Abstract
Rev3 polymerase and Mph1 DNA helicase participate in error-prone and error-free pathways, respectively, for the bypassing of template lesions during DNA replication. Here we have investigated the role of these pathways and their genetic interaction with recombination factors, other nonreplicative DNA helicases, and DNA damage checkpoint components in the maintenance of genome stability, viability, and sensitivity to the DNA-damaging agent methyl methanesulfonate (MMS). We find that cells lacking Rev3 and Mph1 exhibit a synergistic, Srs2-dependent increase in the rate of accumulating spontaneous, gross chromosomal rearrangements, suggesting that the suppression of point mutations by deletion of REV3 may lead to chromosomal rearrangements. While mph1Δ is epistatic to homologous recombination (HR) genes, both Rad51 and Rad52, but not Rad59, are required for normal growth of the rev3Δ mutant and are essential for survival of rev3Δ cells during exposure to MMS, indicating that Mph1 acts in a Rad51-dependent, Rad59-independent subpathway of HR-mediated lesion bypass. Deletion of MPH1 helicase leads to synergistic DNA damage sensitivity increases in cells with chl1Δ or rrm3Δ helicase mutations, whereas mph1Δ is hypostatic to sgs1Δ. Previously reported slow growth of mph1Δ srs2Δ cells is accompanied by G2/M arrest and fully suppressed by disruption of the Mec3-dependent DNA damage checkpoint. We propose a model for replication fork rescue mediated by translesion DNA synthesis and homologous recombination that integrates the role of Mph1 in unwinding D loops and its genetic interaction with Rev3 and Srs2-regulated pathways in the suppression of spontaneous genome rearrangements and in mutation avoidance.
Nonreplicative DNA helicases play an important role in the maintenance of genome stability from bacteria to humans, most likely by affecting the formation and/or resolution of recombination intermediates and by facilitating replication fork progression through chromosomal regions with a propensity to adopt unusual DNA structures or those bound by proteins. In Saccharomyces cerevisiae, this group of DNA helicases includes the 3′-to-5′ helicases Sgs1 and Srs2 and the 5′-to-3′ DNA helicase Rrm3. In the absence of any two of these three helicases, unresolved recombination intermediates accumulate and lead to extremely slow growth that is fully suppressed by deletion of genes encoding early homologous recombination (HR) factors (4, 6, 17, 20, 37, 46). In the absence of Sgs1, cells exhibit increased rates of mitotic recombination, frequent chromosome missegregation, accumulation of extrachromosomal ribosomal DNA (rDNA) circles, and increased rates of gross chromosomal rearrangements (GCRs) involving nonhomologous chromosomes (5, 24, 25, 38, 40, 43, 49, 50). Based on the increased crossover frequency during HO endonuclease-induced double-strand breaks (DSBs) in cells lacking Sgs1, it has also been proposed that Sgs1 may function in decatenation of Holliday junctions (HJs) to yield noncrossovers (12, 22). Like Sgs1, Srs2 acts to favor noncrossover outcomes during DSB repair but appears to act earlier than Sgs1 in regulating recombination outcomes through its ability to dislodge Rad51 from recombinogenic 3′ overhangs, thereby promoting a noncrossover synthesis-dependent single-strand annealing (SDSA) pathway (12, 33, 35). In contrast, Rrm3 has not been implicated in DNA repair but is thought to be important for avoidance of recombination substrate formation by removal of DNA protein complexes in certain chromosomal locations, such as chromosome ends and replication fork barriers at the rDNA locus, thus facilitating replication fork progression (13, 14).
In addition to Sgs1, Rrm3, and Srs2, the yeast genome encodes two other nonreplicative DNA helicases with proposed functions in DNA repair, Mph1 and Chl1. Mph1 possesses 3′-to-5′ helicase activity, and its ATPase activity requires a relatively long fragment of single-stranded DNA (ssDNA) (≥40 nucleotides [nt]) for full activity in vitro (32). Mph1 is also necessary for resistance to the DNA damaging agents methyl methanesulfonate (MMS) and 4-nitroquinoline-1-oxide (4-NQO) and suppresses spontaneous mutations toward canavanine resistance (3, 41). The modest mutator phenotype of the mph1Δ mutant is enhanced by additional mutations in base excision repair (apn1Δ and apn2Δ) and is suppressed by mutations in translesion DNA synthesis (TLS) (rev3Δ) (36, 41). These findings, in combination with the observation of an epistatic relationship between mph1Δ and homologous recombination mutations, have led to the proposal that Mph1 may act in Rad52-dependent, error-free bypassing of DNA lesions (41). Like the 3′-to-5′ DNA helicases Sgs1 and Srs2, Mph1 was recently shown to affect crossover frequency during repair of an HO endonuclease-induced DNA DSB, favoring noncrossovers as the outcome (33). The authors showed that Mph1 can unwind intermediates of homologous recombination in vitro, specifically D loops that are thought to form early during homologous recombination when a homoduplex is invaded by a Rad51 filament. While Srs2 has been shown to be able to disassemble Rad51 filaments in vitro, it does not appear to possess Mph1's ability to dissociate D loops once they have formed (19, 47).
Although Chl1 has been shown to be required for the establishment of sister chromatid cohesion, a possible role in DNA repair by homologous recombination has also been proposed (11, 28, 30, 42). While Chl1 possesses a conserved helicase domain, helicase activity has so far been shown only for its putative human homolog, hCHLR1 (10).
To further elucidate the functional interaction between nonreplicative DNA helicases and DNA repair pathways, we generated a series of mutants with combinations of mph1Δ, chl1Δ, rrm3Δ, srs2Δ, and sgs1Δ mutations and mutations in translesion DNA synthesis (TLS), base excision repair (BER), homologous recombination (HR), and DNA damage checkpoints. In addition to synthetic fitness defects due to aberrant HR and checkpoint activation, we identified epistatic and synergistic relationships with regard to fitness, the accumulation of gross chromosomal rearrangements (GCRs), and sensitivity to DNA damage. We propose that Mph1 functions in a Rad51-dependent, Rad59-independent pathway of HR for DNA lesion bypass and interacts genetically with REV3 in the suppression of gross chromosomal rearrangements.
MATERIALS AND METHODS
Yeast strains and media.
