Abstract
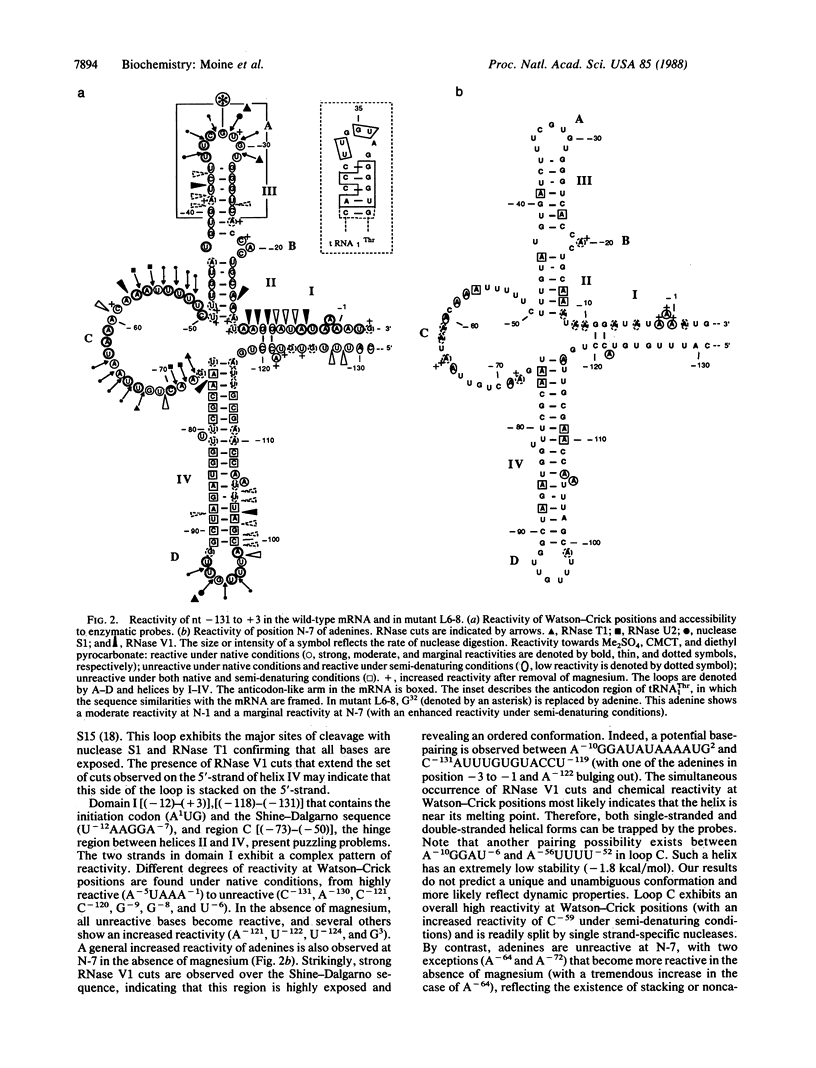
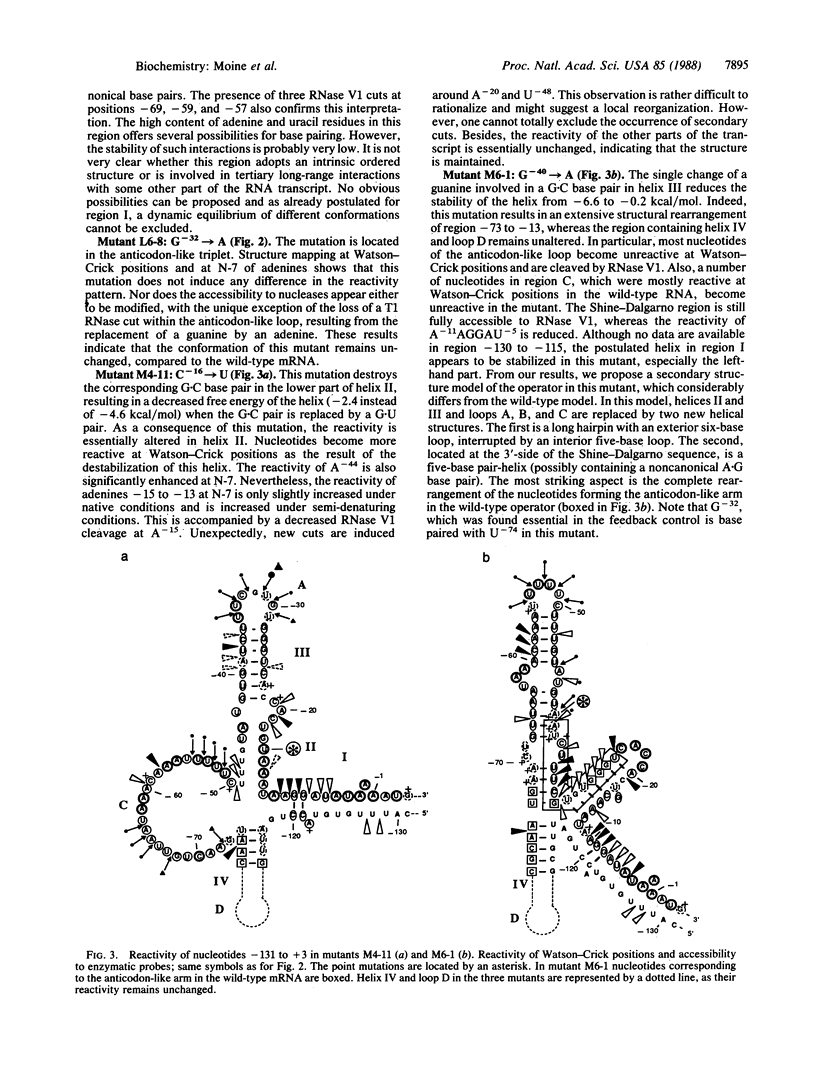
Previous work showed that the expression of the Escherichia coli threonine:tRNAThr ligase (EC 6.1.1.3)-encoding gene (thrS) is negatively autoregulated at the translational level and that a region called the operator that is located between 10 and 50 base pairs upstream of the translation initiation codon of the thrS gene is directly involved in that control. The conformation of an in vitro synthesized RNA fragment extending over the thrS regulatory region has been investigated using chemical and enzymatic probes. This study shows that the RNA folds into four well-defined secondary-structure domains, one of them displaying structural similarities to the anticodon arm of tRNAThr. The conformation of three constitutive mutants containing single base changes in the operator region leading to the loss of the regulatory control was also investigated. The replacement of a base in the anticodon-like loop does not induce any conformational change, suggesting that the residue concerned is directly involved in the regulatory process. However, single mutations in or close to the anticodon-like stem result in a partial or complete reorganization of the structure of the operator region. These rearrangements should affect the binding of the ligase to the operator, leading to loss of the regulatory process.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akins R. A., Lambowitz A. M. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987 Jul 31;50(3):331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- Baudin F., Ehresmann C., Romby P., Mougel M., Colin J., Lempereur L., Bachellerie J. P., Ebel J. P., Ehresmann B. Higher-order structure of domain III in Escherichia coli 16S ribosomal RNA, 30S subunit and 70S ribosome. Biochimie. 1987 Oct;69(10):1081–1096. doi: 10.1016/0300-9084(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Springer M., Dondon J., Grunberg-Manago M. Posttranscriptional autoregulation of Escherichia coli threonyl tRNA synthetase expression in vivo. J Bacteriol. 1986 Jan;165(1):198–203. doi: 10.1128/jb.165.1.198-203.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climie S. C., Friesen J. D. Feedback regulation of the rplJL-rpoBC ribosomal protein operon of Escherichia coli requires a region of mRNA secondary structure. J Mol Biol. 1987 Dec 5;198(3):371–381. doi: 10.1016/0022-2836(87)90287-7. [DOI] [PubMed] [Google Scholar]
