Abstract
The P2 aminopurine transporter, encoded by TbAT1 in African trypanosomes in the Trypanosoma brucei group, carries melaminophenyl arsenical and diamidine drugs into these parasites. Loss of this transporter contributes to drug resistance. We identified the genomic location of TbAT1 to be in the subtelomeric region of chromosome 5 and determined the status of the TbAT1 gene in two trypanosome lines selected for resistance to the melaminophenyl arsenical, melarsamine hydrochloride (Cymelarsan), and in a Trypanosoma equiperdum clone selected for resistance to the diamidine, diminazene aceturate. In the Trypanosoma brucei gambiense STIB 386 melarsamine hydrochloride-resistant line, TbAT1 is deleted, while in the Trypanosoma brucei brucei STIB 247 melarsamine hydrochloride-resistant and T. equiperdum diminazene-resistant lines, TbAT1 is present, but expression at the RNA level is no longer detectable. Further characterization of TbAT1 in T. equiperdum revealed that a loss of heterozygosity at the TbAT1 locus accompanied loss of expression and that P2-mediated uptake of [3H]diminazene is lost in drug-resistant T. equiperdum. Adenine-inhibitable adenosine uptake is still detectable in a ΔTbat1 T. b. brucei mutant, although at a greatly reduced capacity compared to that of the wild type, indicating that an additional adenine-inhibitable adenosine permease, distinct from P2, is present in these cells.
Organisms of the genus Trypanosoma cause a range of infectious diseases, including human African trypanosomiasis (HAT), nagana in cattle, and dourine in equines (2), and are of major public health and economic importance in sub-Saharan Africa. Chemotherapy is used against many of the human- and animal-infective parasites. However, all drugs currently registered for use carry significant problems related to administration, toxicity, increasing incidence of treatment failure, and in the case of animal trypanosomiasis, drug resistance (1). Previous work has shown that defects in the P2 aminopurine transporter, encoded by the TbAT1 gene (the same gene in Trypanosoma equiperdum is referred to as TeAT1 [Trypanosoma equiperdum AT1] in this article), are linked to drug resistance in Trypanosoma brucei brucei (11, 21), Trypanosoma brucei gambiense (22), and Trypanosoma brucei rhodesiense (28), as well as in Trypanosoma evansi (24, 32) and T. equiperdum (3), very close phylogenetic relatives of T. brucei (27). The P2 transporter has been shown to be capable of carrying both melaminophenyl arsenical (11) and diamidine (10, 14, 15, 18) classes of drug into African trypanosomes in the T. brucei group. Furthermore, a series of drug-resistant parasites from both the laboratory and the field have all been shown to be defective in P2-mediated transport using a novel fluorescence test (28), and a restriction fragment length polymorphism (RFLP)-based approach has been used to infer the presence of drug-resistant alleles in populations of human-infective parasites from Uganda (22). Previous work had indicated that two independent genetic mechanisms, namely, introduction of point mutations (21) and deletion of the TbAT1 gene (21, 18), are associated with the acquisition of drug resistance, which implies that several mechanisms can alter P2 transporter activity, causing drug resistance. A more complete understanding of these mechanisms will be essential to underpinning the development of better molecularly based diagnostic tools to monitor drug resistance.
Here we have used three isogenic pairs of resistant lines to investigate alterations within the TbAT1 gene that result in a loss of P2 expression and associated drug resistance. STIB 247 (Swiss Tropical Institute Basel 247), a T. b. brucei line, and STIB 386, a T. b. gambiense line, were selected independently for resistance to the melaminophenyl arsenical drug melarsamine hydrochloride (Cymelarsan). They are resistant to the maximum tolerated dose of melarsamine hydrochloride in animals (25), and the STIB 247 line is cross resistant to diminazene aceturate (26). The third line is a diminazene-resistant T. equiperdum line, PBR, derived from the parental sensitive line T. equiperdum P (33). A previous biochemical study investigated the role of the P2 transporter in the resistance of T. equiperdum PBR to trypanocidal drugs (3), finding that the overall rate of adenosine uptake was reduced. Inhibition studies indicated that the majority of the lost activity was due to a decrease in P2 activity. However, since some adenine-inhibitable adenosine uptake was still present, it was postulated that the change in P2 activity was due to a loss in the affinity for the drugs as substrates. This mechanism of drug resistance, where point mutations to a transporter have altered the substrate specificity and conferred drug resistance, has also been reported in Leishmania donovani (30).
With the identification of TbAT1 encoding P2 activity (21), it was important to determine what changes, if any, occurred to this gene during selection of diminazene resistance in T. equiperdum and melarsamine hydrochloride resistance in T. brucei and what effect these changes have had on transcript availability and transporter activity. The TbAT1 gene does not, however, appear in the published sequence (6) or version 4 of the T. brucei genome sequence, suggesting that a region of the sequence is incomplete. As this gene and possibly surrounding regions are important for our understanding of the mechanisms of resistance to the melaminophenyl arsenicals and diamidines, we determined the sequence of a bacterial artificial chromosome (BAC) clone covering the region of the genome surrounding the TbAT1 gene.
MATERIALS AND METHODS
Trypanosome strains and cultivation.
