Abstract
We are interested in asparagine-linked glycans (N-glycans) of Plasmodium falciparum and Toxoplasma gondii, because their N-glycan structures have been controversial and because we hypothesize that there might be selection against N-glycans in nucleus-encoded proteins that must pass through the endoplasmic reticulum (ER) prior to threading into the apicoplast. In support of our hypothesis, we observed the following. First, in protists with apicoplasts, there is extensive secondary loss of Alg enzymes that make lipid-linked precursors to N-glycans. Theileria makes no N-glycans, and Plasmodium makes a severely truncated N-glycan precursor composed of one or two GlcNAc residues. Second, secreted proteins of Toxoplasma, which uses its own 10-sugar precursor (Glc3Man5GlcNAc2) and the host 14-sugar precursor (Glc3Man9GlcNAc2) to make N-glycans, have very few sites for N glycosylation, and there is additional selection against N-glycan sites in its apicoplast-targeted proteins. Third, while the GlcNAc-binding Griffonia simplicifolia lectin II labels ER, rhoptries, and surface of plasmodia, there is no apicoplast labeling. Similarly, the antiretroviral lectin cyanovirin-N, which binds to N-glycans of Toxoplasma, labels ER and rhoptries, but there is no apicoplast labeling. We conclude that possible selection against N-glycans in protists with apicoplasts occurs by eliminating N-glycans (Theileria), reducing their length (Plasmodium), or reducing the number of N-glycan sites (Toxoplasma). In addition, occupation of N-glycan sites is markedly reduced in apicoplast proteins versus some secretory proteins in both Plasmodium and Toxoplasma.
Animals, fungi, and plants synthesize Asn-linked glycans (N-glycans) by means of a lipid-linked precursor containing 14 sugars (dolichol-PP-Glc3Man9GlcNAc2) (26). Recently we used bioinformatics and experimental methods to show that numerous protists are missing sets of glycosyltransferases (Alg1 to Alg14) and so make truncated N-glycan precursors containing 0 to 11 sugars (46). For example, Entamoeba histolytica, which causes dysentery, makes N-glycan precursors that contain seven sugars (Man5GlcNAc2) (33). Giardia lamblia, a cause of diarrhea, makes N-glycan precursors that contain just GlcNAc2 (41). N-glycan precursors may be identified by metabolic labeling with radiolabeled mannose (Entamoeba) or glucosamine (Giardia) (46). Unprocessed N-glycans of each protist may be recognized by wheat germ agglutinin 1 (WGA-1) (GlcNAc2 of Giardia) or by the antiretroviral lectin cyanovirin-N (Man5GlcNAc2 of Entamoeba) (2, 33, 41).
N-glycans are transferred from lipid-linked precursors to sequons (Asn-Xaa-Ser or Asn-Xaa-Thr, where Xaa cannot be Pro) on nascent peptides by an oligosaccharyltransferase (OST) (28). For the most part, transfer of N-glycans by the OST is during translocation, although there are human and Trypanosoma OSTs that transfer N-glycans after translocation (34, 45).
N-glycan-dependent quality control (QC) systems for protein folding and endoplasmic reticulum (ER)-associated degradation (ERAD), which are present in most eukaryotes, are missing from Giardia and a few other protists that make truncated N-glycans (5, 26, 53). There is positive Darwinian selection for sequons (sites of N-glycans) that contain Thr in secreted and membrane proteins of organisms that have N-glycan-dependent QC (12). This selection occurs for the most part by an increased probability that Asn and Thr will be present in sequons rather than elsewhere in secreted and membrane proteins. In contrast, there is no selection on sequons that contain Ser, and there is no selection on sequons in the secreted proteins of organisms that lack N-glycan-dependent QC.
For numerous reasons, we are interested in the N-glycans of Plasmodium falciparum and Toxoplasma gondii, which cause severe malaria and disseminated infections, respectively.
(i) There has been controversy for a long time as to whether Plasmodium makes N-glycans. While some investigators identified a 14-sugar Plasmodium N-glycan resembling that of the human host (29), others identified no N-glycans (6, 22).
(ii) There is also controversy concerning whether the N-glycans of Toxoplasma, after removal of Glc by glucosidases in the ER lumen, contain either 7 sugars (Man5GlcNAc2), like Entamoeba (32, 33), or 11 sugars (Man9GlcNAc2), like the human host (16, 19, 26). If it is Man5GlcNAc2, then Toxoplasma uses the dolichol-PP-linked glycan predicted by its set of Alg enzymes (32, 46). If it is Man9GlcNAc2, then Toxoplasma uses the dolichol-PP-linked glycan of the host cell (16, 19, 26).
(iii) Both Plasmodium and Toxoplasma are missing proteins involved in N-glycan-dependent QC of protein folding (5).
(iv) We hypothesize that there may be negative selection against N-glycans in Plasmodium and Toxoplasma, because the N-glycans added in the ER lumen during translocation will likely interfere with threading of nucleus-encoded apicoplast proteins into a nonphotosynthetic, chloroplast-derived organelle called the apicoplast (21, 35, 37, 48, 52, 54). Nucleus-encoded apicoplast proteins have a bipartite signal at the N terminus, which targets proteins first to the lumen of the ER and second to lumen of the apicoplast. This bipartite signal has been used in transformed plasmodia where green fluorescent protein (GFP) is targeted to the apicoplast with the bipartite signal of the acyl carrier protein (ACPleader-GFP), to the secretory system with the signal sequence only (ACPsignal-GFP), and to the cytosol with the organelle-targeting transit peptide only (ACPtransit-GFP) (55). Similar constructs have been used to characterize signals that target nucleus-encoded proteins of Toxoplasma to the apicoplast (11, 25).
Here we use a combination of bioinformatic, biochemical, and morphological methods to characterize the N-glycans of Plasmodium and Toxoplasma and to test our hypothesis that there is negative selection against N-glycans in protists with apicoplasts.
MATERIALS AND METHODS
Bioinformatic predictions.
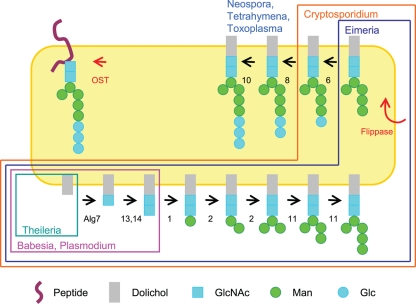
Predicted proteins of Plasmodium, Toxoplasma, Cryptosporidium, Eimeria, Babesia, Neospora, and Theileria, which are present in the NR database at NCBI and/or databases at GeneDB, PlasmoDB, ToxoDB, and CryptoDB, were searched using BLASTP and Saccharomyces Alg enzymes that make precursors to Asn-linked glycans and OST peptides that transfer the N-glycan to the nascent peptide (1, 3, 4, 7, 9, 15, 17, 18, 26, 28, 38, 46, 49). Because protein prediction is difficult for Toxoplasma and Eimeria, which contain many introns in their genes, TBASTN was also used to search the contigs and/or expressed sequence tags (ESTs) of these organisms (17, 49). Figure 1 was drawn based upon the predicted sets of N-glycans made by these apicomplexan parasites, rather than those that are experimentally determined (which might include host N-glycan precursors in the case of Toxoplasma) (16, 19).
Fig. 1.
The present diversity of N-glycan precursors among apicomplexan parasites is likely due to secondary loss of Alg enzymes. Metazoans, fungi, plants, and algae have a complete set of Alg enzymes and so make an N-glycan precursor with 14 sugars (Glc3Man9GlcNAc2) (26, 46). Toxoplasma, Neospora, and the related ciliate Tetrahymena (15) are missing Alg enzymes that add four Man residues in the ER lumen and so make an N-glycan precursor with 10 sugars (Glc3Man5GlcNAc2). Cryptosporidium is also missing Alg8 and Alg10 and so makes an N-glycan precursor with eight sugars (Glc1Man5GlcNAc2). Eimeria is also missing Alg6 and so makes an N-glycan precursor with seven sugars (Man5GlcNAc2). Plasmodium falciparum and P. vivax, as well as Babesia, are missing all the enzymes that add Man and Glc to N-glycan precursors and so add just two sugars (GlcNAc2). Theileria is missing all of the Alg enzymes, as well as the oligosaccharyltransferase (OST), and so makes no N-glycans. With the exception of Theileria, all of the apicomplexa have four OST peptides.
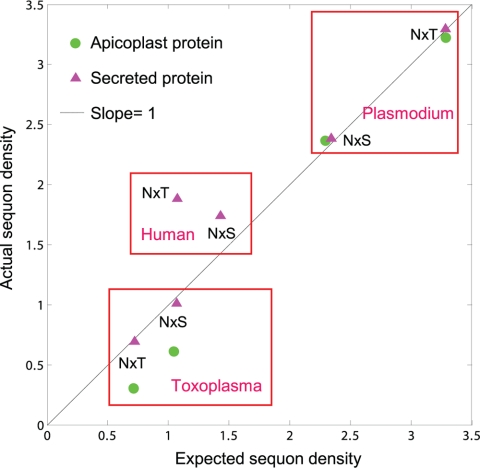
Sequons (potential sites of N-linked glycosylation) were identified in secreted and membrane proteins, which contain an N-terminal signal peptide or transmembrane helices, using previously described methods (12, 27). About 500 predicted nucleus-encoded apicoplast proteins of Plasmodium were downloaded from the PlasmoDB website (4, 11), while 18 experimentally confirmed apicoplast proteins of Toxoplasma were obtained from Omar Harb (11, 25). Selection on sequons was determined by plotting the actual sequon density per 500 amino acids of secreted and membrane proteins on the y axis versus the expected sequon density of these same proteins based upon the concentrations of Asn, Ser, Thr, and Pro (see Fig. 2) (12).