All strains used in this study are derived from Saccharomyces cerevisiae strain S288C and are listed in Table 1. For GCR rate measurements, desired gene deletions were introduced into KHSY802 (MATa ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2Bgl hom3-10 ade2Δ1 ade8 hxt13::URA3), RDKY5027 (MATα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2Bgl hom3-10 ade2Δ1 ade8 hxt13::URA3), or RDKY6678 (MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph) by HR-mediated integration of PCR products by the lithium acetate method (9). All haploid strains, including single mutants, for GCR rate measurements, growth analysis, and fluorescence-activated cell sorting (FACS) were obtained by sporulating diploids heterozygous for the desired mutation(s). Spores were genotyped on selective media or by PCR. For tetrad dissection, desired mutations were introduced by HR-mediated integration of PCR fragments in the strain background RDKY2666 (MATa ura3-52 trp1Δ63 his3Δ200) or RDKY2664 (MATα ura3-52 trp1Δ63 his3Δ200).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| RDKY3615a | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 |
| RDKY2666a | MATaura3-52 trp1Δ63 his3Δ200 |
| RDKY2664a | MATα ura3-52 trp1Δ63 his3Δ200 |
| RDKY6678a | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph |
| RDKY6795a | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph mph1::HIS3 |
| KHSY883 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rad51::HIS3 |
| KHSY1258 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rad52::HIS3 |
| KHSY1399 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rrm3::kanMX6 |
| KHSY1557 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 mph1::HIS3 |
| KHSY1561 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 chl1::HIS3 |
| KHSY1598 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rad52::HIS3 mph1::HIS3 |
| KHSY1600 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 sgs1::TRP1 mph1::HIS3 |
| KHSY1630 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 sgs1::TRP1 |
| KHSY1702 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 srs2::kanMX6 mph1::HIS3 |
| KHSY1713 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rrm3::TRP1 mph1::HIS3 |
| KHSY1725 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 chl1::HIS3 mph1::HIS3 |
| KHSY1872 | MATa/α ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 MPH1/mph1::HIS3 SRS2/srs2::TRP1 |
| KHSY1878 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 mec3::kanMX6 mph1::HIS3 |
| KHSY1889 | MATa/α ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 MPH1/mph1::HIS3 MRE11/mre11::URA3 |
| KHSY1894 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 mph1::HIS3 mec1::HIS3 sml1::TRP1 |
| KHSY1932 | MATa/α ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 MPH1/mph1::URA3 SRS2/srs2::HIS3 MEC3/mec3::TRP1 |
| KHSY1935 | MATa/α ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 MPH1/mph1::HIS3 SRS2/srs2::TRP1 RAD51/rad51::kanMX6 |
| KHSY1954 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rev3::TRP1 |
| KHSY1957 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 apn1::TRP1 |
| KHSY1970 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 apn1::TRP1 mph1::HIS3 |
| KHSY1976 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rev3::TRP1 mph1::HIS3 |
| KHSY2020 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rad51::HIS3 srs2::kanMX6 mph1::HIS3 |
| KHSY2038 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 mec3::HIS3 srs2::kanMX6 mph1::TRP1 |
| KHSY2226 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rev3::TRP1 srs2::kanMX6 mph1::HIS3 |
| KHSY2416 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rev3::TRP1 rad52::HIS3 mph1::HIS3 |
| KHSY2420 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 rev3::TRP1 rad52::HIS3 |
| KHSY2492 | MATaura3-52 trp1Δ63 his3Δ200 leu2Δ1 lys2ΔBgl hom3-10 ade2Δ1 ade8 hxt13::URA3 mph1::HIS3 rad51::HIS3 |
| KHSY3042 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph mec3::TRP1 mph1::HIS3 |
| KHSY3056 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph rev3::TRP1 mph1::HIS3 |
| KHSY3065 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph mec3::TRP1 |
| KHSY3067 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph srs2::HIS3 |
| KHSY3101 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph srs2::HIS3 mph1::HIS3 |
| KHSY3123 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph srs2::HIS3 mph1::HIS3 rev3::TRP1 |
| KHSY3126 | MATaura3-52 leu2Δ1 trp1Δ63 his3Δ200 lys2ΔBgl hom3-10 ade2Δ1 ade8 can1::hisG yel072w::CAN1/URA3 iYEL072::hph srs2::HIS3 mph1::HIS3 mec3::TRP1 |
Obtained from Richard Kolodner (University of California—San Diego).
Media for propagating strains have been previously described (2).
GCR analysis.
GCR rates were determined exactly as previously described (39). Initially, GCR rates were derived from 10-ml cultures of two or three independent strain isolates. For mutants with low GCR rates, up to 75 cultures, ranging from 10 to 50 ml in volume, were analyzed per mutant. To determine the statistical significance of differences between median GCR rates, 95% confidence intervals (α < 0.05) for all median GCR rates were calculated according to the method of Nair (26). GCR rates were measured in the standard GCR strain background RDKY3615 and a modified GCR strain background, RDKY6678 (both strains were kindly provided by Richard Kolodner, University of California—San Diego). In RDKY3615, the CAN1 gene is in its wild-type location on chromosome V and a URA3 cassette was used to replace the HXT13 gene on chromosome V (34, 39). In RDKY6678, the CAN1 gene is deleted (can1::hisG) and a URA3/CAN1 cassette is inserted into YEL072W, located telomeric of HXT13 on chromosome V (34).
Tetrad analysis.
Diploids were sporulated in 1% potassium acetate for 5 days at 30°C, washed, digested with zymolase (500 μg/ml in 1 M sorbitol), and dissected on yeast extract-peptone-dextrose (YPD) agar plates using a micromanipulator mounted on an Axioskop 40 microscope (Zeiss). The YPD plates were incubated for 2 days at 30°C and photographed.
Doubling time measurement.
Overnight cultures of independent isolates were diluted in 5 ml of YPD to an optical density at 600 nm (OD600) of 0.1 to 0.2, and the OD600 was measured in 60-min or 120-min intervals for 6 to 8 h. Doubling times are reported in minutes and are presented as the average doubling time of two or three independent strains for each genotype, with error bars showing the standard deviations.
DNA content analysis.
Cells were grown overnight at 30°C in YPD medium. Cultures were diluted in YPD to an OD600 of 0.2, and incubation was continued until cultures reached an OD600 of 0.6 to 0.8. Cells were then fixed in 70% ethanol for 1 h at room temperature and sonicated in 50 mM sodium citrate (pH 7). The cells were washed once in 50 mM sodium citrate (pH 7), and RNase A was added to a final concentration of 250 μg/ml. After overnight incubation at 37°C, the cells were washed twice in 50 mM sodium citrate. To stain the DNA, Sytox green (Molecular Probes) was added to a final concentration of 1 μM and the cells were incubated in the dark at room temperature for 1 h immediately prior to fluorescence-activated cell sorting (FACS) on a BD LSR II analyzer. The distribution of cells throughout the cell cycle phases was quantified with the FlowJo v8.3.3 software program. The mean obtained from measurements of at least three cultures and standard deviation are reported for every strain.
MMS sensitivity.