The T. b. brucei STIB 247 cell line was originally isolated from a hartebeest in 1971 in the Serengeti National Park, Tanzania (17). The melarsamine hydrochloride-resistant 247Mr line was derived by serial passage in mice treated with subcurative doses of melarsamine hydrochloride. The resulting clone was found to be 130-fold less sensitive to melarsamine hydrochloride and 16-fold less sensitive to diminazene aceturate in vivo (25). The STIB 386 type 2 T. b. gambiense line was originally isolated from a man in Daloa in the Ivory Coast. The melarsamine hydrochloride-resistant 386Mr line was derived by serial passage in mice treated with subcurative doses of melarsamine hydrochloride. T. b. gambiense strain 386Mr is 20-fold less sensitive to melarsamine hydrochloride in vivo (25) and has been shown to have lost P2-mediated pentamidine transport (9). T. equiperdum BoTat 1 (Bordeaux trypanozoon antigenic type 1) P originated from a stock held at the Pasteur Institute, Paris. The diminazene aceturate (Berenil)-resistant line T. equiperdum PBR was derived from the parental line by serial passage through mice treated with subcurative doses of diminazene aceturate (33). This resistant line is 35.6-fold less sensitive to diminazene aceturate and 4-fold less sensitive to melarsamine hydrochloride in vitro. In vivo, the resistant line is insensitive to diminazene aceturate up to the maximum tolerated dose in Swiss mice and 4-fold less sensitive to melarsamine hydrochloride. The ΔTbat1 (P2) null mutant clone was previously described and constructed by sequential replacement of TbAT1 with the neomycin and puromycin resistance markers in T. b. brucei 427 MiTat 1.2 (BS221). The knockout line was 4-fold less sensitive to melarsoprol and melarsen oxide and 18.6-fold less sensitive to diminazene aceturate (23).
Molecular biology.
Bloodstream-form trypanosomes were harvested from culture. RNA was extracted using TRIzol reagent (Life Technologies) or a Qiagen RNeasy kit, according to the manufacturer's protocol. Genomic DNA was extracted from cells using a Qiagen DNeasy kit.
To determine the genomic status of TbAT1 in the various lines, the TbAT1 open reading frame (ORF) was amplified by PCR using Taq polymerase and the primers AT1F (5′ ATG CTC GGG TTT GAC TCA GC 3′) and AT1R (5′ CTA CTT GGG AAG CCC CTC AT 3′). Positive-control primers for amplifying a region of the triose phosphate isomerase (TIM) genes TIM-C and TIM-D were as previously described (9). Thermal cycling was carried out as follows: 1 cycle of 95°C for 2 min and 30 cycles of 95°C for 50 s, 58°C for 50 s, and 65°C for 2 min.
To determine the expression of TbAT1 in T. equiperdum PBR, Northern blots were prepared by standard methods (20), probed with the full-length TbAT1 open reading frame, and radioactively labeled with 32P using the Prime-It II kit (Stratagene). Blots were washed 3× for 30 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS) at 65°C before exposure to autoradiography for 1 to 3 days. To detect the presence of transcripts in the 247mr line, first-strand cDNA was synthesized using the Omniscript reverse transcriptase PCR (RT-PCR) kit (Qiagen) with oligo(dT). Total RNA was DNase treated with Turbo DNase (Ambion) prior to cDNA synthesis, and cDNA was amplified by PCR, using Taq polymerase and primers AT1F (see above) and AT1seq2 (5′ CAT ACT TGT AGT ACT CGA TG). Primers TIM-E and TIM-F were used as controls (19). Thermal cycling was carried out as follows: 1 cycle of 95°C for 2 min and 35 cycles of 95°C for 50 s, 50°C for 50 s, and 65°C for 1 min.
To sequence the TbAT1 ORF from the T. equiperdum wild-type and PBR lines, PCR was conducted using the primers P2F (5′ CAT GCG CTT TGG TGG AGG) and P2R (5′ TTG GCG AAT CGG TGT ACG), both of which fall outside the open reading frame (ORF) of TbAT1. All PCRs for sequencing were conducted in the presence of Pfu polymerase. PCR products, once purified, were cloned into the pGEM-T Easy vector and sequenced. Both strands were fully sequenced in all cases, using internal primers to give complete coverage of the ORF. For RFLP analysis of TbAT1 from the T. equiperdum wild-type and PBR lines, PCR was conducted with the primers AT1F and AT1R in the presence of Taq polymerase. The resultant PCR products were cleaned using the Qiagen PCR purification kit prior to restriction digestion with the enzymes described in the figure legends and text.
Rapid amplification of cDNA 3′ ends (3′ RACE) was carried out using the 5′/3′ RACE kit from Roche, according to manufacturer's instructions, and a gene-specific primer (5′-CTTCGTTGGCGCCATGTTCGC) for the first amplification reaction. The products of a nested PCR using the anchor primer and a second nested gene-specific primer (5′-GCTGTCAATGAGGGGCTTCCC) were ligated into pGEM-T Easy vector and sequenced with both T7 and SP6 oligos to provide double-stranded reads. The 3′ untranslated regions (3′ UTR) of T. equiperdum strains P and PBR were amplified using a primer (5′-GGACCTTCACACGTTTAAACAAGCG) which anneals beyond the indicated polyadenylation site and a primer (5′-CAAATAGTAACTAGTGGCGAGTAGGC) which anneals at 100 bp into the 3′ UTR. The PCR products were purified, cloned into pGEM-T Easy, and sequenced as previously described.
Identification of the genomic position and surrounding sequence of TbAT1.