Fig. 2.
Negative selection against sequons (sites of N-glycans) in Toxoplasma occurs by two mechanisms. First, because Asn is encoded by AAT/C and the coding sequences of Toxoplasma are AT poor, the density of sequons in its secreted proteins (red triangles) and nucleus-encoded apicoplast proteins (green circles) is low (12). Conversely, the AT content of Plasmodium is high, and the density of sequons in its secreted proteins and apicoplast proteins is high. Humans have an intermediate AT content and therefore an intermediate sequon density in their secreted proteins. Second, there is a decreased probability that Asn, Ser, and Thr will be positioned to form sequons rather than elsewhere in nucleus-encoded apicoplast proteins of Toxoplasma. This is shown by plotting the calculated sequon density based upon amino acid composition (x axis) versus actual sequon density (y axis) for secreted proteins and apicoplast proteins of Plasmodium and Toxoplasma. In most cases, there is no selection on sequons, so points fall on the line with a slope of 1. There is, however, negative selection against sequons with Ser or Thr in apicoplast proteins of Toxoplasma, so that the actual sequon density is about one-half the calculated density. In contrast, there is positive selection for sequons with Thr in secreted proteins of humans, so that the actual sequon density is about twice the value calculated by amino acid composition (12). Because the Toxoplasma genome is AT poor and there is additional negative selection against sequons in apicoplast proteins, nearly half of the nucleus-encoded apicoplast proteins have no sequons and so cannot contain N-linked glycans (see Fig. S1 in the supplemental material). In contrast, <10% of Plasmodium secreted and apicoplast proteins have no sequons, while ∼20% of human secreted proteins have no sequons (12).
Sources of parasites, GFP constructs, and antibodies.
The 3D7 (original genome project) strain of Plasmodium falciparum was grown in human red blood cells (RBCs), for the most part without synchronization (36). When necessary (for example, for metabolic labeling), plasmodia were synchronized with sorbitol. Transformed plasmodia with GFP targeted to the apicoplast (ACPleader-GFP), the parasitophorous vacuole (ACPsignal-GFP), or the cytosol (ACPtransit-GFP) were obtained from MR4 (55). Antibodies to Plasmodium proteins which are present in the ER (Pf39), food vacuole (DPAP1), and plasma membrane (MSP1-19) were obtained from MR4 (30, 51). An antibody to RhopH3 was a generous gift from J. F. Dubremetz (14).
The RH strain of Toxoplasma gondii was grown in human foreskin fibroblasts (44). A vector for Toxoplasma with yellow fluorescent protein (YFP) targeted to the apicoplast (ACP-YFP) was a generous gift from Boris Striepen, University of Georgia (35). Vectors for Toxoplasma with YFP targeted to the ER (P30-YFP-HDEL) and to the Golgi apparatus (GRASP55-YFP) were generous gifts from Graham Warren, Yale University (24, 39). Vectors were transiently transfected into Toxoplasma by electroporations. Antibodies to Toxoplasma proteins which are present in the rhoptry (ROP1; monoclonal antibody [MAb] Tg49) (47) and plasma membrane (SAG1; MAb DG52) (8) were generous gifts from Peter Bradley (UCLA) and Jeroen Saeij (MIT), respectively.
Metabolic labeling of Plasmodium and isolation of N-glycans and precursors.
Plasmodia were labeled in vitro using methods described previously for labeling of glycosylphosphatidylinositol (GPI) anchors (22). RBCs were infected with Plasmodium, synchronized with sorbitol, and grown to a density of 12 to 14%. Depending upon when Plasmodium-infected RBCs were harvested, trophozoites, which are mid-stage, or schizonts, which are later stage, predominated. RBCs containing trophozoites and schizonts were radiolabeled with 500 μCi/ml [46-3H]Man or [28-3H]GlcN in a Glc-free medium for 15 min in a final volume of 250 μl. Infected RBCs were isolated on Percoll gradients, and dolichol-PP-glycans were extracted and separated by thin-layer chromatography (TLC). Controls included Giardia, which makes dolichol-PP-GlcNAc2; Saccharomyces, which makes dolichol-PP-Glc3Man9GlcNAc2; and uninfected RBCs, which cannot make N-glycans in the absence of the ER and Golgi. Alternatively, plasmodia were released from RBCs by saponin lysis, proteins were delipidated by extractions with chloroform-methanol-water, and N-glycans were released by treatment with 500 units of peptide:N-glycanase F (PNGaseF) (New England Biolabs) for 16 h at 37°C (46).
Size exclusion chromatography of radiolabeled glycans, after release from dolichol with mild acid or release from proteins with PNGaseF, was performed on a 1.5- by 100-cm Bio-Gel P-4 column with 0.1 M acetic acid–1% (vol/vol) n-butanol as the mobile phase. Internal standards for Plasmodium glycans were GlcNAc and GlcNAc2 (commercially available). Other standards from Saccharomyces Δalg3, Δalg5, and Δalg3 Δalg5 mutants included Glc3Man5GlcNAc2, Man9GlcNAc2, and Man5GlcNAc2, respectively. Radioactivity was measured by scintillation counting.
WGA affinity purification of Plasmodium glycoproteins and characterization by lectin blotting after SDS-PAGE and by mass spectroscopy (MS).
Late-stage plasmodia were released from RBCs with saponin, and washed plasmodia were treated with 0.1% Triton X-100 in the presence of EDTA-free Complete protease inhibitor cocktail (Roche). Insoluble material was removed by centrifugation (>12,000 × g), and soluble proteins were applied to a WGA-1-Sepharose column (EY Laboratories, Inc.) (56). Because there may be nonspecific binding to this column, Plasmodium glycoproteins were eluted with 50 mM diacetylchitotriose rather than with SDS. WGA-1-purified Plasmodium proteins were run on SDS-PAGE with a 4 to 20% gradient of acrylamide (Bio-Rad). In parallel lanes were Plasmodium proteins solubilized with 0.1% Triton X-100 but not applied to the WGA-1 column. Alternatively, Plasmodium proteins were denatured and incubated with 1,000 units PNGaseF for 4 h at 37°C in NEB G7 phosphate buffer prior to loading for SDS-PAGE. After SDS-PAGE, proteins either were fixed and stained with silver or Coomassie blue or were transferred to nitrocellulose membranes by electroporation, incubated with horseradish peroxidase (HRP)-conjugated GSL-II, and developed with ECL chemiluminescent substrate (Pierce).
Alternatively, WGA-1-bound Plasmodium glycoproteins were run 0.5 cm into SDS-PAGE, excised, digested with trypsin, and identified by liquid chromatography-tandem MS (LC-MS-MS), using methods that we have previously used to identify the cyst proteins of Giardia (41). Briefly, reversed-phase chromatography was carried out using a nano-high-pressure liquid chromatography (nano-HPLC) pump and autosampler (Surveyor and MicroAS; Thermo Finnigan, San Jose, CA) on a 10-cm by 100-μm (inner diameter) Magic C18 reversed-phase capillary column (Michrom, Auburn, CA) at the Boston University Proteomics Core Facility or at the MIT Center for Cancer Research Biopolymers Laboratory. Peptides were separated using gradients of 5% to 90% acetonitrile over 30 to 120 min in the presence of 0.5% acetic acid (57). Peptides were analyzed using an LTQ ProteomeX ion trap mass spectrometer (Thermo Finnigan, San Jose, CA), and mass spectra were compared to tryptic digests of predicted Plasmodium proteins using SEQUEST and theGPM software (www.thegpm.org). All searches were conducted on a reverse database to ensure that the false-positive rate for protein identification was kept below 2%. Tryptic peptides with a SEQUEST XCorr score of >1.5, 2.5, or 3.5 for Z = 1, 2, or 3, respectively, and a peptide score of <0.05 were considered a match. GPM-based searches were scored based on having a loge value greater than a cutoff threshold of 2% false positives, typically loge < −10. Proteins with one or more high-scoring fully tryptic peptides were considered present.
Three-dimensional high-resolution fluorescence microscopy.
RBCs infected with plasmodia were washed once in phosphate-buffered saline (PBS), fixed for 30 min at room temperature (RT) in 4% paraformaldehyde and 0.0075% glutaraldehyde in PBS, and washed once in PBS. Except in those circumstances where we wished to label only the surface of infected RBCs, Triton X-100 was added to a final concentration of 0.1% to permeabilize the organisms for 10 min at RT. Cells were washed with PBS, and unreacted aldehydes were quenched with 0.1-mg/ml sodium borohydride in PBS for 10 min at RT. After washing with PBS, we incubated cells with 3% bovine serum albumin (BSA) in PBS for 30 min prior to staining with lectins or antibodies. To label the plasma membranes of plasmodia, RBCs which were infected with late-stage plasmodia were lysed with 0.15% saponin prior to fixing and labeling.