Overnight cultures were diluted in YPD to an OD600 of 0.2 and grown at 30°C to an OD600 of 0.6. A series of 10-fold dilutions was prepared for every yeast culture, and 5 μl was spotted on YPD and on YPD containing the appropriate levels of MMS. Colony growth was recorded after 24 h, 48 h, and 72 h of incubation at 30°C. The 48-h time point is shown.
RESULTS
Translesion DNA synthesis suppresses GCR accumulation in mph1Δ cells.
Deletion of MPH1 has been shown to cause an increase in spontaneous base substitutions at CAN1, which can be suppressed by disrupting error-prone translesion DNA synthesis (41). To determine how spontaneous DNA lesions are processed if they cannot be bypassed by Mph1 or TLS, we deleted MPH1 and REV3 in a yeast strain that has been modified to allow the detection of gross chromosomal rearrangements, such as translocations, large deletions, and de novo telomere additions (18, 39). We found that the rev3Δ mph1Δ mutant showed a statistically significant increase in the GCR rate over that of the single mutants, as indicated by the nonoverlapping 95% confidence intervals (α < 0.05) for the median GCR rates (Table 2). This may indicate that the avoidance of point mutations by deletion of the error-prone DNA polymerase Rev3 occurs at the expense of increased formation of chromosomal rearrangements, suggesting that as long as Rev3 is present, spontaneous DNA lesions in the mph1Δ mutant are preferentially taken care of by TLS, whereas an alternative repair pathway preferentially utilized in the rev3Δ mph1Δ mutant is prone to GCR formation. To test the possibility that Srs2, a DNA helicase that has been shown to regulate homologous recombination by disrupting recombinogenic Rad51-filaments (19, 47), may channel DNA lesions into this alternative DNA repair pathway, we determined the effect of an srs2Δ mutation on GCR formation in the rev3Δ mph1Δ mutant and found that GCR formation was eliminated (Table 2). That the viability of the rev3Δ mph1Δ mutant was significantly reduced upon introduction of the srs2Δ mutation (Fig. 1A) is consistent with previous reports of reduced fitness for the mph1Δ srs2Δ mutant (33, 45) and suggests that spontaneous DNA lesions in the rev3Δ mph1Δ mutant may become substrates for homologous recombination pathways that need to be regulated by Srs2 to prevent cell death. In contrast to the mph1Δ mutant, the rev3Δ mutant does not require Srs2 for normal growth (Fig. 1A).
Table 2.
Effect of defects in DNA lesion bypass, homologous recombination, DNA helicases, and the DNA damage checkpoint on accumulation of gross chromosomal rearrangements in the standard GCR strain background RDKY3615
| Relevant genotype | Strain | GCR rate (Canr 5-FOAr) (× 10−10)a | 95% CIb (Canr 5-FOAr) (× 10−10) | Fold increase over wild-type level |
|---|---|---|---|---|
| Wild type | RDKY3615 | 3.5c | 1 | |
| mph1 | KHSY1557 | 20 | 5–34 | 6 |
| rev3 | KHSY1954 | 10 | 5–21 | 3 |
| rev3 mph1 | KHSY1976 | 56 | 44–71 | 16 |
| rev3 mph1 srs2 | KHSY2226 | <14 | <11–18 | <4 |
| srs2 | RDKY5557 | 2d | <2–11 | 0.6 |
| mph1 srs2 | KHSY1702 | 1.2 | <2–6 | 0.3 |
| mph1 mec3 | KHSY1878 | 55 | 24–73 | 16 |
| mph1 mec3 srs2 | KHSY2038 | 56 | 30–68 | 16 |
| mec1 sml1 | KHSY895 | 471 | 209–859 | 135 |
| mec1 sml1 mph1 | KHSY1894 | 290 | 154–467 | 83 |
| apn1 | KHSY1957 | 19 | 14–41 | 5 |
| apn1 mph1 | KHSY1970 | 15 | <15–51 | 4 |
| rad52 | KHSY1258 | 435 | 317–520 | 124 |
| rad52 mph1 | KHSY1598 | 275 | 131–467 | 79 |
| sgs1 | KHSY1630 | 220 | 144–276 | 64 |
| sgs1 mph1 | KHSY1600 | 239 | 162–528 | 68 |
| chl1 | KHSY1561 | 14 | <14–94 | 4 |
| chl1 mph1 | KHSY1725 | 40 | <10–202 | 11 |
| rrm3 | KHSY1399 | 14d | 5–28 | 4 |
| rrm3 mph1 | KHSY1713 | 21 | <17–48 | 6 |
5-FOA, 5-fluoroorotic acid.
Ninety-five percent confidence intervals (CIs) were calculated according to the method of Nair (26), with nonoverlapping confidence intervals indicating statistically significant differences (α < 0.05) between median GCR rates.
GCR rate from reference 2.
GCR rate from reference 38.
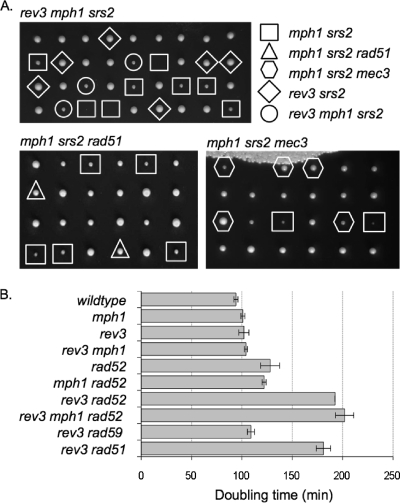
Fig. 1.
Genetic interactions between rev3Δ, mph1Δ, srs2Δ, and HR mutations were assessed by testing the fitness of mutants. (A) Tetrads from diploids heterozygous for rev3Δ, mph1Δ, and srs2Δ; mph1Δ, srs2Δ, and rad51Δ; or mph1Δ, srs2Δ, and mec3Δ were dissected on rich medium and genotyped by spotting on selective medium or by PCR to determine the presence of gene deletions. In contrast to the mph1Δ mutant, the rev3Δ mutant does not require SRS2 for normal growth. Deletion of MEC3 or disruption of HR rescues the slow growth of the mph1Δ srs2Δ mutant. (B) Doubling times of mutant strains and appropriate controls in rich medium (YPD) are shown with standard deviations. Cells lacking Rev3 require Rad51 and Rad52 but not Rad59 for normal growth, and these growth defects are unaffected by Mph1.