The genome sequence of the Trypanosoma brucei line TREU 927/4 (Trypanosomiasis Research Edinburgh University) (GPAL/KE/70/EATRO 1534) single variant antigen type (VAT) derivative GUTat 10.1 has been previously published (6). To identify a clone containing the TbAT1 gene from the T. brucei bacterial artificial chromosome (BAC) library RPCI93 for sequencing, the TbAT1 gene was amplified from a sheared genomic DNA clone (47M12.TF; EMBL accession number AQ947989). The PCR product was used as a probe to screen a high-density filter containing the gridded BAC library. The fingerprinting pattern of several positive BAC clones confirmed that these BACs are anchored in the same genomic region as BACs 25N21 and 29K2, which overlap in this region of sequence release 4 of the chromosome 5 BAC tile path. The presence of the TbAT1 gene was confirmed by PCR, and BAC 26D11 was chosen for sequencing by random sequencing of small insert libraries using the BigDye Terminator cycle sequencing kit from Applied Biosystems. Sequence readings were assembled using Phrap (P. Green, unpublished data; www.phrap.org). Manual base calling and finishing were carried out using Gap4 software (8). Gaps and low-quality regions of the sequence were resolved by primer walking and targeted PCRs. Additional PCRs (primers used: r1, 5′-GATTCGGTGGGAGGACTG, paired with r2, 5′-CTGGTAAGGGAAGCAATAAG; r3, 5′-GAAGGGGAAGCAGCTATG, paired with r4, 5′-CAAAGGCGTGTAAAAACTTC; r5, 5′-CGTCTGTCGTTGTCATTTC, paired with r6, 5′-CCAACTCTACGAGGAACG; r7, 5′-GGTTTTCTTCACCACATTTC, paired with r8, 5′-CAGGTCAACGGAGAAACAC; r9, 5′-CATGACGGTGACATACAATG, paired with r10, 5′-GCTGTTGTCGTTGTTGTGC) were designed to confirm consistency between the assembled BAC sequence and the genomic DNA.
The assembled contiguous BAC sequence was annotated using Artemis software (7). Protein-coding sequences were predicted, and putative functions were assigned as previously described (6). Pseudogenes were annotated using a Blastx search against the UniProtKB database (29). This identified sequences with translations of high sequence similarity to known proteins but which are interrupted by stop codons and/or frameshifts. The full annotation of BAC 26D11 can be viewed and searched via GeneDB (http://www.genedb.org/). Sequence comparisons (tBlastx) between version 4 of T. brucei chromosome 5 and the 26D11 BAC sequence were visualized using the Artemis Comparison Tool (12).
Transport assays.
Parasites were separated from blood using a DEAE cellulose column. Uptake assays were performed using the centrifugation through oil procedure essentially as previously described (10, 14, 15, 18).
The uptake of 20 μM diminazene and 10 μM adenosine was measured over 10 min at 25°C. [3H]diminazene and [3H]adenosine were used as described in reference 15. Concentrations (1 mM) of adenine, inosine, and hypoxanthine, either singly or together, were combined in 100 μl of CBSS (25 mM HEPES, 120 mM NaCl, 5.4 mM KCl, 0.55 mM CaCl, 0.4 mM MgSO4, 5.6 mM Na2HPO4, 11.1 mM d-glucose; pH 7.4) containing radiolabeled substrate. A total of 100 μl of trypanosomes at 2 × 108 cells/ml was added to the tubes, which were then incubated for 30, 60, 120, 300, or 600 s at 25°C. The cells were then spun through the oil layer to form a pellet separated from the excess radioactivity in the aqueous phase. The entire reaction tube was flash frozen in liquid nitrogen, the pellet was cut off into a scintillation vial, and the radioactivity was measured in a scintillation counter. Each experiment was conducted in triplicate.
Nucleotide sequence accession number.
The sequence data from this study have been submitted to EMBL under accession number FM160648.
RESULTS
Genomic location of TbAT1.
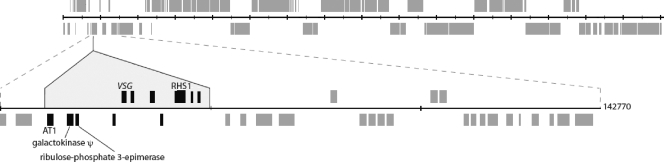
Sequence release 4 of the Trypanosoma brucei 927 genome lacks the TbAT1 gene, and so, clones from a genomic DNA BAC library were screened with a TbAT1-specific probe. One of the hybridizing clones, clone 26D11, was chosen for sequencing. The assembled sequence totals 142,770 nucleotides and encompasses 34 genes, the majority of which are syntenic with the left end of the published chromosome 5 sequence between nucleotide positions 71500 and 161900 (Fig. 1). However, in addition to the common regions, the BAC carries a further 43,453 nucleotides of novel sequence (indicated by the shaded areas in Fig. 1). In addition to the TbAT1 protein, this segment encodes a number of other proteins, including six proteins of unknown function, and one of which (Tb927.5.292b) has a predicted signal sequence, suggestive of surface targeting. No functional clues were discerned from sequence similarity or protein domain search results for the other coding sequences. The remaining genes include a galactokinase pseudogene, a variant surface glycoprotein-related gene, and genes encoding a retrotransposon hot spot protein family member, as well as a putative ribulose-phosphate 3-epimerase. These data now provide a corrected version of the chromosome 5 sequence and confirm the location of the TbAT1 gene on this chromosome.
Fig. 1.
Genomic location of TbAT1. Alignment of BAC 26D11 against T. brucei chromosome 5. Diagrammatic representation of chromosome 5, with gray boxes indicating directional gene clusters. The region of BAC 26D11 homologous to chromosome 5 is shown schematically below. The region of sequence unique to 26D11 is shaded, genes shared by both the BAC and chromosome sequences are indicated in gray, while the novel genes, including TbAT1, are colored in black. VSG, variant surface glycoprotein; RHS1, retrotransposon hot spot 1.
Three drug-resistant trypanosome lines are defective in the P2 transporter.