Ulex europaeus agglutinin 1 (UEA-1) was labeled with Alexafluor 488 (Molecular Probes), which uses the same filter sets as fluorescein but quenches less easily. Griffonia simplicifolia lectin (GSL-II) was labeled with Alexafluor 594, which uses the same filter sets as rhodamine. Plasmodium-infected RBCs were colabeled with 2 μg/ml each of GSL-II and UEA-1 in PBS plus 3% BSA for 60 min at RT and then washed twice in PBS. Alternatively, transformed plasmodia expressing GFP targeted to the cytosol, apicoplast, or parasitophorous vacuole were incubated with GSL-II. As a control for accessibility to exogenous probes, transformed plasmodia expressing GFP were incubated with anti-GFP antibodies labeled with Alexafluor 594 instead of GSL-II. In other experiments, nontransformed plasmodia were incubated with GSL-II and Alexafluor 488-labeled antibodies to the rhoptries, ER, food vacuole, or plasma membrane.
Toxoplasma tachyzoites released from human foreskin fibroblasts were fixed, permeabilized, and washed, as described for Plasmodium. Toxoplasma tachyzoites were incubated with Alexafluor-labeled cyanovirin-N, scytovirin, UEA-1, or Maclura pomifera agglutinin (MPA-1) (2). Alternatively, transformed Toxoplasma tachyzoites expressing GFP or YFP targeted to the apicoplast, ER, or Golgi were incubated with cyanovirin-N. As a control for accessibility to exogenous probes, transformed Toxoplasma tachyzoites expressing GFP were incubated with anti-GFP antibodies instead of cyanovirin-N. In other experiments, nontransformed Toxoplasma tachyzoites were labeled with cyanovirin-N and Alexafluor-labeled antibodies to rhoptries or ER.
The nuclei of labeled Plasmodium and Toxoplasma cells were labeled with 2 μg/ml DAPI (4′,6′-diamidino-2-phenylindole), and SlowFade antifade solution (Invitrogen) was added. Slides were examined by three-dimensional multiple-wavelength fluorescence microscopy using an Olympus IX70 microscope equipped for Deltavision deconvolution (Applied Precision). This system employs restorative as well as deconvolution techniques. Images were collected at 0.2-mm optical sections for the indicated wavelengths and were subsequently deconvolved using SoftWoRx (Applied Precision). Data were examined either as optical sections or as a projection of the entire stack.
Use of cyanovirin and scytovirin to estimate the relative amount of high-mannose N-glycans in Toxoplasma.
High-mannose oligosaccharides, including biosynthetic Man5, were synthesized via a linear strategy as described previously (42). To facilitate carbohydrate microarray fabrications, a thiol-terminated ethylene glycol linker was synthetically incorporated on the reducing end of the mannosides in place of GlcNAc2 (2). Glycans were robotically printed onto maleimide-functionalized amine-coated slides using established protocols (2). The carbohydrate arrays were probed with Alexafluor-labeled cyanovirin, scytovirin, WGA-1, and concanavalin A. Alexafluor-labeled cyanovirin-N and scytovirin were used to label the surface of wild-type Saccharomyces, which makes N-glycans based upon a precursor composed of Man9GlcNAc2. These same lectins were applied to the Saccharomyces Δalg3 mutant, which makes N-glycans based upon a precursor composed of Man5GlcNAc2 (26). Yeast cells were examined with the deconvolving microscope and were analyzed with a BD FACSCalibur flow cytometer. In parallel, Toxoplasma cells labeled with cyanovirin-N and scytovirin were analyzed with the flow cytometer. The relative amounts of Man5GlcNAc2 and Man9GlcNAc2 in Toxoplasma N-glycans were estimated using the assumption that cyanovirin-N binds to both N-glycans, while scytovirin binds only to Man9GlcNAc2.
RESULTS
Overview.
Because we studied two different organisms (Plasmodium and Toxoplasma) in parallel using multiple methods, a “road map” to Results is presented here. Bioinformatic predictions concerning the length of N-glycans made by each organism (see Fig. 1) and the density of sites of N-linked glycans (sequons) on secreted and membrane proteins (see Fig. 2) are presented first. Experimental demonstrations of the actual N-glycans made by Plasmodium (see Fig. 3 and 4) and Toxoplasma (see Fig. 5) are presented next. Finally, colocalizations of the N-glycans of Plasmodium and Toxoplasma with the apicoplast and secretory compartments of each organism (see Fig. 6 and 7, respectively) are presented.
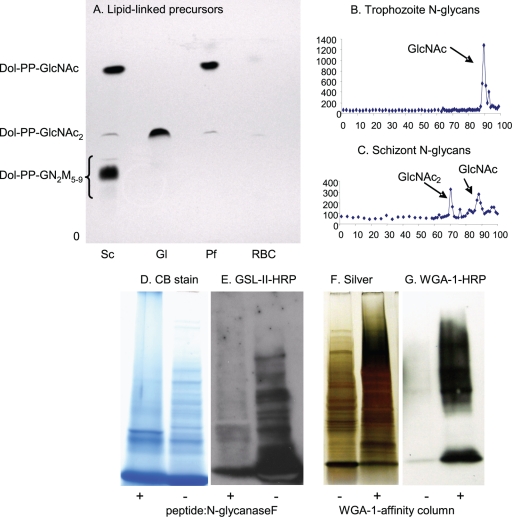
Fig. 3.
Plasmodia metabolically labeled with tritiated glucosamine (GlcN) make N-glycan precursors and N-glycans composed of a single GlcNAc and of GlcNAc2. (A) TLC shows that Plasmodium (Pf) trophozoites (early stages) make predominantly dolichol-PP-GlcNAc with a slight amount of dolichol-PP-GlcNAc2. For comparison, Giardia (Gl), which has the same Alg enzymes as Plasmodium (Fig. 1) (46), makes predominantly dolichol-PP-GlcNAc2, while Saccharomyces (Sc) makes a mixture of dolichol-PP-GlcNAc2 and high-mannose dolichol-PP-glycans. (B) N-glycans released by PNGaseF from glycoproteins of Plasmodium trophozoites are composed predominantly of a single GlcNAc. (C) N-glycans of Plasmodium schizonts (later stages) are a mixture of GlcNAc and GlcNAc2. (D to G) Additional biochemical evidence for short N-glycans of Plasmodium is shown by blots with the GlcNAc-binding lectin GSL-II after treatment of proteins with PNGaseF and by WGA-1 blots of WGA-1-affinity-purified proteins. PNGaseF-treated Plasmodium proteins, which are stained with Coomassie blue (D), show a marked decrease in binding GSL-II (E). This result shows that GSL-II is binding to N-glycans. WGA-1-affinity markedly enriches Plasmodium proteins, which are stained with silver (F), that bind to the lectin (G). Proteins identified by mass spectroscopy after a representative WGA-1 affinity experiment are shown in Table 1.
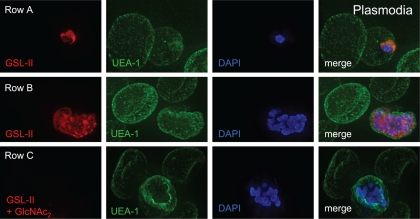
Fig. 4.
Deconvolving micrographs of Plasmodium-infected RBCs show that the GlcNAc-binding lectin GSL-II (red) (59) is specific for plasmodia, while the fucose-binding lectin UEA-1 (green) is specific for the surface of RBCs. Row A, an early trophozoite of Plasmodium has a single nucleus stained with DAPI (blue in all micrographs) and a cup-shaped secretory system stained with GSL-II after permeabilization with nonionic detergent. Row B, schizonts (later stages) of Plasmodium have multiple nuclei and much more widespread staining with GSL-II. Row C, the labeling by GSL-II of late stage plasmodia is blocked by coincubation with GlcNAc2 (diacetyl-chitobiose). GSL-II does not bind to cytosolic proteins (see Fig. S2A in the supplemental material), but GSL-II binds to the surface of plasmodia which have been released from RBCs with saponin (see Fig. S2B).
Fig. 5.