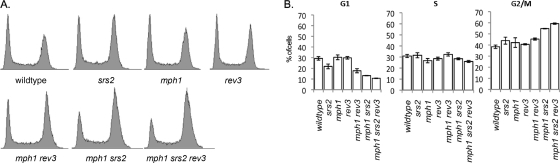
To test whether slow growth of the mph1Δ srs2Δ and rev3Δ mph1Δ srs2Δ mutants was due to increased G2/M arrest or to slowed progression through S phase resulting from impaired DNA lesion bypass, cell cycle profiles were obtained (Fig. 2A) and the fraction of cells in each cell cycle phase from three independent cultures was quantified (Fig. 2B). We found that the mph1Δ srs2Δ and rev3Δ mph1Δ srs2Δ mutants showed increased arrest in G2/M compared to the corresponding single and double mutants, but the mutations lacked any discernible affect on S phase. That the fraction of cells in S phase was largely unaffected indicates that impairment of DNA lesion bypass does not hinder the timely completion of genome replication but may instead cause the formation of DNA structures that later in the cell cycle impair progress through mitosis.
Fig. 2.
Effect of rev3Δ and srs2Δ mutations on cell cycle progression of cells lacking Mph1. Asynchronous cultures were grown to mid-log phase, fixed, and stained with Sytox green to measure DNA content by FACS. (A) Cell cycle profiles reveal that mph1Δ srs2Δ cells accumulate in G2/M phase, which is enhanced further by a rev3Δ mutation. (B) Quantification of cell fractions in G1, S, and G2/M phases, determined by FACS analysis of three cell cultures for each strain, using FlowJo v8.3.3. Error bars indicate the standard deviations.
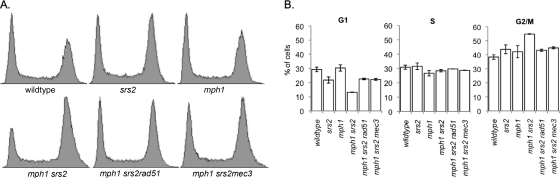
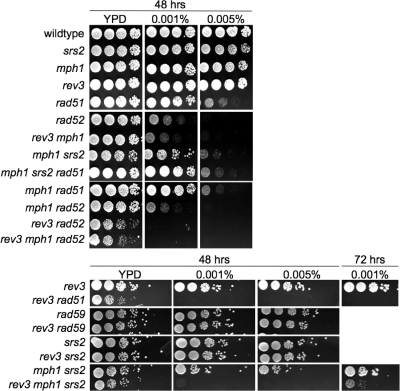
Deletion of RAD51, which had previously been shown to improve colony growth of the mph1Δ srs2Δ mutant and other DNA helicase double mutants (4, 6, 17, 20, 33, 37, 46), abolished the G2/M arrest of mph1Δ srs2Δ cells and allowed them to progress through the cell cycle as did the srs2Δ single mutant (Fig. 3A). In addition to disrupting homologous recombination, we found that disruption of the DNA damage checkpoint clamp by MEC3 deletion also improved growth of the mph1Δ srs2Δ mutant (Fig. 1A) and had the same effect on viability and cell cycle progression as the deletion of RAD51 (Fig. 3). In contrast, introduction of a rev3Δ mutation into the mph1Δ srs2Δ mutant did not affect growth (Fig. 1A) but led to a small increase in the fraction of G2/M-arrested cells (Fig. 2). Similarly, sensitivity of the mph1Δ srs2Δ mutant to MMS was aggravated further by a rev3Δ mutation but was alleviated by a rad51Δ mutation (Fig. 4). In the presence of MMS, rev3Δ mph1Δ srs2Δ cells emerged only after incubation for >72 h. This strong synergistic increase in MMS sensitivity of the triple mutant compared to that of any of the double mutants suggests that all three genes mediate independent pathways for survival in the presence of DNA damage. Taken together, these findings suggest that deleting the error-prone DNA polymerase Rev3 in mph1Δ cells while effectively avoiding points mutations causes the appearance of a different mutation type, i.e., gross chromosomal rearrangements, which activates the DNA damage checkpoint in G2/M and causes cell death if Srs2 is not present to regulate HR-dependent DNA lesion bypass.
Fig. 3.
The G2/M arrest of mph1Δ srs2Δ cells is suppressed by disrupting homologous recombination or the DNA damage checkpoint. Cell cycle profiles (A) and quantification (B) of the fractions of cells in G1, S, and G2/M phases show that the rad51Δ and mec3Δ mutations are equally effective at decreasing cell accumulation in G2/M, showing an increase in the fraction of cells in G1. Neither mutation affects the fraction of cells in S phase. The DNA content of Sytox green-stained cells from at least three mid-log-phase cultures of every strain was analyzed by FACS.
Fig. 4.
Effect of mutations affecting translesion DNA synthesis and homologous recombination on sensitivity to MMS. Tenfold dilutions of exponentially growing cultures were spotted on YPD (viable cell count) or YPD containing 0.001% or 0.005% MMS. Colony growth after 48 h (and 72 h for selected mutants) of incubation at 30°C is shown.
Suppression of genome rearrangements by srs2Δ depends on functional DNA damage checkpoint.
Genome instability in cells lacking Sgs1 helicase is suppressed by the DNA damage checkpoint, as demonstrated by synergistic GCR rate increases upon introduction of the mec3Δ, rad24Δ, mec1Δ, rad53Δ or rad9Δ mutation into the sgs1Δ mutant (40). As demonstrated by overlapping 95% confidence intervals, no statistically significant changes in the GCR rate of the mph1Δ mutant were observed upon introduction of checkpoint mutations (mec3Δ and mec1Δ) (Table 2), suggesting that the DNA damage checkpoint is not required for the suppression of GCRs in the mph1Δ mutant. However, when we introduced the mec3Δ mutation into the mph1Δ srs2Δ mutant, which itself exhibited wild-type levels of GCRs, the GCR rate increased to that of the mph1Δ mec3Δ mutant (Table 2), thus suggesting that, in contrast to the case with the rev3Δ mph1Δ and mph1Δ mutants, GCR formation in checkpoint-deficient mutants is not dependent on Srs2. Similarly, the GCR rate of the mec3Δ mutant did not change upon introduction of an srs2Δ mutation. This ability of the srs2Δ mutation to suppress GCR formation in checkpoint-proficient cells but not in checkpoint-deficient cells suggests that the Mec3 checkpoint detects the aberrant HR intermediates that form in the absence of Srs2, leading to G2/M arrest and avoidance of GCRs, whereas in the absence of the checkpoint, these aberrant HR intermediates go on to form GCRs.
Lack of Rev3 and Mph1 causes synergistic GCR rate increase in new GCR strain susceptible to duplication-mediated rearrangements.