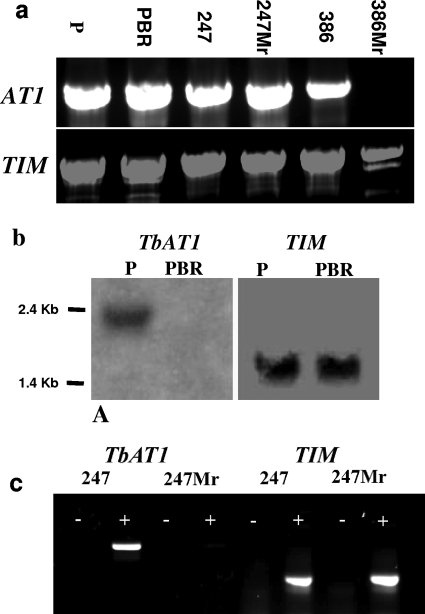
T. b. brucei 247Mr and T. b. gambiense 386Mr, both selected for resistance to melarsamine hydrochloride, and T. equiperdum PBR, selected for resistance to diminazene, have previously been shown by a fluorescence-based assay to lack a functional P2/TbAT1 transporter (28). This lack has also been shown for T. equiperdum PBR (3) and T. b. gambiense 386Mr (9) through studies of P2-mediated transport. As it has been shown that the 427 arsenical drug-resistant line has deleted the TbAT1 gene, we investigated the status of the TbAT1 gene in these lines, along with their drug-sensitive parental cell lines, by PCR analysis of genomic DNA. Results are shown in Fig. 2a. An amplicon of the predicted size using DNA from all three of the drug-sensitive parental lines and in the T. b. brucei 247Mr and the T. equiperdum PBR resistant lines is observed, showing that the gene is present and that deletion is not the mechanism of resistance in these cases. In contrast, the 386Mr line shows no amplification (Fig. 2a), although the control gene is amplified, thus showing that the TbAT1 gene has been deleted.
Fig. 2.
TbAT1 gene and its expression in wild-type and drug-resistant trypanosome lines. (a) TbAT1 was amplified from genomic DNA prepared from T. equiperdum P, T. equiperdum PBR, STIB 247 (247), 247Mr, STIB 386 (386), and 386Mr. (b) Northern blot assay of T. equiperdum strains P and PBR, probed with the TbAT1 ORF and subsequently with a control probe for triosephosphate isomerase (TIM) as a loading control. (c) Reverse transcriptase PCR with cDNA from STIB 247 and 247Mr, confirming the lack of product in the drug-resistant derived STIB 247 line. A minus sign indicates a no-reverse-transcriptase control. TIM primers were used as positive controls.
As both the 247Mr and T. equiperdum PBR lines had previously been shown to lack P2 transporter activity, we tested whether these lines had lost expression of the TbAT1 gene. Northern blot analysis of RNA from the drug-sensitive and -resistant lines of T. equiperdum showed that TbAT1 RNA was no longer stably expressed in the PBR line (Fig. 2b). RT-PCR analysis of RNA from the 247Mr line also showed the loss of stable expression of the transporter gene (Fig. 2c). In both sets of analyses, the parental sensitive lines were shown to express TbAT1, and transcripts of the positive-control (TIM) gene were detected in the preparations from the resistant lines. Thus, it can be concluded that selection for resistance results in a loss of stable RNA expression of the TbAT1 gene. This demonstrates a novel mechanism of drug resistance associated with the P2 transporter.
TeAT1 ORF and 3′ UTR of T. equiperdum P/PBR.
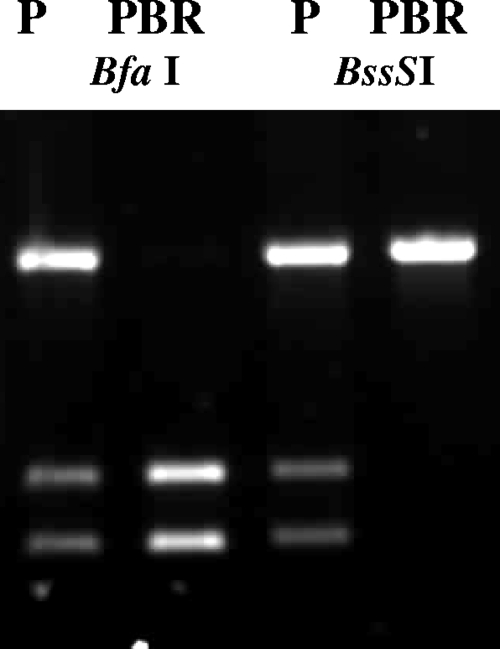
To investigate the possible basis of the loss of transcript in the PBR resistant lines, the TeAT1 ORF was sequenced from both the sensitive and resistant lines. The entire TeAT1 ORF (1,389 bp) and short flanking regions (PCR product size, ∼1,500 bp) were amplified from both T. equiperdum lines P and PBR. Multiple independent PCR products were cloned and then sequenced from each line in order to identify the presence of any mutations or deletions in the gene. All 10 T. equiperdum PBR TeAT1 clones had identical sequences, and three T. equiperdum P clones also had sequences identical to those in the resistant line. However, six other clones from the sensitive line had identical sequences, except for two nucleotide changes, compared to those of the other clones from this line. Comparison of these sequences with the 927 reference sequence identified eight single nucleotide polymorphisms (SNPs), three of which lead to nonsynonymous substitutions (Table 1). The sequence data indicate that the wild-type T. equiperdum P line is heterozygous for the TeAT1 locus, whereas T. equiperdum PBR has lost heterozygosity at this locus and become homozygous for one of the alleles (Table 1). To confirm this finding, the A/C SNP present at position 627 was investigated by PCR-RFLP using the diagnostic enzymes BfaI (cutting the “A” allele) and BssSI (cutting the “C” allele). Using genomic DNA, TeAT1 was PCR amplified from the wild-type line, and the amplicon was digested with BfaI and BssSI to generate three fragments, with sizes of 1,392 bp (uncut allele), 626 bp, and 766 bp (Fig. 3), indicating the presence of two alleles. Restriction digestion with BfaI from the amplicon derived from the PBR line, however, lacked the 1,392-bp band, whereas the two lower bands were present, indicating the presence of only the A allele. Digestion with BssSI yielded the converse result, where only the uncut 1,392-bp band was present in the resistant line (Fig. 3). This indicated that BssSI did not cut PBR TeAT1, and therefore, the C allele is absent. Thus, loss of heterozygosity at the TeAT1 locus has accompanied selection of resistance in T. equiperdum and may be associated with a loss of expression of this gene.