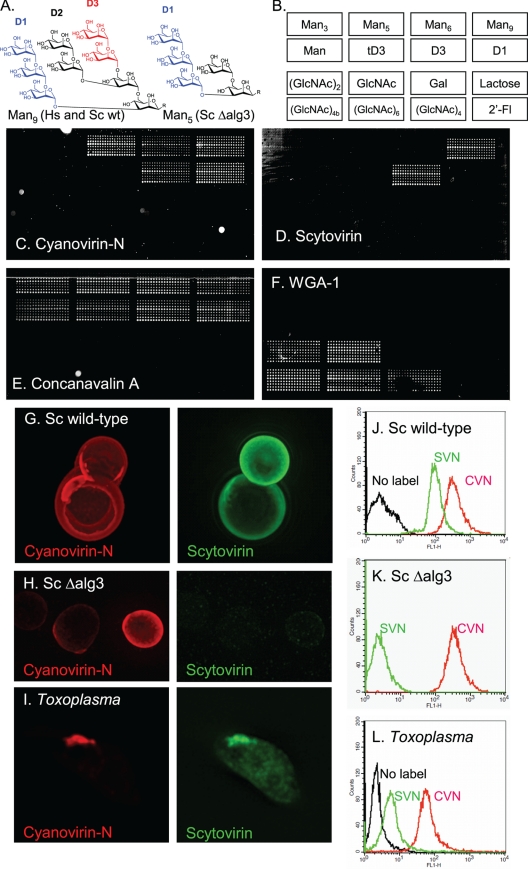
Use of antiretroviral lectins cyanovirin-N and scytovirin, which have distinct N-glycan specificities, to argue for the presence of N-glycans derived from the parasite and the host in apical secretory vesicles of Toxoplasma. (A) Man9GlcNAc2, which is the N-glycan present on glycoproteins of host cells and wild-type Saccharomyces after Glc residues are removed by glucosidases, has three arms, which are labeled D1 to D3. Man5GlcNAc2, which is the glycan made from N-glycan precursors of Toxoplasma and Saccharomyces ΔAlg3 mutants after Glc residues are removed by glucosidases, has a single D1 arm. In chemically synthesized Man9 and Man5, the two GlcNAcs are replaced by a linker that binds the sugars to the glass slide (2, 42). (B) Map of spotted sugars in panels C to F. (C) Alexafluor-labeled cyanovirin-N binds to biosynthetic Man5, Man9, and both D1 and D3 arms of Man9, which have been applied to glass slides. (D) In contrast, scytovirin only binds to Man9 and its D3 arm but does not bind to Man5. (E) The plant lectin concanavalin A binds to the entire set of mannose sugars, which are arrayed on a glass slide. (F) WGA-1, which is specific for GlcNAc, fails to bind to mannose sugars. (G) Wild-type Saccharomyces, which makes N-glycans based upon a Man9GlcNac2 precursor, is labeled well by both cyanovirin-N (red) and scytovirin (green). (H) In contrast, the Saccharomyces ΔAlg3 mutant, which makes N-glycans based upon a Man5GlcNac2 precursor, is labeled with cyanovirin-N (red) but not with scytovirin (green). (I) Cyanovirin-N and scytovirin both label the apical end of a Toxoplasma tachyzoite which has been released from a monolayer of human foreskin fibroblasts. (J to L) The binding of cyanovirin-N and scytovirin to wild-type Saccharomyces (J), the Saccharomyces ΔAlg3 mutant (K), and Toxoplasma (L) were measured using flow cytometry. These results suggest that Toxoplasma N-glycans are composed of a mix of Man5GlcNAc2 and Man9GlcNAc2, as recently proposed (19).
Fig. 6.
Deconvolving micrographs of Plasmodium-infected RBCs show that GSL-II, which binds to Plasmodium N-glycans, colocalizes with rhoptries (row A) and the ER (row B) but does not colocalize with apicoplasts (row C), the parasitophorous vacuole (row D), or the food vacuole (row E). In each case GSL-II is labeled red with Alexafluor 594, while antibodies labeled green with Alexafluor 488 are against RhopH3 (rhoptries), Pf39 (ER), and food vacuole (DPAP1) (14, 30, 51). Alternatively, GFP is targeted to apicoplasts (ACPleader-GFP) and the parasitophorous vacuole (ACPsignal-GFP) (55). Control experiments with anti-GFP antibodies (see Fig. S4 in the supplemental material) show that both apicoplasts and parasitophorous vacuoles are accessible to exogenous probes.
Fig. 7.
Deconvolving micrographs of Toxoplasma tachyzoites show that cyanovirin-N, which binds to Toxoplasma N-glycans, colocalizes with rhoptries (row A), the ER (row B), and the plasma membrane (row C) but does not colocalize with apicoplasts (row D) and the Golgi apparatus (row E). In each case cyanovirin-N is labeled red with Alexafluor 594, while antibodies labeled green with Alexafluor 488 are against ROP1 (rhoptries) and SAG1 (plasma membrane). Alternatively, fluorescent proteins are targeted to apicoplasts (ACP-YFP), the ER (P30-GFP-HDEL), and the Golgi apparatus (GRASP55-YFP). Control experiments with anti-GFP antibodies (see Fig. S5 in the supplemental material) show that both apicoplasts and the Golgi apparatus are accessible to exogenous probes.
Extensive secondary loss of Alg enzymes that make N-glycan precursors from vector-borne apicomplexans (Plasmodium, Babesia, and Theileria).
When we previously examined Alg enzymes of diverse eukaryotes, the enzymes that add the second GlcNAc to the N-glycan precursor (Alg13 and Alg14) were not identified, and whole-genome sequences of Theileria, Babesia, Eimeria, and Neospora were not available (7, 10, 17, 26, 38, 46, 49). Here we found that secondary loss among Alg genes of apicomplexan parasites is even more extensive than previously demonstrated (Fig. 1) (46). While Alg enzymes of metazoans, plants, and most fungi predict an N-glycan precursor with 14 sugars (Glc3Man9GlcNAc2), Alg enzymes of Toxoplasma, Neospora, and Tetrahymena predict an N-glycan precursor composed of 10 sugars (Glc3Man5GlcNAc2). The predicted N-glycan precursors of Cryptosporidium and Eimeria consist of eight sugars (Glc1Man5GlcNAc2) and seven sugars (Man5GlcNAc2), respectively (1). Plasmodium spp. (including P. falciparum and P. vivax) and Babesia spp. have genes encoding Alg7, Alg13, and Alg14, as well as four OST peptides, and so they are predicted to make an N-glycan precursor with two sugars (GlcNAc2), the same as that of Giardia that we have recently extensively studied (9, 18, 41, 46). Theileria is missing all of the Alg enzymes and OST peptides and so makes no N-glycans (46). The predicted N-glycans of Plasmodium and Toxoplasma were experimentally tested as described below.
Apparent selection against sites for N-linked glycosylation in apicoplast proteins of Toxoplasma.
Relative to Plasmodium, Toxoplasma has greatly reduced densities of sites of N glycosylation (sequons) in its secreted and nucleus-encoded apicoplast proteins (Fig. 2) (11, 12, 25). Because of the reduced sequon density in Toxoplasma, nearly half of the apicoplast proteins lack sequons and so cannot contain N-glycans (see Fig. S1 in the supplemental material). In contrast, <10% of Plasmodium secreted and nucleus-encoded apicoplast proteins lack sequons, while ∼20% of human secreted proteins lack sequons (12).
Sequon density is reduced in Toxoplasma secreted and apicoplast proteins by two different mechanisms. First, because Asn is encoded by AAT or AAC, codon usage bias has a strong effect on the density of N-glycan sites in secreted proteins (12). Toxoplasma, which is 41% AT in its coding sequences, has the lowest density of N-glycan sites in its secreted proteins of 33 eukaryotes examined (12). Plasmodium, which is 76% AT in its coding sequences, has the third-highest density of N-glycan sites in its secreted proteins (Fig. 2). Humans, which are 48% AT in their coding sequences, have an intermediate density of sequons in their secreted proteins (12).
Second, there is a decreased probability that Asn, Ser, and Thr will be present in sequons rather than elsewhere in apicoplast proteins of Toxoplasma. This type of negative selection is shown by plotting the actual density of sequons (y axis) versus the calculated density of sequons based upon the composition of Asn, Ser, Thr, and Pro in these proteins (x axis) (Fig. 2) (12). There is no selection on sequons in secreted proteins of Toxoplasma or secreted and apicoplast proteins of Plasmodium, so all of these points fall on the line with the slope of 1 where actual sequon density equals calculated sequon density (Fig. 2). Points below the line with slope of 1 (sequons of apicoplast proteins of Toxoplasma) reflect negative selection against sequons where actual sequon density is less than the calculated sequon density. In contrast, there is positive selection for sequons with Thr in human secreted proteins (Fig. 2), so this point falls above the line with the slope of 1 (the actual sequon density is twice the calculated sequon density) (12).
Evidence for very short N-glycans of Plasmodium.
Because N-glycan synthesis by Plasmodium has been so controversial (6, 22, 29), we used multiple biochemical and morphological methods to determine whether asexual stages of Plasmodium falciparum grown in red blood cells (RBCs) make the predicted N-glycan (GlcNAc2). Evidence for the very short N-glycans of Plasmodium included the following.
(i) Plasmodia metabolically labeled with tritiated GlcN make lipid-linked N-glycan precursors and N-glycans composed of GlcNAc or GlcNAc2, depending upon the stage (Fig. 3A to C). In contrast, there is no labeling of N-glycan precursors or N-glycans with tritiated Man, which labels Plasmodium GPI anchors (data not shown) (6, 22). Using the same methods, Giardia N-glycans were found to be predominantly GlcNAc2 (46).