Putnam et al. (34) recently showed that the rate of GCR accumulation depends significantly on chromosomal features in the breakpoint region. For example, while GCRs in the standard GCR strain background (RDKY3615) are due largely to single-copy-sequence-mediated rearrangements, GCRs in a newly designed strain (RDKY6678) are duplication mediated due to the presence of imperfect homology between the HXT13-DSF1 sequence in the breakpoint region on chromosome V and sequences on chromosomes IV, X, and XIV (34). This new GCR strain accumulates chromosomal rearrangements at an increased rate compared to that for the standard GCR strain, with wild-type cells having a 56-fold-higher GCR rate than the standard strain (34). To assess the effect of DNA lesion bypass defects on GCR formation in this new strain, rev3Δ, mph1Δ, srs2Δ, and mec3Δ mutations were introduced into RDKY6678 (Table 3). Consistent with our observations with the standard GCR strain background (Table 2), the rev3Δ mph1Δ double mutant exhibited a synergistic GCR rate increase compared to results for the mph1Δ and rev3Δ single mutants. Interestingly, the significantly greater synergistic effect of combining the rev3Δ and mph1Δ mutations in the new GCR strain background (Table 3; 167-fold increase over rates for the RDKY6678 wild type) compared to results with the standard GCR strain background (Table 2, 16-fold increase over rates for the RDKY3615 wild type) indicates that alternative pathways utilized for DNA lesion bypass in the rev3Δ mph1Δ mutant may be more prone to duplication-mediated than to single-copy-sequence mediated genome rearrangements. As in the standard GCR strain (Table 2), deletion of SRS2 in the new GCR strain led to a significant decrease in the GCR rate of the rev3Δ mph1Δ mutant to the level of the srs2Δ mutant, suggesting that viable GCR formation depends on the antirecombinase Srs2 despite the different breakpoint regions in the two GCR strain backgrounds and the different GCR types that are likely to arise from them. The fact that deletion of SRS2 did not cause a GCR rate increase in the mph1Δ mec3Δ mutant in the standard GCR background (Table 2) but led to a significant GCR rate increase in the new GCR background (Table 3) is likely due to the greater requirement of Srs2 for GCR suppression in the new GCR strain background (Table 3, srs2Δ: 26-fold increase over wild-type level) than in the standard GCR strain (Table 2, srs2Δ: 0.6-fold increase over wild-type level).
Table 3.
Effects of mph1, rev3, srs2, and mec3 deletions on the accumulation of chromosomal rearrangements in a new GCR strain background (RDKY6678) that is prone to duplication-mediated rearrangementsa
| Relevant genotype | Strain | GCR rate (Canr 5-FOAr) (× 10−8)b | 95% CIc (Canr 5-FOAr) (× 10−8) | Fold increase over wild-type level |
|---|---|---|---|---|
| Wild type | RDKY6678 | 2d | 1 | |
| mph1 | RDKY6795 | 18 | 10–31 | 9 |
| rev3 | KHSY3110 | 13 | 8–20 | 7 |
| rev3 mph1 | KHSY3056 | 334 | 237–389 | 167 |
| rev3 mph1 srs2 | KHSY3123 | 57 | 41–67 | 29 |
| srs2 | KHSY3067 | 31 | 21–54 | 16 |
| mph1 srs2 | KHSY3101 | 40 | 32–61 | 20 |
| mec3 | KHSY3065 | 44 | 30–59 | 22 |
| mph1 mec3 | KHSY3042 | 43 | 30–98 | 22 |
| mph1 mec3 srs2 | KHSY3126 | 142 | 116–191 | 71 |
See reference 34.
5-FOA, 5-fluoroorotic acid.
Ninety-five percent confidence intervals (CIs) were calculated according to the method of Nair (26), with nonoverlapping confidence intervals indicating statistically significant differences (α < 0.05) between median GCR rates.
GCR rate from reference 34.
Genetic interactions between MPH1 and other DNA helicases.
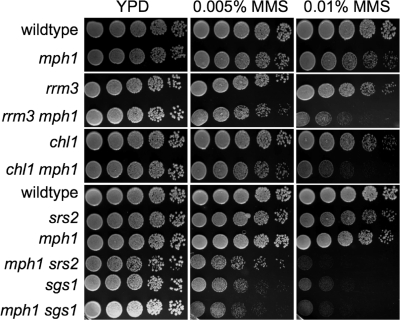
Negative genetic interactions between any two of the DNA helicases Sgs1, Srs2, and Rrm3 have previously been shown to be caused by the accumulation of aberrant intermediates of homologous recombination (6, 29, 37, 46). Since such a negative, HR-dependent genetic interaction has now also been established between mph1Δ and srs2Δ, we tested mph1Δ mutants with deletions of other confirmed or putative DNA helicase genes (sgs1Δ, rrm3Δ, and chl1Δ) for growth defects, GCR accumulation, and sensitivity to MMS. We observed that unlike the case with the mph1Δ srs2Δ mutant, the meiotic products of diploids heterozygous for the mph1Δ mutation and either the sgs1Δ, rrm3Δ, or chl1Δ mutation grew normally. In addition to a synergistic increase in sensitivity to MMS for the mph1Δ srs2Δ mutant (1), we also identified synergistic increases in sensitivity for the mph1Δ rrm3 and mph1Δ chl1Δ mutants but not for the mph1Δ sgs1Δ mutant, which appeared as sensitive as the sgs1Δ single mutant (Fig. 5). This indicates that Mph1, Chl1, Rrm3, and Srs2 contribute independently to survival during exposure to MMS, while Mph1 appears to be hypostatic to Sgs1. Since Schurer et al. (41) reported a synergistic increase in mitotic homologous recombination at three markers for the mph1Δ sgs1Δ mutant and therefore suggested that Mph1 may play an antirecombinogenic role in the sgs1Δ mutant, we tested whether Mph1 also interacted with Sgs1 or other DNA helicases in the suppression of GCRs. However, we found that the mph1Δ sgs1Δ mutant accumulates GCRs at the same rate as the sgs1Δ mutant, indicating no genetic interaction between MPH1 and SGS1 in the suppression of chromosomal rearrangements (Table 2). Deletion of MPH1 also failed to induce significant changes in the accumulation of GCRs in srs2Δ, chl1Δ, and rrm3Δ mutants, as indicated by the overlap between 95% confidence intervals (Table 2).
Fig. 5.
Effect of an mph1Δ mutation on MMS sensitivity of mutants lacking various other confirmed (Sgs1, Rrm3, and Srs2) or putative (Chl1) DNA helicases. Tenfold dilutions of exponentially growing cultures were spotted on YPD or YPD containing 0.01% or 0.005% MMS. Colony growth after 48 h of incubation at 30°C is shown.
Rad52/Rad51, but not Rad59, are essential for DNA damage tolerance and normal growth in the absence of translesion DNA synthesis.
