FPLC
RNAs are purified from crude transcription reactions using an AKTA prime FPLC system equipped with a 50 mL superloop and three 5 mL HiTrap DEAE-sepharose FF columns (GE Healthcare) connected in series. The DEAE columns are equilibrated with three column volumes of buffer A (50 mM sodium phosphate [pH 6.5], 150 mM sodium chloride, and 0.2 mM EDTA) at room temperature. Buffer B contains the same components with 2 M sodium chloride. Both buffers can be prepared in large quantities, sterile filtered, and stored at 4°C (buffer A) or room temperature (buffer B) to avoid precipitation of sodium chloride. The stopped transcription reaction (10–40 mL) is loaded into the 50 mL superloop and weak anion-exchange chromatography is performed using the following gradient, while collecting 10 mL fractions in sterile 15 mL plastic tubes: 0–70 mL (0% B at 1 mL/min) to load the sample onto the DEAE columns, 70–100 mL (0%–10% B at 2 mL/min) to wash remaining rNTPs off the column, 100–380 mL (10%–30% B at 2 mL/min) to separate small abortive transcripts, the desired RNA product, and the plasmid DNA template, 380–410 mL (30%–100% B at 4 mL/min), 410–455 mL (100% B at 4 mL/min), and 455–485 mL (100%–0% B at 4 mL/min) to wash and equilibrate the column for the next purification. For small-scale transcriptions below 1 mL, the reaction mixture is diluted to 2 mL with buffer A to ensure complete loading into the superloop and chromatography performed using a single 1-mL HiTrap DEAE-sepharose FF column and the same gradient profile with buffer volumes reduced to 1/15 collecting 2 mL fractions. Fractions are analyzed by denaturing PAGE (8% acrylamide, 8 M urea) loading 5 μL of each fraction. In case the RNA elution profile displays more than one peak along the gradient indicating alternative conformations or multimerization of the RNA, each fraction should be analyzed by gel filtration on an HPLC system using a SEC-125 column (Bio-Rad) and buffer A. Structurally homogeneous RNA fractions are then pooled, concentrated, and equilibrated into the appropriate buffers for subsequent experiments using 15-mL Centriprep centrifugal devices.
ACKNOWLEDGMENTS
We thank all members of the Lukavsky group for helpful suggestions. This work was supported by a grant from HFSP (RGP0024/2008) to P.J.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1862210.
REFERENCES
- Anderson AC, Scaringe SA, Earp BE, Frederick CA. HPLC purification of RNA for crystallography and NMR. RNA. 1996;2:110–117. [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Kieft JS. Improved native affinity purification of RNA. RNA. 2007;13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR. Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res. 1992;20:4515–4523. doi: 10.1093/nar/20.17.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA. Preparation of homogeneous ribozyme RNA for crystallization. Methods Mol Biol. 1997;74:365–370. doi: 10.1385/0-89603-389-9:365. [DOI] [PubMed] [Google Scholar]
- Grodberg J, Dunn JJ. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988;170:1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern JA, Davis RH. Application of solution equilibrium analysis to in vitro RNA transcription. Biotechnol Prog. 1997;13:747–756. doi: 10.1021/bp970094p. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Batey RT. A general method for rapid and nondenaturing purification of RNAs. RNA. 2004;10:988–995. doi: 10.1261/rna.7040604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, McKenna SA, Viani Puglisi E, Puglisi JD. Rapid purification of RNAs using fast performance liquid chromatography (FPLC) RNA. 2007;13:289–294. doi: 10.1261/rna.342607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- Lim F, Peabody DS. Mutations that increase the affinity of a translational repressor for RNA. Nucleic Acids Res. 1994;22:3748–3752. doi: 10.1093/nar/22.18.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Puglisi JD. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD. Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat Protoc. 2007;2:3270–3277. doi: 10.1038/nprot.2007.480. [DOI] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikovskaya O, Serganov AA, Polonskaia A, Serganov A, Patel DJ. Preparation and crystallization of riboswitch-ligand complexes. Methods Mol Biol. 2009;540:115–128. doi: 10.1007/978-1-59745-558-9_9. [DOI] [PubMed] [Google Scholar]
- Price SR, Ito N, Oubridge C, Avis JM, Nagai K. Crystallization of RNA-protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol. 1995;249:398–408. doi: 10.1006/jmbi.1995.0305. [DOI] [PubMed] [Google Scholar]
- Serano TL, Cohen RS. A small predicted stem–loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Shields TP, Mollova E, Ste Marie L, Hansen MR, Pardi A. High-performance liquid chromatography purification of homogenous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA. 1999;5:1259–1267. doi: 10.1017/s1355838299990945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck OC. Keeping RNA happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]