(ii) The GlcNAc-binding Griffonia simplicifolia lectin (GSL-II) binds to numerous Plasmodium glycoproteins, which have been separated on SDS-PAGE and blotted to polyvinylidene difluoride (PVDF) membranes (Fig. 3D and E) (59). Pretreatment of Plasmodium glycoproteins with peptide:N-glycanaseF (PNGaseF) substantially decreases the binding of the GSL-II.
(iii) WGA-1-affinity chromatography and release with excess acetyl-chitotetrose (GlcNAc4) markedly enriches glycoproteins of plasmodia isolated from RBCs by saponin lysis (Fig. 3F and G) (56).
(iv) Mass spectroscopy of Plasmodium proteins shows that WGA-1-affinity chromatography enriches secreted proteins, including those from rhoptries and the ER (Table 1) (57). Because the peptide coverage was limited, we were not able to identify glycopeptides containing N-linked GlcNAc.
Table 1.
Most-abundant proteins identified by mass spectroscopy before or after WGA-1 affinity purification of Plasmodium glycoproteinsa
| Proteinb | % of totalc |
|---|---|
| Before WGA-1 affinity purification | |
| MSP1 | 12 |
| SAM synthetase | 8 |
| Hsp70 (secreted) | 8 |
| Glyceraldehyde 3-phosphate dehydrogenase | 7 |
| EF1-alpha | 7 |
| Lactate dehydrogenase | 4 |
| Enolase | 3 |
| Hsp70-1 (cytosolic) | 3 |
| Fructose-bisphosate aldolase | 3 |
| Hsp86 (cytosolic) | 2 |
| Hsp70-2 (cytosolic) | 1 |
| After WGA-1 affinity purification | |
| MSP1 | 17 |
| RAP1 | 14 |
| RAP2 | 8 |
| Glyceraldehyde 3-phosphate dehydrogenase | 8 |
| Endoplasmin | 5 |
| RAP3 | 4 |
| Acid phosphatase | 1 |
| MDR | 1 |
| Pyruvate kinase | 1 |
| Ornithine aminotransferase | 1 |
| Plasmepsin | 0.6 |
Results for a representative experiment are shown.
Proteins in bold are secreted or membrane proteins. While cytosolic proteins dominate prior to WGA-1 affinity purification, secreted proteins dominate after WGA-1-purification.
Relative protein abundance was estimated from the area/height results using SEAQUEST.
(v) The GlcNAc-binding lectin GSL-II labels well N-glycans of Plasmodium, while the fucose-binding lectin Ulex europaeus agglutinin 1 (UEA-1) binds to RBCs (Fig. 4, rows A and B). GSL-II labeling of plasmodia is inhibited by excess diacetyl-chitobiose (GlcNAc2) (Fig. 4, row C), and there is no overlap between GSL-II binding and GFP targeted to the cytosol (see Fig. S2, row A, in the supplemental material). While GSL-II labeling of the surface of RBCs infected with plasmodia is very low or absent (data not shown), GSL-II labels the surface of trophozoites and schizonts, which have been released from RBCs with saponin and costained with antibodies to MSP-1 (see Fig. S2, row B). GSL-II, which also binds to N-glycans of Giardia (see Fig. S3A), was used because WGA-1 (used to affinity purify glycoproteins of Plasmodium and Giardia) (41) binds to sialic acid residues on RBCs (see Fig. S3B).
Use of antiretroviral lectins to argue for the presence of both Man5GlcNAc2 and Man9GlcNAc2 in the N-glycans of Toxoplasma.
Using biochemical methods, Garénaux et al. (19) recently concluded that Toxoplasma uses its own N-glycan precursor (Glc3Man5GlcNAc2) as well as that of the host (Glc3Man9GlcNAc2) to make N-glycans. In contrast, other investigators identified N-glycans based upon a Glc3Man5GlcNAc2 precursor (32) or a Glc3Man9GlcNAc2 precursor (16). The goal here was to use antiretroviral lectins which bind to Man9GlcNAc2 only (scytovirin) or bind to both Man5GlcNAc2 and Man9GlcNAc2 (cyanovirin-N) (2) to localize N-glycans of Toxoplasma and so rule out the possibility that Toxoplasma N-glycans based upon a Glc3Man9GlcNAc2 precursor were the result of host glycoprotein contamination.
The specificity of the antiretroviral lectins (scytovirin and cyanovirin-N) for particular N-glycans was demonstrated in three ways. First, we prepared a carbohydrate microarray containing a series of synthetic high-mannose oligosaccharides, which were prepared with a chemical linker in place of the GlcNAc2 on the reducing end (2, 42). With the array, we confirmed that cyanovirin-N binds to Man5 and Man9, as well as to the D1 arm (present in Man5) and the D3 arm (absent in Man5) (Fig. 5A to C). Scytovirin, which binds to Man9 and the D3 arm, does not bind to either biosynthetic Man5 or the D1 arm (Fig. 5D). In contrast, the plant lectin concanavalin A, which has previously been used to visualize Toxoplasma glycoproteins with N-glycans (16, 32), binds to any mannose-containing glycan (Fig. 5E). Second, we used fluorescence microscopy to show that cyanovirin-N and scytovirin both bind to wild-type Saccharomyces, which builds its N-glycans on a precursor that contains nine Man residues (Fig. 5G) (26). In contrast, cyanovirin-N but not scytovirin binds to a Saccharomyces Δalg3 mutant, which builds its N-glycans on a precursor that contains five Man residues (Fig. 5H) (26). Third, we used flow cytometry to measure the relative binding of cyanovirin-N and scytovirin to wild-type Saccharomyces and to Saccharomyces Δalg3 mutants (Fig. 5J and K). As expected, we found that cyanovirin-N binds to both wild-type and Δalg3 Saccharomyces, while scytovirin binds only to wild-type Saccharomyces.
The presence of Toxoplasma N-glycans composed of Man9GlcNAc2, which derive from host cell N-glycan precursors, was confirmed in two ways (19). First, scytovirin (specific for Man9GlcNAc2) and cyanovirin-N (which binds to both Man5GlcNAc2 and Man9GlcNAc2) label the same apical structures in Toxoplasma, which were fixed and permeabilized (Fig. 5I). Second, although cyanovirin-N labels Toxoplasma more strongly than does scytovirin, the scytovirin labeling is well above background with no label, as measured by flow cytometry (Fig. 5L). A rough calculation made from the ratios of cyanovirin-N binding to scytovirin binding to Toxoplasma suggests that ∼2/3 of the Toxoplasma N-glycans derive from the endogenous precursor (Glc3Man5GlcNAc2) and 1/3 derive from the host precursor (Glc3Man9GlcNAc2). In summary, the results here argue for the presence of N-glycans which derive from both protist and host precursors in apical glycoproteins of Toxoplasma, as recently suggested (19).
GSL-II strongly labels the Plasmodium rhoptry and weakly labels the ER and surface but does not label the apicoplast, food vacuole, or parasitophorous vacuole.
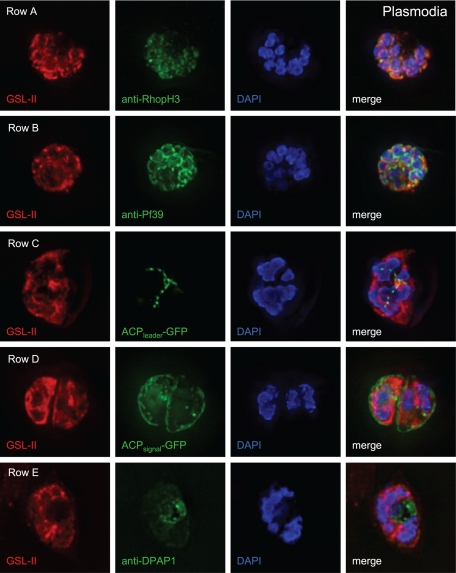
For colocalization studies with Plasmodium N-glycans (labeled red with GSL-II), either we examined transfected plasmodia expressing GFP targeted to various subcellular structures (55) or we labeled nontransformed plasmodia with antibodies to proteins associated with particular subcellular structures (Fig. 6). Both methods worked very well, and unsynchronized cultures were used to sample various stages of development. GSL-II labeling overlaps best with the binding of antibodies to rhoptry protein 3 (anti-RhopH3) (Fig. 6, row A) (14). Rhoptry proteins were also enriched after binding Plasmodium glycoproteins to the WGA-1 resin (Table 1).
GSL-II overlaps to a lesser degree with binding of antibodies to Pf39, which is a membrane-associated calcium-binding protein with a C-terminal ER retention signal (Fig. 6, row B) (51). In contrast, there is no overlap between GSL-II and GFP targeted to the apicoplasts (ACPleader-GFP) (Fig. 6, row C), parasitophorous vacuole (ACPsignal-GFP) (Fig. 6, row D), and cytosol (ACPtransit-GFP) (see Fig. S2A in the supplemental material) (55). Control antibodies to GFP label the apicoplasts and parasitophorous vacuole, showing that accessibility does not prevent labeling of these compartments by GSL-II (see Fig. S4A and S4B, respectively). For the most part, GSL-II does not overlap with antibodies to dipeptidyl aminopeptidase I (anti-DPAP1), which is a food vacuole protein (Fig. 6, row E) (30). We conclude that occupation of N-glycan sites is markedly reduced in apicoplast proteins compared to some secretory proteins in Plasmodium.