Although the rev3Δ mph1Δ mutant exhibits a synergistic increase in the GCR rate and in sensitivity to MMS, it grows unimpaired in the absence of MMS, with a doubling time indistinguishable from that of the single mutants (Fig. 1B). However, sporulation of diploids heterozygous for the rev3Δ and rad52Δ mutations revealed slower growth for the rev3Δ rad52Δ mutant that was unaffected by deletion of MPH1 (Fig. 1B). That the rev3Δ rad52Δ mutant does grow, albeit slowly, could mean that spontaneous DNA lesions needing to be bypassed during DNA replication are rare and/or that alternative, yet minor, pathways for lesion bypass exist in addition to HR and TLS. To distinguish between these possibilities, the ability of the HR-deficient rev3Δ mutant to grow in the presence of MMS was assessed (Fig. 4). While the rev3Δ mutant was no more sensitive than wild-type cells, consistent with previous findings (41), the rev3Δ rad52Δ mutant was significantly more sensitive than the rad52Δ mutant. In fact, not a single colony emerged in repeated experiments, even after a >72-h incubation time on 0.001% MMS, for strains lacking both REV3 and RAD52, lending support to the proposal that besides HR and TLS, no other pathways exist in yeast for the bypassing of induced DNA lesions. To determine whether Rad51- or Rad59-dependent branches of homologous recombination are essential for rev3Δ survival, the viability of spores obtained from diploids heterozygous for rev3Δ and either the rad51Δ or rad59Δ mutation was assessed, revealing normal growth for the rev3Δ rad59Δ mutant while the rev3Δ rad51Δ mutant grew slowly (Fig. 1B). Moreover, the rev3Δ rad59Δ mutant was no more sensitive than the single mutants, whereas the rev3Δ rad51Δ mutant could not form any colonies in the presence of MMS (Fig. 4), similar to the case with the rev3Δ rad52Δ mutant. Thus, although rev3Δ exhibits synergistic increases in sensitivity to MMS when combined with mph1Δ, mph1Δ srs2Δ, rad52Δ, or rad51Δ, the normal growth of the rev3Δ mph1Δ mutant as opposed to the impaired growth of the rev3Δ rad51Δ and rev3Δ rad52Δ mutants suggests that in addition to Mph1-dependent HR, other, Mph1-independent, Rad51-dependent HR pathways exist for DNA lesion bypass.
DISCUSSION
We have investigated genetic interactions between genes involved in DNA lesion bypass (MPH1 and REV3), homologous recombination (RAD52, RAD51, RAD59, and SRS2), and the DNA damage checkpoint (MEC3 and MEC1) with regard to fitness, MMS sensitivity, and suppression of genome instability. We find that suppression of point mutations that arise in an mph1Δ mutant as a result of the error-prone Rev3 polymerase replicating across a template lesion results in the appearance of GCRs. This finding may suggest that mutations are not actually avoided but are simply shifted toward a different mutation type. Synergistic GCR rate increases in two strain backgrounds, each designed to accumulate different GCR spectra (34), demonstrate that REV3 and MPH1 interact genetically to suppress various types of spontaneous GCRs but are especially effective at suppressing GCRs in the newly designed GCR strain background. For this new GCR strain, Putnam et al. (34) determined that GCRs accumulate largely as a result of nonallelic homologous recombination (NAHR) between DNA sequences in the breakpoint region on chromosome V and similar regions on chromosomes IV, X, and XIV. Hence, the greater synergistic GCR rate increase identified here in this new GCR background compared to that for the standard GCR strain suggests greater roles for MPH1 and REV3 in the suppression of such NAHR-mediated GCRs than in the suppression of single-copy-sequence-mediated rearrangements. The requirement of Srs2, which regulates the outcomes of HR by antagonizing strand invasion, for the formation of viable chromosomal rearrangements further supports a prominent role of HR in the formation of GCRs when Mph1 and Rev3 are absent for DNA lesion bypass. We further show that the negative genetic interaction between the mph1Δ and srs2Δ mutations, coupled with accumulation of cells in G2/M and further exacerbation of the G2/M arrest by disruption of REV3, is suppressed by disrupting the DNA damage checkpoint. Synergism in MMS sensitivity was observed for mph1Δ mutants lacking CHL1, RRM3, SRS2, or REV3, whereas epistasis was observed for mph1Δ mutants lacking RAD52, RAD51, or SGS1. Combined with our observation that the rev3Δ mutant required RAD51 and RAD52 but not RAD59 or MPH1 for normal growth, this suggests that Rev3 (TLS) and RAD51 (HR) are the two pathways for bypass of spontaneous DNA lesions, with Mph1 defining only one Rad51 subpathway. While the rad51Δ mutation appeared to suppress MMS sensitivity of the mph1Δ srs2Δ mutant to the level exhibited by a rad51Δ single mutant, the rev3Δ mutation led to a further synergistic increase, suggesting that Mph1, Srs2, and Rev3 contribute to bypass and/or repair of induced DNA lesions independently.
Our observation of suppression of the G2/M arrest of the mph1Δ srs2Δ mutant by mec3Δ, in addition to the recently reported suppression by rad51Δ (33), suggests that cells lacking Mph1 and Srs2 are overwhelmed with HR intermediates that do not impair S phase but activate the DNA damage checkpoint prior to mitosis. That Srs2 is essential for normal growth in the absence of Mph1 could mean that DNA lesions, normally bypassed by the Mph1 pathway, will enter another HR pathway that is potentially lethal if it is not regulated by Srs2. According to recent findings by Prakash et al. (33), Srs2, Mph1, and Sgs1 independently promote noncrossover pathways during mitotic DSB repair. They suggest that Srs2 diverts DNA lesions away from crossover events that can result from double Holliday junction (dHJ) resolution into the noncrossover SDSA pathway by preventing second-strand invasion. Accumulation of dHJs due to the absence of Srs2 could overwhelm resolution pathways, especially when alternative pathways for DNA lesion bypass, such as TLS, are absent. In addition to its ability to inhibit crossover formation during repair of an HO-induced DSB, Mph1 has also been reported to unwind D loops in vitro (33). It has therefore been proposed that Mph1 promotes SDSA and may reverse strand invasion events before they can form dHJs. Thus, the overall burden of lesions that are committed to HR pathways and could potentially go on to form dHJs would be expected to increase in the absence of Mph1 and even further when Rev3 is also absent.
Recent findings suggest how FANCM, a human homolog of Mph1, could perform a role in error-free bypass of DNA lesions. FANCM is part of the eight-component Fanconi anemia core complex, which is involved in the repair of intrastrand cross-links and is associated with Fanconi anemia (15, 16, 23, 27, 48). FANCM can branch migrate three- and four-way junctions and, like Mph1, unwind D loops (7, 8). Combining these two activities, it has been proposed that FANCM may stall and remodel replication forks to promote repair of an approaching DNA lesion, thus preventing the fork from encountering the lesion and collapsing (7). Without FANCM, forks would collapse, leading to broken chromatids and increased gross chromosomal rearrangements, both hallmarks of Fanconi cells (44).