Table 1.
Point mutations in the TbAT1 nucleotide sequence and changes to the amino acid sequence in the T. equiperdum P and PBR cell lines compared to the T. brucei 927 sequencea
| Type of sequence | Position | Point mutation in indicated cell line |
Related mutation (position of mutation) | ||
|---|---|---|---|---|---|
| 927 | P | PBR | |||
| Nucleotide | 21 | C | T | T | |
| 151 | A | C/A | A | K/Q (51) | |
| 542 | G | A | A | G/E (181) | |
| 625 | T | C | C | ||
| 627 | A | C/A | A | ||
| 699 | T | C | C | ||
| 716 | A | G | G | D/G (239) | |
| 1122 | A | G | G | ||
| Amino acid | 51 | K | Q/K | K | C/A (151) |
| 181 | G | E | E | G/A (542) | |
| 239 | D | G | G | A/G (716) | |
Six of nine T. equiperdum P clones contained the additional point mutations at positions 151 and 627. SNPs that code for nonsynonymous changes are as follows: position 151, A = K and C = Q; position 542, G = G and A = E; and position 716, A = D and G = G.
Fig. 3.
Loss of heterozygosity in drug-resistant T. equiperdum. PCR-amplified products of TbAT1 ORF from T. equiperdum strains P and PBR digested with BfaI and BssSI.
3′ RACE was performed on T. equiperdum P (wild-type) cDNA. Of 8 clones from the nested PCR product, 5 indicated that polyadenylation springs from an A residue at 719 bp from the termination codon. Two further clones point to polyadenylation sites within 30 bp downstream of this site, and a final clone appears to indicate that polyadenylation may occur at 640 bp from the termination codon. This last site coincides with a poly(A) stretch in the 3′ UTR and could be an artifact due to mispriming of the oligo(dT) anchor primer on this poly(A) stretch. The double-stranded sequencing also indicated a heterozygosity in the 3′ UTR of T. equiperdum P, which has been lost in the PBR line. To investigate this further, we amplified and sequenced a 750-bp fragment of the 3′ UTR from both T. equiperdum lines P and PBR covering this area of heterozygosity. Of 9 T. equiperdum P clones, 6 carried an A residue at a site that is 514 bp into the 3′ UTR, and the remaining 3 carried a G residue. Sequencing of the T. equiperdum PBR 3′ UTR once again indicates that a loss of heterozygosity has accompanied loss of expression, as all 10 clones carried the A residue at this site.
The coding sequences of the TbAT1 genes from T. brucei 247 and 247Mr were amplified by PCR, and the amplicons were cloned and sequenced from both lines (data not shown). Comparison of the sequences (two clones from each line) showed that they were identical, which indicates that the genes in both lines are homozygous and identical.
Diminazene uptake is lost in T. equiperdum-selected PBR.
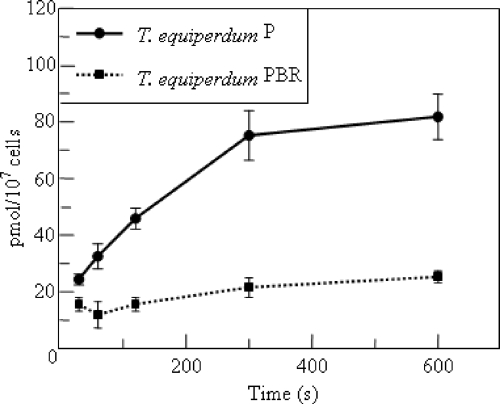
Previous studies have shown the presence of a P2-like adenine-inhibitable adenosine transport function in the T. equiperdum PBR cell line (3). However, by fluorescence assay, P2 transport function itself has been shown to be absent (28). Furthermore, there is no detectable transcript for TbAT1 in this cell line (Fig. 2b). As diminazene has been shown to be accumulated in T. brucei predominantly via the P2 transporter (15), uptake of 20 μM diminazene was measured in both wild-type and diminazene-resistant T. equiperdum to resolve this apparent contradiction. While diminazene uptake in wild-type T. equiperdum is robust, uptake in T. equiperdum PBR is detectable only at low levels over 10 min, indicating that P2 transport is indeed lost (Fig. 4). The low level of uptake indicates the presence of another low-capacity diminazene transporter. Indeed, while the T. equiperdum PBR line is considered diminazene resistant, it is still sensitive to low micromolar concentrations of the drug, as is the ΔTbat1 line, indicating that a secondary route of uptake is probably important for diminazene, which is analogous to the situation reported for furamidine (18).
Fig. 4.
Uptake of diminazene in T. equiperdum strains P and PBR. Accumulation of 20 μM diminazene was measured over time in wild-type T. equiperdum and its diminazene-resistant derivative, PBR.