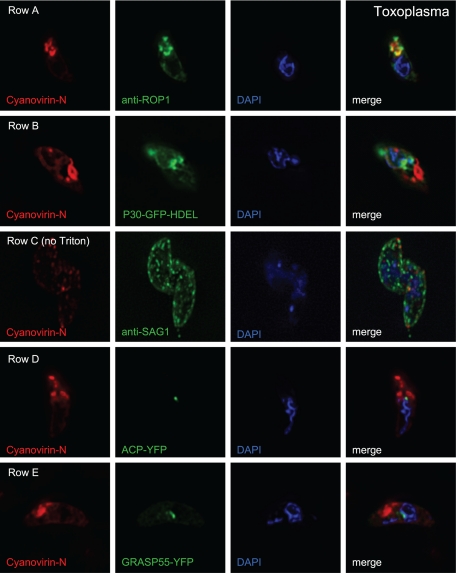
Cyanovirin-N strongly labels the Toxoplasma rhoptry and weakly labels the ER and plasma membrane but does not label the apicoplast and Golgi apparatus.
Cyanovirin-N, which binds N-glycans of Toxoplasma, colocalizes extensively but not exclusively with antibodies to rhoptry protein 1 (anti-ROP1; MAb Tg49) (Fig. 7, row A) (47). Cyanovirin-N also colocalizes in apical secretory organelles with the GalNAc-binding plant lectin Maclura pomifera agglutinin 1 (MPA-1), which binds to O-linked glycans of Toxoplasma, as well as the fucose-binding lectin UEA-1 (data not shown) (50). Cyanovirin-N weakly colocalizes with GFP targeted to the ER (P30-GFP-HDEL) (Fig. 7, row B) and with antibodies to surface antigen 1 (anti-SAG1; MAb DG52) (Fig. 7, row C) (8, 24). In contrast, there is no overlap between cyanovirin-N and yellow fluorescent protein fused to the acyl carrier protein (ACP-YFP), which labels apicoplasts of Toxoplasma (Fig. 7, row D) (35). There is also no labeling of the apicoplast with MPA-1 and UEA-1 (data not shown). Control antibodies to GFP label Toxoplasma apicoplasts, showing that lack of accessibility does not prevent labeling of the organelle by cyanovirin-N and the plant lectins (see Fig. S5B in the supplemental material). There is no overlap between the Golgi marker (GRASP55) and cyanovirin-N (Fig. 7, row E) (40). This result may be secondary to modifications of N-glycans in the Golgi apparatus of Toxoplasma which block cyanovirin-N binding. Finally, cyanovirin-N does not bind to cisternae of the inner membrane complex (23). We conclude that glycoproteins containing unmodified N-glycans (recognized by cyanovirin-N) are present in the apical secretory vesicles but absent from the apicoplast.
DISCUSSION
Development of novel probes for N-glycans of Plasmodium and Toxoplasma.
The biochemical methods used here showed that Plasmodium makes the N-glycan precursor predicted by its Alg enzymes (dolichol-PP-GlcNAc2), as well as dolichol-PP-GlcNAc (Fig. 1, 3, and 4) (26, 46). We did not find any host (high-mannose) N-glycans associated with Plasmodium, as previously suggested (29). The very short Plasmodium N-glycans were likely missed by other investigators because they made the reasonable assumption that Plasmodium N-glycans contain mannose residues, as was the case for all N-glycans studied to that date (6, 22, 26, 46).
Because WGA-1, which was used to localize GlcNAc2 on N-glycans of Giardia (41), cross-reacts with sialic acid residues on RBC glycoproteins, we used GSL-II to localize Plasmodium N-glycans (Fig. 3, 4, and 6; see Fig. S2 and S3 in the supplemental material) (59). As GSL-II, like WGA-1 and other GlcNAc-binding lectins, may have a weaker affinity for a single GlcNAc rather than GlcNAc polymers, we may have failed to localize some Plasmodium N-glycans containing a single GlcNAc. Still, GSL-II brightly labels numerous secretory compartments of plasmodia and is an excellent “counterstain” for organelles localized with GFP or antibodies (Fig. 6) (14, 21, 30, 51, 52). Indeed GSL-II labeling provides three-dimensional images of plasmodia using deconvolution methods, while interference contrast microscopy, which is commonly used in association with GFP or antiparasite antibodies, provides information in only two dimensions. GSL-II labeling of plasmodia, in conjunction with UEA-1 labeling of RBCs and DAPI labeling of RBCs (Fig. 4), could be used to facilitate diagnosis of malaria.
The antiretroviral lectins cyanovirin-N and scytovirin used here to localize Toxoplasma N-glycans have much greater specificity for high-mannose N-glycans than the plant lectin concanavalin A, which was previously used to localize Toxoplasma N-glycans (Fig. 5 and 7) (2, 16, 32). With scytovirin, we confirmed the remarkable finding of Garénaux et al. (19) that Toxoplasma N-glycans derive from precursors predicted by the parasite Alg enzymes (Man5GlcNAc2), as well as those of the host (Man9GlcNAc2) (26, 32, 46). While recent studies demonstrate membrane fusion between the host cell ER and the parasitophorous vacuole (20), it is not clear how lipid-pyrophosphate-linked N-glycan precursors move from the parasitophorous vacuole to the protist ER, where the OST adds N-glycans to nascent proteins (28, 45). Regardless, scytovirin should be a useful tool for studies of mechanisms of membrane fusion between the host ER and the parasitophorous vacuole.
Summary of the evidence for selection against N-glycans in Plasmodium and Toxoplasma.
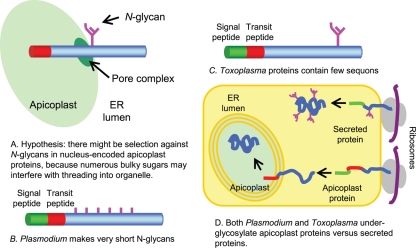
The present experiments support our hypothesis that there is Darwinian selection against N-glycans in nucleus-encoded apicoplast proteins, which traverse through the ER prior to threading into the organelle (Fig. 8A). (11, 21, 25, 35, 37, 39, 48, 52, 55). Negative selection against N-glycans appears to occur by multiple mechanisms.
Fig. 8.
Model summarizing our hypothesis and results with Plasmodium and Toxoplasma. (A) We hypothesized that bulky N-glycans might interfere with threading of nucleus-encoded apicoplast proteins into the organelle. (B) We found that Plasmodium markedly reduces the length of its N-glycans without reducing the density of sites of N-linked glycosylation. (C) In contrast, Toxoplasma has longer N-glycans but dramatically reduces the density of sites of N-glycans (sequons), so that nearly half of the Toxoplasma nucleus-encoded apicoplast proteins have no sequons and so contain no N-glycans. (D) While secreted proteins are N glycosylated, sites of N-glycans (sequons) on nucleus-encoded apicoplast proteins of Plasmodium and Toxoplasma remain unoccupied.
(i) There is extensive secondary loss of Alg enzymes that make N-glycan precursors in apicomplexans, which are transmitted via an arthropod vector. Plasmodium and Babesia are capable of making dolichol-PP-GlcNAc2, while Theileria makes no N-glycan at all (Fig. 1 and 8B).
(ii) In Plasmodium, early blood stage parasites make lipid-linked N-glycan precursors which are composed predominantly of a single GlcNAc, while later blood stage parasites make N-glycan precursors which are a mixture of GlcNAc and GlcNAc2 (Fig. 3). This result suggests that Plasmodium is underutilizing the few Alg enzymes that the organism has (Fig. 1).
(iii) The density of sequons (sites of N-linked glycosylation) in Toxoplasma secreted and apicoplast proteins is extremely low, which correlates with the marked GC-rich codon usage of this protist (Fig. 2 and 8C) (12). In addition, there is decreased probability that Asn, Ser, and Thr will be in sequons rather than elsewhere in nucleus-encoded apicoplast proteins of Toxoplasma, so that nearly half of these proteins lack any sites of N-linked glycosylation. Positive selection for sequons with Thr in secreted proteins of eukaryotes, which use N-glycans for quality control of protein folding, also occurs by changes in the probability that Asn and Thr will be in sequons rather than elsewhere in the protein (12).
(iv) There appears to be a marked reduction in the occupation of N-glycan sites (sequons) in nucleus-encoded apicoplast proteins of Plasmodium and Toxoplasma in comparison to secreted proteins of each protist (Fig. 6, 7, and 8D). In the case of Plasmodium, nucleus-encoded apicoplast proteins have the same high density of sequons as secreted proteins (Fig. 2), so we had expected strong GSL-II labeling of the Plasmodium apicoplast. In the case of Toxoplasma, nucleus-encoded apicoplast proteins have about half the density of N-glycan sites as secreted proteins. While previous investigators concluded that concanavalin A colocalizes with the Toxoplasma apicoplast, the pictures shown suggest that concanavalin A stains the ER adjacent to the apicoplast (16).