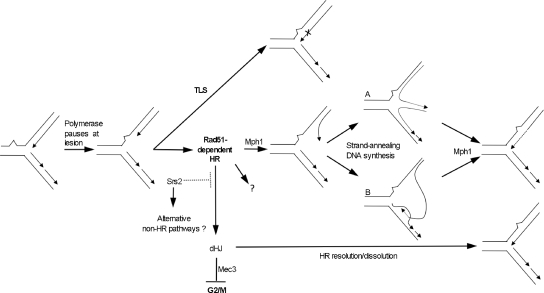
The recruitment of Srs2 to the replisome when PCNA becomes sumoylated in the presence of DNA damage (31) and the ability of the human Mph1 homolog FANCM to remodel replication forks in vitro lead us to propose a model in which Mph1 and Srs2 perform their roles in recombination directly at the fork to restart replication after encountering a DNA lesion (Fig. 6). Based on the slow growth of Rad52/Rad51-deficient rev3Δ cells and the inability of the rev3Δ rad52Δ and rev3Δ rad51Δ mutants to form any colonies in the presence of MMS, we propose that in wild-type cells, DNA lesions may be bypassed by either Rev3-mediated TLS or Rad51-dependent HR, with Mph1 being involved in only one subpathway of Rad51-dependent HR. Rev3-mediated TLS is prone to errors but not GCR formation, while properly regulated HR pathways, including the Mph1 pathway, are error free. Mph1 may act at a stalled replication fork by unwinding the leading strand from its template, a scenario which has been suggested to resemble unwinding of a D loop (7). Mediated by HR proteins, the leading strand may then anneal with the lagging strand, forming a chicken foot (Fig. 6, structure A), or invade the sister chromatid, forming a D loop (Fig. 6, structure B), followed by DNA synthesis at the 3′ end. Based on the ability of Mph1 to reverse D loops in vitro, it also seems possible that Mph1 acts to resolve these HR intermediates. For example, Mph1 could unwind the D loop formed by HR proteins after limited DNA synthesis or dissolve the chicken-foot structure by reverse branch migration. Reannealing of the daughter strands with their templates would then result in error-free bypass of the DNA lesion in the template strand and resumption of replication. While FANCM has been shown to migrate three- and four-way junctions in vitro (7, 8), as proposed in this model, this remains to be determined for Mph1. The recent report of a physical interaction between Mph1 and RPA (1) could suggest that Mph1 is recruited to stalled replication forks via RPA-bound regions of ssDNA that are generated when the replication machinery stalls at a lesion in the template. While Mph1 can unwind 40 bp by itself, it requires RPA to unwind duplexes that are 100 bp and fails on those that are 500 bp (32). This rather modest helicase activity could ensure that Mph1 does not expose unnecessarily large regions of ssDNA at stalled forks while at the same time being sufficiently strong to unwind the leading strand from its template needed for D-loop/chicken-foot formation and/or to reverse HR-mediated invasion of the sister chromatid. Moreover, the ATPase activity of Mph1 requires a relatively long stretch of ssDNA (≥40 nt) for full activation in vitro (32), which could help to ensure that Mph1 is active only on replication forks that have stalled because they are likely to contain longer regions of ssDNA than unperturbed forks. In our model, Srs2 is recruited to damaged replication forks to suppress dHJ formation, thereby promoting Mph1-mediated fork rescue. Such a role for Srs2 at the replication fork is consistent with recent findings (21, 31). Hence, in the absence of Srs2, an increasing number of forks would enter dHJ pathways for rescue, overwhelming dHJ resolution pathways and leading to aberrant and/or unresolved intermediates and eventually G2/M arrest. This accumulation of srs2Δ cells in G2/M accelerates as more DNA lesions become substrates for dHJ pathways upon elimination of Mph1 and Rev3. Unresolved or aberrant DNA structures may not be the only cause for Mec3-dependent cell cycle arrest of mph1Δ srs2Δ cells. According to findings by Prakash et al. (33), HR intermediates during DSB repair are increasingly resolved as crossovers when Srs2 and Mph1 are absent, possibly due to increased HR and impairment of single-strand annealing pathways, such as SDSA. Thus, not only is increased dHJ formation likely to overwhelm dHJ resolution pathways, it is also likely to increase the number of crossovers, which could be dangerous for haploid mitotic cells and contribute to diminished cell proliferation. Although formation and unwinding of D-loop-like structures could be envisaged at replication forks and the recently proposed role of Mph1 in SDSA repair of DSBs could be likened to reversing chicken-foot/D-loop structures at stalled forks, it remains to be tested whether Mph1 can branch migrate three- or four-way junctions to reverse these HR intermediates and does indeed function at the replication fork.
Fig. 6.
Model for the role of Mph1 in the maintenance of genome stability. DNA lesions arise spontaneously during DNA replication and are bypassed by an error-free, Mph1-mediated, noncrossover pathway of homologous recombination (HR). Mph1 may unwind the leading strand from its template, suggested to resemble a D loop (7), followed by Rad51/52-mediated chicken-foot formation (A) and then resolution by reverse branch migration. A D-loop structure could also form when the leading strand switches template (B), and Mph1 could dissolve this D loop by reverse branch migration. In the absence of Mph1, lesions are bypassed by error-prone, Rev3-mediated TLS or they are channeled by Srs2 into alternative bypass pathways that can result in GCRs. If TLS is disrupted in the mph1Δ mutant, point mutations from TLS are avoided, but GCRs arise as a consequence of aberrant repair, most likely nonallelic HR. In the absence of both Mph1 and Srs2, cells accumulate at G2/M and lose viability due to Rad51-mediated accumulation of dHJs and Mec3-mediated checkpoint activation. In the absence of the checkpoint, cells continue through the cell cycle in the presence of DNA lesions. The dotted line emerging from Srs2 indicates that Srs2 tightly regulates the levels of dHJ formation at paused replication forks by inhibiting Rad51-mediated strand invasion.
ACKNOWLEDGMENTS
We thank Charly Szekeres (USF College of Medicine) for FACS analysis and Richard Kolodner (University of California—San Diego) for RDKY yeast strains.
We declare that there are no conflicts of interest.
Footnotes
Published ahead of print on 11 December 2009.