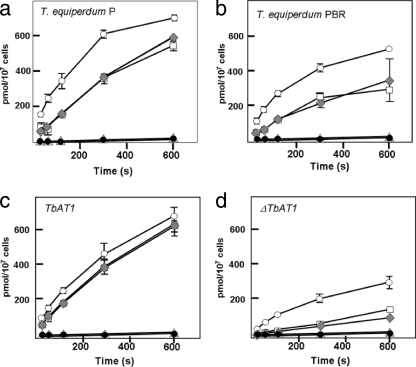
A residual adenine-inhibitable adenosine uptake component has been previously identified in T. equiperdum PBR, yet all other data indicate a functional loss of TeAT1/P2 transport. Therefore, the question remains as to the identity of this residual adenine-sensitive adenosine uptake component. To investigate this, we compared the uptake of adenosine in both wild-type and diminazene-resistant T. equiperdum lines and also in the T. b. brucei ΔTbat1 line, along with the wild-type parental line from which it was derived (23). The uptake of 10 μM [3H]adenosine in the presence of various inhibitors was followed. A total of 1 mM inosine was used to block the P1 transporter, which takes up adenosine with a high affinity. A total of 1 mM adenine was used to block any P2 or P2-like adenosine transport, and 1 mM hypoxanthine was used to block any transport of adenosine mediated by the H2 nucleobase transporter (31). The results of these uptake assays are shown in Fig. 5. The uptake of adenosine is clearly reduced in both the PBR line (Fig. 5a and b) and the ΔTbat1 line (Fig. 5c and d) compared to that in their respective wild-type parental lines. Blockage of adenosine uptake with either 1 mM inosine or inosine and hypoxanthine leads to a similar reduction in adenosine uptake in both the wild-type and mutant lines but did not abolish uptake completely, thus showing that additional routes of uptake were present. Addition of adenine (in the presence of inosine and hypoxanthine) completely inhibited adenosine uptake in all four lines, indicating that both the PBR and ΔTbat1 lines retain a P2-like uptake activity that is not encoded by the TbAT1 gene.
Fig. 5.
Adenosine uptake via a P2-independent adenine-inhibited carrier. Inhibition of 10 μM adenosine uptake in T. equiperdum P (a), T. equiperdum PBR (b), TbAT1 (wild type) (c), and ΔTbAT1 (d). Bars indicate standard deviations (n = 3). Open circles show uptake in the absence of inhibitor, open squares show uptake in the presence of 1 mM inosine (which blocks the P1 transporter), gray diamonds represent uptake in 1 mM inosine and 1 mM hypoxanthine (blocking the P1 adenosine transporter and the H2 hypoxanthine carrier), filled circles show 1 mM inosine and adenine, which inhibits both the P1 and P2 transporters, and open triangles represent uptake with all three of the aforementioned inhibitors.
DISCUSSION
In this study, we have characterized various genetic changes associated with the TbAT1 gene in trypanosomes selected for resistance to the diamidine drug diminazene and the melaminophenyl arsenical melarsamine hydrochloride, as well as correction of the genome sequence of chromosome 5.
The original T. brucei genome assembly did not contain TbAT1. We therefore identified a BAC (26D11) containing the gene in order to identify its genomic localization. The assembly strategy for chromosome 5 was based on the mapping of a seed BAC from the RPCI93 library, followed by identification of overlapping and outwardly extending BACs through sequence similarity searches against a database of BAC end sequences. A number of candidate BACs were subsequently chosen for fingerprinting and selected for sequencing only if the fingerprint data were consistent and shared by others in the set. The region of chromosome 5 to which BAC 26D11 is homologous is located toward the extreme 5′ end of the assembly, where the consensus was generated based on two BACs (25N21 and 29K2) that overlap in this region. The sequence unique to BAC 26D11 is flanked by ∼7.5 kb of duplicated sequence, encompassing an adenylyl cyclase gene. Given that the genetic data showed this locus to be homozygous and diploid, it is possible that BACs 25N21 and 29K2 each contain a deletion around the TbAT1 locus. An alternative explanation is that BACs 25N21 and 29K2 encoded the 5′ and 3′ ends of the duplicon, respectively, and hence, neither of these BACs carries the sequence unique to 26D11 including the TbAT1 gene. Whether the location of TbAT1 in the subtelomeric region of chromosome 5 in fact contributes to a more rapid generation of resistant cells under subcurative drug pressure due to the greater rates of recombination in these areas (4) is a matter for future investigation.
Deletion of the TbAT1 gene is associated with the selection of resistance to melarsamine hydrochloride in T. b. gambiense strain 386 (25), as identified by PCR analysis. Gene deletion was previously reported in a field isolate of arsenical-resistant T. b. rhodesiense (22) and also in a T. b. brucei line selected for resistance to the diamidine furamidine (18). In the case of T. b. brucei 247, a similar melarsamine hydrochloride-driven selection procedure (25) led to derivation of a line in which the gene is still identified by PCR, but in which no transcript was detected by RT-PCR and from which P2 transport activity is lacking. A further strain, T. equiperdum PBR selected for resistance to diminazene, has also retained the TeAT1 gene, as detected by PCR, but in this line as well, no transcript was identified in Northern hybridization experiments. We went on to investigate the status of TeAT1 in the paired T. equiperdum lines.
Of the six nucleotide differences identified here between the 927 sequence and the PBR line, three of these mutations have also been identified in STIB 777R, a laboratory-derived, melarsen oxide-resistant cell line (21). Two of these mutations code for changes in the amino acid sequence of TbAT1, and one is a silent mutation. These same three mutations also occur, among others, in the TbAT1 gene in T. b. gambiense cerebrospinal fluid isolates from patients in Uganda, in a T. b. rhodesiense isolate (STIB 871) that shows some resistance to diminazene aceturate and melarsoprol, and in a T. b. gambiense stock (K 003) from Angola (22). Further investigation showed that TeAT1 in the T. equiperdum wild-type cell line is heterozygous, but the T. equiperdum PBR line contains only one of these alleles, implying that the loss of P2 transport may be linked to a loss of heterozygosity at the TeAT1 locus. This finding was also upheld in the 3′ UTR, where heterozygosity is apparent in the T. equiperdum P line and is lost in the T. equiperdum PBR line.