(v) Our N-glycan occupancy results are consistent with recent studies of protein targeting to the apicoplast that (i) demonstrate a pore (Tic20 homolog) through which Toxoplasma apicoplast proteins are threaded (54) and (ii) suggest the possibility that apicoplast proteins are rapidly dislocated from the ER lumen by an ERAD-type mechanism prior to targeting to the organelle (13, 21).
(vi) How apicoplast-targeted proteins avoid N glycosylation is unclear. One possibility is that the nucleus-encoded apicoplast proteins are shuttled out of the ER and into the organelle while the proteins are being translocated and so before N-glycans are added (28, 39, 48). An alternative possibility is that oligosaccharyltransferases (OSTs) of apicomplexans do not transfer N-glycans to nascent proteins during translocation. Instead, parasite OSTs may transfer N-glycans to proteins after translocation into the ER, as has been recently described for some human and Trypanosoma OSTs (34, 45). This suggests the possibility that apicoplast proteins remain without N-glycans, because they are removed from the lumen of the ER prior to addition of the N-glycans by the posttranslational OST.
Alternative interpretations and limitations of the evidence.
Alternative interpretations and limitations of the evidence are as follows.
(i) Our results show that apicomplexans transmitted by arthropods (Plasmodium, Babesia, and Theileria) have very short or no N-glycans, while apicomplexans which have an oocyst form and are transmitted orally (Toxoplasma, Neospora, Cryptosporidium, and Eimeria) have longer N-glycan precursors (Fig. 1). Because apicomplexans transmitted by an arthropod vector are phylogenetically more similar to each other than to apicomplexans with oocyst forms, it is not clear whether differences in N-glycan length are the result of differences in mode of transmission or the result of common ancestry (31).
(ii) Extensive secondary loss of Alg enzymes also occurs in Giardia, which makes the same very short N-glycans as Plasmodium, and in microsporidia, which make no N-glycans (12, 41, 46). Because these organisms, which are unrelated to Plasmodium and Theileria, do not have an apicoplast, it is likely that very short N-glycans have other benefits for parasitic protists. For example, the very short N-glycan GlcNAc2 is not immunogenic, while longer GlcNAc polymers, such as those present in chitin, are immunogenic (43).
(iii) A strong GC-rich codon bias also occurs in Leishmania, which does not have an apicoplast (12). While a high GC content is necessary for genome stability in thermophilic bacteria (58), the forces driving high GC content in other organisms are not well understood.
(iv) As discussed above, it is likely that the lectins used here do not identify all of the N-glycans present on glycoproteins of Plasmodium and Toxoplasma. In the case of plasmodia, there may be apicoplast and secreted proteins with N-glycans composed of a single GlcNAc that are missed by GSL-II. In the case of Toxoplasma, there may be complex N-glycans, which have been modified in the Golgi apparatus, that are not recognized by cyanovirin-N or scytovirin (2).
(v) In addition to apicoplasts, other secretory compartments of Plasmodium (e.g., the food vacuole, parasitophorous vacuole, and RBC surface) are not labeled with GSL-II (Fig. 6). Similarly, in addition to apicoplasts, other secretory compartments of Toxoplasma (the Golgi apparatus, inner membrane complex, and plasma membrane) are not labeled by cyanovirin-N (Fig. 7). These results suggest that there is a complicated relationship between N-linked glycosylation and protein targeting in these apicomplexans that we do not yet understand.
(vi) This relative lack of specificity with regard to selection against N-glycans in apicomplexans is in contrast to the great specificity of positive selection for sites of N-linked glycosylation (sequons) in the vast majority of eukaryotes (12). Positive selection for sequons occurs only in organisms with N-glycan-dependent QC of protein folding, is for secreted proteins but not cytosolic proteins, is for sequons with Thr but not those with Ser, and is based upon the increased probability that Asn and Thr are present in sequons rather than elsewhere in secreted proteins.
(vii) There is a built-in redundancy or inconsistency with regard to the apparent selection against N-glycans in these apicoplexans. If apicoplast proteins of both Plasmodium and Toxoplasma are selectively underglycosylated (Fig. 6, 7, and 8D), why is there also apparent selection against N-glycan length in Plasmodium (Fig. 1 and 8B) and N-glycan density in Toxoplasma (Fig. 2 and 8C)?
Because of the alternative explanations of the data and because we do not have a mechanism for the reduced occupancy of N-glycan sites of Plasmodium and Toxoplasma, we think that these studies provide suggestive evidence for our hypothesis that there is selection against N-glycans in nucleus-encoded apicoplast proteins (Fig. 8A). A strength of the hypothesis is that it makes sense of disparate facts concerning these parasites, which are otherwise unexplained (e.g., secondary loss of Alg enzymes in apicomplexans [Fig. 1 and 8B] and marked decrease in sites of N-linked glycosylation in Toxoplasma [Fig. 2 and 8C]). Experiments to test our hypothesis and to further explore the complex relationship between protein-targeting and glycosylation in these apicomplexans are in progress.
ADDENDUM IN PROOF
Recent work by Boris Striepen and colleagues (S. Agrawal, G. G. van Dooren, W. L. Beatty, and B. Striepen, J. Biol. Chem. 284:33683–33691, 2009) shows that an endosymbiont-derived endoplasmic reticulum-associated degradation(ERAD) system functions in the import of apicoplast proteins. Their results suggest that apicoplast proteins may by removed from the lumen of the ER prior to addition of N-glycans.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants AI048082 (to J.S.), AI057919 (to M.T.D.), and GM31318 (to P.W.R.). M.T.D. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. Support for D.M.R. was provided by the Training Program in Host Pathogen Interactions (T32 AI052070). Support for M.J.G. was provided by American Heart Association Scientist Development grant 0635480N and a March of Dimes Basil O'Connor Starter Scholar Research Award.
We thank Dick Cook of MIT for some mass spectroscopy data.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., et al. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445 [DOI] [PubMed] [Google Scholar]
- 2.Adams E. W., Ratner D. M., Bokesch H. R., McMahon J. B., O'Keefe B. R., Seeberger P. H. 2004. Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology; glycan-dependent gp120/protein interactions. Chem. Biol. 11:875–881 [DOI] [PubMed] [Google Scholar]
- 3.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., et al. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37:D539–D543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee S., Vishwanath P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W., Samuelson J. 2007. Evolution of quality control of protein-folding in the ER lumen. Proc. Natl. Acad. Sci. U. S. A. 104:11676–11681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berhe S., Gerold P., Kedees M. H., Holder A. A., Schwarz R. T. 2000. Plasmodium falciparum: merozoite surface proteins 1 and 2 are not post-translationally modified by classical N- or O-glycans. Exp. Parasitol. 94:194–197 [DOI] [PubMed] [Google Scholar]
- 7.Brayton K. A., Lau A. O., Herndon D. R., Hannick L., Kappmeyer L. S., et al. 2007. Genome sequence of Babesia bovis and comparative analysis of apicomplexan hemoprotozoa. PLoS Pathog. 3:1401–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bülow R., Boothroyd J. C. 1991. Protection of mice from fatal Toxoplasma gondii infection by immunization with p30 antigen in liposomes. J. Immunol. 147:3496–3500 [PubMed] [Google Scholar]
- 9.Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., et al. 2008. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455:757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chantret I., Dancourt J., Barbat A., Moore S. E. 2005. Two proteins homologous to the N- and C-terminal domains of the bacterial glycosyltransferase MurG are required for the second step of dolichyl-linked oligosaccharide synthesis in S. cerevisiae. J. Biol. Chem. 280:9236–9242 [DOI] [PubMed] [Google Scholar]
- 11.Chen Z., Harb O. S., Roos D. S. 2008. In silico identification of specialized secretory-organelle proteins in apicomplexan parasites and in vivo validation in Toxoplasma gondii. PLoS One 3:e3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui J., Smith T., Robbins P. W., Samuelson J. 2009. Darwinian selection for sequons (sites of Asn-liked glycosylation) with Thr in phylogenetically disparate eukaryotes and viruses. Proc. Natl. Acad. Sci. U. S. A. 106:13421–13428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeRocher A. E., Coppens I., Karnataki A., Gilbert L. A., Rome M. E., Feagin J. E., Bradley P. J., Parsons M. 2008. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot. Cell 7:1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doury J. C., Goasdoue J. L., Tolou H., Martelloni M., Bonnefoy S., Mercereau-Puijalon O. 1997. Characterization of the binding sites of monoclonal antibodies reacting with the Plasmodium falciparum rhoptry protein RhopH3. Mol. Biochem. Parasitol. 85:149–159 [DOI] [PubMed] [Google Scholar]