REFERENCES
- 1.Banerjee S., Smith S., Oum J. H., Liaw H. J., Hwang J. Y., Sikdar N., Motegi A., Lee S. E., Myung K. 2008. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol. 181:1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C., Umezu K., Kolodner R. D. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2:9–22 [DOI] [PubMed] [Google Scholar]
- 3.Entian K. D., Schuster T., Hegemann J. H., Becher D., Feldmann H., Guldener U., Gotz R., Hansen M., Hollenberg C. P., Jansen G., Kramer W., Klein S., Kotter P., Kricke J., Launhardt H., Mannhaupt G., Maierl A., Meyer P., Mewes W., Munder T., Niedenthal R. K., Ramezani Rad M., Rohmer A., Romer A., Hinnen A., et al. 1999. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 262:683–702 [DOI] [PubMed] [Google Scholar]
- 4.Fabre F., Chan A., Heyer W. D., Gangloff S. 2002. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. U. S. A. 99:16887–16892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangloff S., Soustelle C., Fabre F. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192–194 [DOI] [PubMed] [Google Scholar]
- 7.Gari K., Decaillet C., Delannoy M., Wu L., Constantinou A. 2008. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U. S. A. 105:16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gari K., Decaillet C., Stasiak A. Z., Stasiak A., Constantinou A. 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29:141–148 [DOI] [PubMed] [Google Scholar]
- 9.Gietz R. D., Woods R. A. 2006. Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313:107–120 [DOI] [PubMed] [Google Scholar]
- 10.Hirota Y., Lahti J. M. 2000. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 28:917–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holloway S. L. 2000. CHL1 is a nuclear protein with an essential ATP binding site that exhibits a size-dependent effect on chromosome segregation. Nucleic Acids Res. 28:3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivessa A. S., Lenzmeier B. A., Bessler J. B., Goudsouzian L. K., Schnakenberg S. L., Zakian V. A. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 12:1525–1536 [DOI] [PubMed] [Google Scholar]
- 14.Ivessa A. S., Zhou J. Q., Schulz V. P., Monson E. K., Zakian V. A. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 16:1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joenje H., Patel K. J. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446–457 [DOI] [PubMed] [Google Scholar]
- 16.Kennedy R. D., D'Andrea A. D. 2005. The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev. 19:2925–2940 [DOI] [PubMed] [Google Scholar]
- 17.Klein H. L. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolodner R. D., Putnam C. D., Myung K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552–557 [DOI] [PubMed] [Google Scholar]
- 19.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305–309 [DOI] [PubMed] [Google Scholar]
- 20.Lee S. K., Johnson R. E., Yu S. L., Prakash L., Prakash S. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286:2339–2342 [DOI] [PubMed] [Google Scholar]
- 21.Liberi G., Chiolo I., Pellicioli A., Lopes M., Plevani P., Muzi-Falconi M., Foiani M. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 19:5027–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo Y. C., Paffett K. S., Amit O., Clikeman J. A., Sterk R., Brenneman M. A., Nickoloff J. A. 2006. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell Biol. 26:4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathew C. G. 2006. Fanconi anaemia genes and susceptibility to cancer. Oncogene 25:5875–5884 [DOI] [PubMed] [Google Scholar]
- 24.Mullen J. R., Kaliraman V., Brill S. J. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154:1101–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myung K., Datta A., Chen C., Kolodner R. D. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113–116 [DOI] [PubMed] [Google Scholar]
- 26.Nair K. R. 1940. Table of confidence intervals for the median in samples from any continuous population. Sankhya 4:551–558 [Google Scholar]
- 27.Niedzwiedz W., Mosedale G., Johnson M., Ong C. Y., Pace P., Patel K. J. 2004. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell 15:607–620 [DOI] [PubMed] [Google Scholar]
- 28.Ogiwara H., Ui A., Lai M. S., Enomoto T., Seki M. 2007. Chl1 and Ctf4 are required for damage-induced recombinations. Biochem. Biophys. Res. Commun. 354:222–226 [DOI] [PubMed] [Google Scholar]
- 29.Ooi S. L., Shoemaker D. D., Boeke J. D. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35:277–286 [DOI] [PubMed] [Google Scholar]
- 30.Petronczki M., Chwalla B., Siomos M. F., Yokobayashi S., Helmhart W., Deutschbauer A. M., Davis R. W., Watanabe Y., Nasmyth K. 2004. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J. Cell Sci. 117:3547–3559 [DOI] [PubMed] [Google Scholar]
- 31.Pfander B., Moldovan G. L., Sacher M., Hoege C., Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436:428–433 [DOI] [PubMed] [Google Scholar]
- 32.Prakash R., Krejci L., Van Komen S., Anke Schurer K., Kramer W., Sung P. 2005. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 280:7854–7860 [DOI] [PubMed] [Google Scholar]
- 33.Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J. E., Sung P., Ira G. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23:67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Putnam C. D., Hayes T. K., Kolodner R. D. 2009. Specific pathways prevent duplication-mediated genome rearrangements. Nature 460:984–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert T., Dervins D., Fabre F., Gangloff S. 2006. Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25:2837–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheller J., Schurer A., Rudolph C., Hettwer S., Kramer W. 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155:1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt K. H., Kolodner R. D. 2004. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 24:3213–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt K. H., Kolodner R. D. 2006. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl. Acad. Sci. U. S. A. 103:18196–18201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt K. H., Pennaneach V., Putnam C. D., Kolodner R. D. 2006. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 409:462–476 [DOI] [PubMed] [Google Scholar]
- 40.Schmidt K. H., Wu J., Kolodner R. D. 2006. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol. Cell. Biol. 26:5406–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schurer K. A., Rudolph C., Ulrich H. D., Kramer W. 2004. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics 166:1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiratori A., Shibata T., Arisawa M., Hanaoka F., Murakami Y., Eki T. 1999. Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast 15:219–253 [DOI] [PubMed] [Google Scholar]
- 43.Sinclair D. A., Mills K., Guarente L. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313–1316 [DOI] [PubMed] [Google Scholar]
- 44.Thompson L. H., Hinz J. M., Yamada N. A., Jones N. J. 2005. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ. Mol. Mutagen. 45:128–142 [DOI] [PubMed] [Google Scholar]
- 45.Tong A. H., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Menard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A. M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C. 2004. Global mapping of the yeast genetic interaction network. Science 303:808–813 [DOI] [PubMed] [Google Scholar]
- 46.Torres J. Z., Schnakenberg S. L., Zakian V. A. 2004. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 24:3198–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309–312 [DOI] [PubMed] [Google Scholar]
- 48.Wang W. 2007. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 8:735–748 [DOI] [PubMed] [Google Scholar]
- 49.Watt P. M., Louis E. J., Borts R. H., Hickson I. D. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253–260 [DOI] [PubMed] [Google Scholar]
- 50.Yamagata K., Kato J., Shimamoto A., Goto M., Furuichi Y., Ikeda H. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. U. S. A. 95:8733–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]