The loss of heterozygosity associated with the TeAT1 locus in PBR may account for the lack of P2 transcript and hence the loss of P2 function in this resistant line. If the A allele in the heterozygous P cell line were to be transcriptionally silent or to produce a nonfunctional transporter, drug resistance could have been acquired solely by a loss of heterozygosity in the PBR line, which carries only the A allele. However, the sequence of the A allele at positions 151 and 627 of the ORF is identical to that of TbAT1 from both the T. brucei 927 (Table 1) and 427 wild-type strains, which have fully functional P2 transport systems and a typical sensitivity to drugs. This would suggest that these mutations within the coding sequence of the gene, followed by loss of heterozygosity alone, are unlikely to bring about drug resistance.
Another possibility for the loss of drug sensitivity is that a silent copy of TbAT1 has been generated by changes up- or downstream of the coding sequence, which regulate the expression of the A allele. In this model, a mutation occurring in one allele (the A allele) might silence that gene, allowing the loss of heterozygosity to lead to silencing of the entire TbAT1 locus. In the PBR cell line, it would appear that the active C allele has been selected against, and a subsequent loss of heterozygosity has resulted in a loss of expression of the TeAT1 gene. Gene expression in trypanosomes is frequently associated with regulatory elements present within the 3′ UTR of genes, including AU-rich elements (AREs) (5, 13, 16). However, the 3′ UTR of T. equiperdum has been established in this study, and the closest canonical AREs in the 3′ sequence occur at a position that is 1,283 bp from the termination codon, well beyond the polyadenylation site of this sequence and, as such, should not be influencing expression in any conventional way. The influence of any other UTRs remains to be elucidated, including the influence of the SNP at position 514 of the UTR. The finding of loss of expression of TbAT1 at the mRNA level is also seen for the T. brucei 247 line selected for melarsamine hydrochloride resistance. This represents a novel genetic mechanism for drug resistance in T. brucei group trypanosomes.
Loss of expression of TeAT1 would be expected to effectively eliminate all P2 transport function, defined as high-affinity adenine-inhibitable adenosine transport (11), in a trypanosome line. However, adenine-sensitive adenosine transport was still present in the drug-resistant T. equiperdum line (3), and this had led to the original hypothesis that drug resistance had arisen from a change in affinity of the P2 transporter for the substrate. The absence of expression of TeAT1 in this line clearly eliminates this hypothesis. Blockage of the uptake of adenosine with inosine and subsequently with adenine shows that there is still an adenine-sensitive adenosine transport component in the T. equiperdum PBR cell line. This activity is also apparent in the ΔTbat1 cell line, which has been confirmed to have no P2 transport activity (23). Thus, our data support the occurrence of a further permease, capable of adenine and adenosine uptake, for which a physiological role is not yet known.
The accumulation of point mutations, gene deletion, loss of heterozygosity, and loss of a stable transcript of the TbAT1 gene are now all shown to be genetic mechanisms leading to net loss of activity of the P2 aminopurine/drug transporter. It remains to be shown whether all these alterations are in play in generating resistance in a field setting, but any test for drug resistance based on genetic profiles will need to incorporate multiple alterations if they are to be of use in identifying drug resistance in the field. We have recently developed a fluorescence-based test that reports P2 transporter activity (28), independent of the genetic basis of its loss. This test appears to hold great promise as a diagnostic tool for the detection of drug resistance in patients treated for human African trypanosomiasis.
ACKNOWLEDGMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council grant (C13486) to M.P.B., R.J.S.B., and H.P.D.K. and the Wellcome Trust grant (074732) to A.T., A.M., and C.M.R.T.
Footnotes
Published ahead of print on 4 December 2009.
REFERENCES
- 1.Barrett M. P., Boykin D. W., Brun R., Tidwell R. R. 2007. Human African trypanosomiasis: pharmacological re-engagement with a neglected disease. Br. J. Pharmacol. 152:1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett M. P., Burchmore R. J., Stich A., Lazzari J. O., Frasch A. C., Cazzulo J. J., Krishna S. 2003. The trypanosomiases. Lancet 362:1469–1480 [DOI] [PubMed] [Google Scholar]
- 3.Barrett M. P., Zhang Z. Q., Denise H., Giroud C., Baltz T. 1995. A diamidine-resistant Trypanosoma equiperdum clone contains a P2 purine transporter with reduced substrate affinity. Mol. Biochem. Parasitol. 73:223–229 [DOI] [PubMed] [Google Scholar]
- 4.Becker M., Aitcheson N., Byles E., Wickstead B., Louis E., Rudenko G. 2004. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 14:2319–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berberof M., Vanhamme L., Tebabi P., Pays A., Jefferies D., Welburn S., Pays E. 1995. The 3′-terminal region of the mRNAs for VSG and procyclin can confer stage specificity to gene expression in Trypanosoma brucei. EMBO J. 14:2925–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 7.Berriman M., Rutherford K. 2003. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 4:124–132 [DOI] [PubMed] [Google Scholar]