- 15.Eisen J. A., Coyne R. S., Wu M., Wu D., Thiagarajan M., et al. 2006. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauquenoy S., Morelle W., Hovasse A., Bednarczyk A., Slomianny C., Schaeffer C., van Dorsselaer A., Tomavo S. 2008. Proteomics and glycomics analyses of N-glycosylated structures involved in Toxoplasma gondii-host cell interactions. Mol. Cell. Proteomics 7:891–910 [DOI] [PubMed] [Google Scholar]
- 17.Gajria B., Bahl A., Brestelli J., Dommer J., Fischer S., et al. 2008. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 36:D553–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner M. J., Hall N., Fung E., White O., Berriman M., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garénaux E., Shams-Eldin H., Chirat F., Bieker U., Schmidt J., Michalski J. C., Cacan R., Gue′rardel Y., Schwarz R. T. 2008. The dual origin of Toxoplasma gondii N-glycans. Biochemistry 47:12270–12276 [DOI] [PubMed] [Google Scholar]
- 20.Goldszmid R. S., Coppens I., Lev A., Caspar P., Mellman I., Sher A. 2009. Host ER-parasitophorous vacuole interaction provides a route of entry for antigen cross-presentation in Toxoplasma gondii-infected dendritic cells. J. Exp. Med. 206:399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gould S. B., Waller R. F., McFadden G. I. 2008. Plastid evolution. Annu. Rev. Plant Biol. 59:491–517 [DOI] [PubMed] [Google Scholar]
- 22.Gowda D. C., Gupta P., Davidson E. A. 1997. Glycosylphosphatidylinositol anchors represent the major carbohydrate modification in proteins of intraerythrocytic stage Plasmodium falciparum. J. Biol. Chem. 272:6428–6439 [DOI] [PubMed] [Google Scholar]
- 23.Gubbels M. J., Wieffer M., Striepen B. 2004. Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol. Biochem. Parasitol. 137:99–110 [DOI] [PubMed] [Google Scholar]
- 24.Hager K. M., Striepen B., Tilney L. G., Roos D. S. 1999. The nuclear envelope serves as an intermediary between the ER and Golgi complex in the intracellular parasite Toxoplasma gondii. J. Cell Sci. 112:2631–2638 [DOI] [PubMed] [Google Scholar]
- 25.Harb O. S., Chatterjee B., Fraunholz M. J., Crawford M. J., Nishi M., Roos D. S. 2004. Multiple functionally redundant signals mediate targeting to the apicoplast in the apicomplexan parasite Toxoplasma gondii. Eukaryot. Cell 3:663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helenius A., Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019–1049 [DOI] [PubMed] [Google Scholar]
- 27.Käll L., Krogh A., Sonnhammer E. L. L. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 28.Kelleher D. J., Gilmore R. 2006. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16:47R–62R [DOI] [PubMed] [Google Scholar]
- 29.Kimura E. A., Couto A. S., Peres V. J., Casal O. L., Katzin A. M. 1996. N-linked glycoproteins are related to schizogony of the intraerythrocytic stage in Plasmodium falciparum. J. Biol. Chem. 271:14452–14461 [DOI] [PubMed] [Google Scholar]
- 30.Klemba M., Gluzman I., Goldberg D. E. 2004. A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J. Biol. Chem. 279:43000–43007 [DOI] [PubMed] [Google Scholar]
- 31.Kuo C. H., Wares J. P., Kissinger J. C. 2008. The Apicomplexan whole-genome phylogeny: an analysis of incongruence among gene trees. Mol. Biol. Evol. 25:2689–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luk F. C., Johnson T. M., Beckers C. J. 2008. N-linked glycosylation of proteins in the protozoan parasite Toxoplasma gondii. Mol. Biochem. Parasitol. 157:169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magnelli P., Cipollo J. F., Ratner D. M., Cui J., Kelleher D., Gilmore R., Costello C. E., Robbins P. W., Samuelson J. 2008. Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. J. Biol. Chem. 283:18355–18364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manthri S., Güther M. L., Izquierdo L., Acosta-Serrano A., Ferguson M. A. 2008. Deletion of the TbALG3 gene demonstrates site-specific N-glycosylation and N-glycan processing in Trypanosoma brucei. Glycobiology 18:367–383 [DOI] [PubMed] [Google Scholar]
- 35.Mazumdar J., Wilson E. H., Masek K., Hunter C. A., Striepen B. 2006. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. U. S. A. 103:13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moll K., Ljungström I., Perlmann H., Scherf A., Wahlgren M. (ed.). 2008. Methods in malaria research, 5th ed., version 5.2.MR4/ATCC, Manassas, VA [Google Scholar]
- 37.Nishi M., Hu K., Murray J. M., Roos D. S. 2008. Organellar dynamics during the cell cycle of Toxoplasma gondii. J. Cell Sci. 121:1559–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pain A., Renauld H., Berriman M., Murphy L., Yeats C. A., et al. 2005. Genome of the host-cell transforming parasite Theileria annulata compared with T. parva. Science 309:131–133 [DOI] [PubMed] [Google Scholar]
- 39.Parsons M., Karnataki A., Feagin J. E., DeRocher A. 2007. Protein trafficking to the apicoplast: deciphering the apicomplexan solution to secondary endosymbiosis. Eukaryot. Cell 6:1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelletier L., Stern C. A., Pypaert M., Sheff D., Ngô H. M., et al. 2002. Golgi biogenesis in Toxoplasma gondii. Nature 418:548–552 [DOI] [PubMed] [Google Scholar]
- 41.Ratner D. M., Cui J., Steffen M., Moore L. L., Robbins P. W., Samuelson J. 2008. Changes in the N-glycome (glycoproteins with Asn-linked glycans) of Giardia lamblia with differentiation from trophozoites to cysts. Eukaryot. Cell 7:1930–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratner D. M., Plante O. J., Seeberger P. H. 2002. A linear synthesis of branched high-mannose oligosaccharides from the HIV-1 viral surface envelope glycoprotein gp120. Eur. J. Org. Chem. 5:826–833 [Google Scholar]
- 43.Reese T. A., Liang H. E., Tager A. M., Luster A. D., Van Rooijen N., Voehringer D., Locksley R. M. 2007. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos D. S., Donald R. G., Morrissette N. S., Moulton A. L. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27–63 [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Canada C., Kelleher D. J., Gilmore R. 2009. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136:272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuelson J., Banerjee S., Magnelli P., Cui J., Kelleher D. J., Gilmore R., Robbins P. W. 2005. The diversity of protist and fungal dolichol-linked precursors to Asn-linked glycans likely results from secondary loss of sets of glycosyltransferases. Proc. Natl. Acad. Sci. U. S. A. 102:1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartzman J. D. 1986. Inhibition of a penetration-enhancing factor of Toxoplasma gondii by monoclonal antibodies specific for rhoptries. Infect. Immun. 51:760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheiner L., Soldati-Favre D. 2008. Protein trafficking inside Toxoplasma gondii. Traffic 9:636–646 [DOI] [PubMed] [Google Scholar]
- 49.Shirley M. W., Ivens A., Gruber A., Madeira A. M., Wan K. L., Dear P. H., Tomley F. M. 2004. The Eimeria genome projects: a sequence of events. Trends Parasitol. 20:199–201 [DOI] [PubMed] [Google Scholar]
- 50.Stwora-Wojczyk M. M., Kissinger J. C., Spitalnik S. L., Wojczyk B. S. 2004. O-glycosylation in Toxoplasma gondii: identification and analysis of a family of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Int. J. Parasitol. 34:309–322 [DOI] [PubMed] [Google Scholar]
- 51.Templeton T. J., Fujioka H., Aikawa M., Parker K. C., Kaslow D. C. 1997. Plasmodium falciparum Pfs40, renamed Pf39, is localized to an intracellular membrane-bound compartment and is not sexual stage-specific. Mol. Biochem. Parasitol. 90:359–365 [DOI] [PubMed] [Google Scholar]
- 52.Tonkin C. J., Foth B. J., Ralph S. A., Struck N., Cowman A. F., McFadden G. I. 2008. Evolution of malaria parasite plastid targeting sequences. Proc. Natl. Acad. Sci. U. S. A. 105:4781–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trombetta E. S., Parodi A. J. 2003. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 19:649–676 [DOI] [PubMed] [Google Scholar]
- 54.van Dooren G. G., Tomova C., Agrawal S., Humbel B. M., Striepen B. 2008. Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proc. Natl. Acad. Sci. U. S. A. 105:113574–113579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Waller R. F., Reed M. B., Cowman A. F., McFadden G. I. 2000. Protein trafficking to the plastid of Plasmodium falciparum is via the secretory pathway. EMBO J. 19:1794–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y., Wu S. L., Hancock W. S. 2006. Approaches to the study of N-linked glycoproteins in human plasma using lectin affinity chromatography and nano-HPLC coupled to electrospray linear ion trap-Fourier transform mass spectrometry. Glycobiology 16:514–523 [DOI] [PubMed] [Google Scholar]
- 57.Yates J. R., III, Carmack E., Hays L., Link A. J., Eng J. K. 1999. Automated protein identification using microcolumn liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 112:553–569 [DOI] [PubMed] [Google Scholar]
- 58.Zavala A., Naya H., Romero H., Musto H. 2002. Trends in codon and amino acid usage in Thermotoga maritima. J. Mol. Evol. 54:563–568 [DOI] [PubMed] [Google Scholar]
- 59.Zhu K., Bressan R. A., Hasegawa P. M., Murdock L. L. 1996. Identification of N-acetylglucosamine binding residues in Griffonia simplicifolia lectin II. FEBS Lett. 390:271–274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.