- Deckman I. C., Draper D. E. S4-alpha mRNA translation regulation complex. II. Secondary structures of the RNA regulatory site in the presence and absence of S4. J Mol Biol. 1987 Jul 20;196(2):323–332. doi: 10.1016/0022-2836(87)90693-0. [DOI] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni A. L., Joshi S., Chapeville F. tRNA-like structures in the genomes of RNA viruses. Prog Nucleic Acid Res Mol Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- Hall T. C. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Hentze M. W., Caughman S. W., Rouault T. A., Barriocanal J. G., Dancis A., Harford J. B., Klausner R. D. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987 Dec 11;238(4833):1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- Herbert C. J., Labouesse M., Dujardin G., Slonimski P. P. The NAM2 proteins from S. cerevisiae and S. douglasii are mitochondrial leucyl-tRNA synthetases, and are involved in mRNA splicing. EMBO J. 1988 Feb;7(2):473–483. doi: 10.1002/j.1460-2075.1988.tb02835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney K. R., Nomura M. Secondary structure of the autoregulatory mRNA binding site of ribosomal protein L1. Mol Gen Genet. 1987 Nov;210(1):60–68. doi: 10.1007/BF00337759. [DOI] [PubMed] [Google Scholar]
- Leibold E. A., Munro H. N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5' untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel M., Eyermann F., Westhof E., Romby P., Expert-Bezançon A., Ebel J. P., Ehresmann B., Ehresmann C. Binding of Escherichia coli ribosomal protein S8 to 16 S rRNA. A model for the interaction and the tertiary structure of the RNA binding site. J Mol Biol. 1987 Nov 5;198(1):91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- Mougel M., Philippe C., Ebel J. P., Ehresmann B., Ehresmann C. The E. coli 16S rRNA binding site of ribosomal protein S15: higher-order structure in the absence and in the presence of the protein. Nucleic Acids Res. 1988 Apr 11;16(7):2825–2839. doi: 10.1093/nar/16.7.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Graffe M., Butler J. S., Grunberg-Manago M. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4384–4388. doi: 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Plumbridge J. A., Butler J. S., Graffe M., Dondon J., Mayaux J. F., Fayat G., Lestienne P., Blanquet S., Grunberg-Manago M. Autogenous control of Escherichia coli threonyl-tRNA synthetase expression in vivo. J Mol Biol. 1985 Sep 5;185(1):93–104. doi: 10.1016/0022-2836(85)90185-8. [DOI] [PubMed] [Google Scholar]
- Thomas M. S., Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987 Apr 10;15(7):3085–3096. doi: 10.1093/nar/15.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Maizels N. tRNA-like structures tag the 3' ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]