- 8.Bonfield J. K., Smith K., Staden R. 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bridges D. J., Gould M. K., Nerima B., Maser P., Burchmore R. J., de Koning H. P. 2007. Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamidine drugs in African trypanosomes. Mol. Pharmacol. 71:1098–1108 [DOI] [PubMed] [Google Scholar]
- 10.Carter N. S., Berger B. J., Fairlamb A. H. 1995Uptake of diamidine drugs by the P2 nucleoside transporter in melarsen-sensitive and -resistant Trypanosoma brucei brucei. J. Biol. Chem. 270:28153–28157 [DOI] [PubMed] [Google Scholar]
- 11.Carter N. S., Fairlamb A. H. 1993Arsenical-resistant trypanosomes lack an unusual adenosine transporter. Nature 361:173–176 [DOI] [PubMed] [Google Scholar]
- 12.Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 13.Colasante C., Robles A., Li C. H., Schwede A., Benz C., Voncken F., Guilbride D. L., Clayton C. 2007. Regulated expression of glycosomal phosphoglycerate kinase in Trypanosoma brucei. Mol. Biochem. Parasitol. 151:193–204 [DOI] [PubMed] [Google Scholar]
- 14.de Koning H. P. 2001. Uptake of pentamidine in Trypanosoma brucei brucei is mediated by three distinct transporters: implications for cross-resistance with arsenicals. Mol. Pharmacol. 59:586–592 [DOI] [PubMed] [Google Scholar]
- 15.de Koning H. P., Anderson L. F., Stewart M., Burchmore R. J., Wallace L. J., Barrett M. P. 2004. The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: additional insights on diamidine resistance in african trypanosomes. Antimicrob. Agents Chemother. 48:1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotz H. R., Hartmann C., Huober K., Hug M., Clayton C. 1997. Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res. 25:3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenni L., Marti S., Schweizer J., Betschart B., Le Page R. W., Wells J. M., Tait A., Paindavoine P., Pays E., Steinert M. 1986. Hybrid formation between African trypanosomes during cyclical transmission. Nature 322:173–175 [DOI] [PubMed] [Google Scholar]
- 18.Lanteri C. A., Stewart M. L., Brock J. M., Alibu V. P., Meshnick S. R., Tidwell R. R., Barrett M. P. 2006. Roles for the Trypanosoma brucei P2 transporter in DB75 uptake and resistance. Mol. Pharmacol. 70:1585–1592 [DOI] [PubMed] [Google Scholar]
- 19.MacLeod A., Turner C. M., Tait A. 1997. Detection of single copy gene sequences from single trypanosomes. Mol. Biochem. Parasitol. 84:267–270 [DOI] [PubMed] [Google Scholar]
- 20.Maniatis T., Sambrook J., Fritsch E. F. 1982. Molecular cloning: a laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Maser P., Sutterlin C., Kralli A., Kaminsky R. 1999. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285:242–244 [DOI] [PubMed] [Google Scholar]
- 22.Matovu E., Geiser F., Schneider V., Maser P., Enyaru J. C., Kaminsky R., Gallati S., Seebeck T. 2001. Genetic variants of the TbAT1 adenosine transporter from African trypanosomes in relapse infections following melarsoprol therapy. Mol. Biochem. Parasitol. 117:73–81 [DOI] [PubMed] [Google Scholar]
- 23.Matovu E., Stewart M. L., Geiser F., Brun R., Mäser P., Wallace L. J., Burchmore R. J., Enyaru J. C., Barrett M. P., Kaminsky R., Seebeck T., de Koning H. P. 2003. Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryot. Cell 2:1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross C. A., Barns A. M. 1996. Alteration to one of three adenosine transporters is associated with resistance to cymelarsan in Trypanosoma evansi. Parasitol. Res. 82:183–188 [DOI] [PubMed] [Google Scholar]
- 25.Scott A. G., Tait A., Turner C. M. 1996. Characterisation of cloned lines of Trypanosoma brucei expressing stable resistance to MelCy and suramin. Acta Trop. 60:251–262 [DOI] [PubMed] [Google Scholar]
- 26.Scott A. G., Tait A., Turner C. M. 1997. Trypanosoma brucei: lack of cross-resistance to melarsoprol in vitro by cymelarsan-resistant parasites. Exp. Parasitol. 86:181–190 [DOI] [PubMed] [Google Scholar]
- 27.Stevens J. R., Noyes H. A., Schofield C. J., Gibson 2001. The molecular evolution of Trypanosomatidae. Adv. Parasitol. 48:2–56 [DOI] [PubMed] [Google Scholar]
- 28.Stewart M. L., Krishna S., Burchmore R. J., Brun R., de Koning H. P., Boykin D. W., Tidwell R. R., Hall J. E., Barrett M. P. 2005. Detection of arsenical drug resistance in Trypanosoma brucei with a simple fluorescence test. Lancet 366:486–487 [DOI] [PubMed] [Google Scholar]
- 29.UniProt Consortium 2008. The universal protein resource (UniProt). Nucleic Acids Res. 36:D190–D195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasudevan G., Carter N. S., Drew M. E., Beverley S. M., Sanchez M. A., Seyfang A., Ullman B., Landfear S. M. 1998. Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc. Natl. Acad. Sci. U. S. A. 95:9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace L. J., Candlish D., de Koning H. P. 2002. Different substrate recognition motifs of human and trypanosome nucleobase transporters. Selective uptake of purine antimetabolites. J. Biol. Chem. 277:26149–26156 [DOI] [PubMed] [Google Scholar]
- 32.Witola W. H., Inoue N., Ohashi K., Onuma M. 2004. RNA-interference silencing of the adenosine transporter-1 gene in Trypanosoma evansi confers resistance to diminazene aceturate. Exp. Parasitol. 107:47–57 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z. Q., Giroud C., Baltz T. 1991. In vivo and in vitro sensitivity of Trypanosoma evansi and T. equiperdum to diminazene, suramin, MelCy, quinapyramine and isometamidium. Acta Trop. 50:101–110 [DOI] [PubMed] [Google Scholar]