Abstract
We use affinity purification of the double bromodomain protein Brd2 to isolate a multicomponent nuclear complex from cultured cells, and apply mass spectrometry/proteomics methods to identify the participants. We then confirm by immunoblot several transcription co-activators and co-repressors, proteins of the Swi/Snf chromatin remodeling complex, which regulate transcription control of cyclin A. This multiprotein complex is likely to contribute to cell cycle control and play a role in proliferation and cancer.
Keywords: bromodomain, multiprotein complexes, affinity chromatography, high pressure liquid chromatography, mass spectral analysis, transcription factors, TATA binding protein-associated factors, chromatin assembly and disassembly, cyclin A
Introduction
Mammalian cells require proper transcriptional control of the cell cycle in order to negotiate the complex processes of DNA synthesis, chromosome duplication, and resolution of the topological problems of duplication. Failure to maintain this control jeopardizes cell survival or promotes malignancy. Proteins that contain a “bromodomain” motif1 regulate chromatin structure2 and epigenetics3 and have been noticed in this connection. The 110-amino acid bromodomain contains a highly conserved four-helix, left-twisted bundle with a hydrophobic cleft between two conserved loops, and acquired its name from the Drosophila brahma protein, wherein the motif was first defined.4 The bromodomain is found in almost all histone acetyltransferases (HATs), in transcriptional coactivators and in components of chromatin-associated multiprotein complexes. Bromodomains bind to acetylated ε-amino groups of nucleosomal histone lysines.5,6 Understanding the dynamic composition of bromodomain protein complexes has illuminated mechanisms of gene regulation, cell cycle control, and malignancy. The composition of complexes that regulate cell cycle genes likely reflects shifting alliances of transcription factors, HATs/HDACs, and nucleosome remodeling machines, dependent on the phase of the cell cycle. Mass spectrometry (MS) and proteomics methods have become powerful new approaches to characterize these large complexes and define their role in cell cycle control.
Some important multiprotein complexes contain the bromodomain protein Brd2. In Brd2, two mutually related bromodomain motifs are arrayed in tandem at the amino terminus.7 We have discovered that Brd2 functions as a transcriptional co-activator and nuclear localized kinase,8 and we are continuing to investigate its activities. Brd2 belongs to the BET (Bromodomain/ExtraTerminal domain) family of co-activators, defined by amino-terminal bromodomains and a protein–protein interaction motif.9 Double bromodomain-containing proteins play a role in cell cycle control,10,11 mitotic processes,12 gene regulation in response to mitogenic signal transduction,8,11,13 development7 and cancer.10,12 Brd2 and its Drosophila ortholog7 female sterile homeotic (fsh) possess double bromodomains most similar in primary sequence to the bromodomain9 of CBP/p300, and in structure to the bromodomains of RNA Polymerase II TATA Box-Associated Factor (TAFII) 250, an essential participant in basal transcription and cyclin gene control.14 Brd2 and Brd4 bind acetylated nucleosomal lysines,15,16 as does their yeast ortholog17 Bdf1, which has TAFII250-like function.18 The high selectivity of Brd2 binding to acetylated lysine 12 of histone H415 may provide a mechanistic basis for the hypothesized “histone code” of acetylation, phosphorylation, methylation and other PTMs of nucleosomal histones that contribute to transcription regulation.19 Many BET proteins are found in transcription complexes,2,3,9 where they may perform “scaffolding” functions20 and participate in chromatin restructuring.2,3,21
We have reported Brd2-dependent transactivation13 of the E2F-regulated cell cycle gene cyclin A, which governs the S phase of mitogenesis. Consistent with this observation, Brd2 nuclear complexes contain both E2F-1 and -213 and a histone H4-directed HAT.11 An ATP-dependent mechanism recruits Brd2 into the complex in all cell types we have studied. This observation argues that Brd2 participation in the complex has functional significance. Both (a) recruitment of E2Fs and (b) chromatin modification with HATs and HDACs regulate the transcription of cyclin A as well as other cyclin genes11 (and literature cited within ref 11), but the details of this coordinated mechanism are insufficiently understood. Mitogenic stimulation induces nuclear translocation of Brd2 protein and leads to its participation in these transcriptional events.22 Other groups have used MALDI of trypsin-digested proteins to show that Brd2-like proteins participate in multiprotein transcription complexes, such as Mediator,23 which contains transcriptional repressors MED14 and MED21 and co-activators MED6 and MED7. The murine Mediator proteins MED1 and CDK8 also associate with Brd2, Brd4, and E2F in an RNA Polymerase II-containing complex.24 We have hypothesized that Brd2 and similar BET proteins provide a scaffolding or platform function for transcription or chromatin remodeling complexes, anchoring them to nucleosomes and recruiting the required activities at the appropriate time.20 The severe phenotypes associated with disruption or deregulation of Brd2, Brd4, TAFII250 or Bdf110,12,14,16,18 could be explained in part by loss of a scaffolding function.
Elevated Brd2 kinase activity in human leukemic lympho-blasts8 led us to express a Brd2 transgene in the lymphoid lineage of mice, under murine immunoglobulin promoter/enhancer (Eμ) control (hereafter “Tg”) to give B cell-restricted, constitutive expression of the Brd2 scaffold. We expected that the mice would develop B cell leukemia or lymphoma, and this was indeed the case.10 Long before Tg mice develop malignancy, unstimulated Tg B cells show upregulated cyclin A transcription. Chromatin immunoprecipitation establishes the physical presence of Brd2 at the cyclin A promoter.11 Taken together, these observations suggest that cyclin A is a direct target of Brd2 action, and that Brd2-associated HAT and E2F activity is implicated in cancer. We decided next to characterize comprehensively the Brd2 complex components using advanced MS and proteomics methods in cultured cells. The generality of the machinery for eukaryotic transcriptional control of cyclin A and the highly conserved components, including histones, suggest that fundamental insights could be gained from the study of a model system.
Large, multiprotein complexes are important because they carry out multifaceted biochemical reactions of mammalian cells. These complexes may include pathway particles that catalyze sequential reactions in a metabolic pathway. The structural and functional characterization of these complexes has been a major goal of the last fifty years of protein biochemistry and has required efforts of heroic proportions, inasmuch as the relative abundances of the specific enzymes are often low and their activities labile. The methods traditionally used to detect rare proteins suffer from low sensitivity and specificity. The advent of high sensitivity MS- and proteomics-based protein identification methods has greatly facilitated the discovery, identification and characterization of biologically significant proteins and their PTMs, and identification of proteins within multiprotein machines. Thus, proteomics is becoming the method of choice for the detection of histone acetylation,25 analysis of tumor proteins and anti-cancer drug discovery.26 Many such machines, including DNA and RNA polymerases, RNA splicing complexes and SWI/SNF chromatin remodeling complexes, require ATP for function. Our studies of the Brd2 complex in fibroblasts, HeLa and lymphoid cells have enjoyed the advantage that complex composition is always ATP-dependent,8,13 enabling us to establish stringent conditions for immunoaffinity chromatography. Control immunoaffinity experiments conducted in buffers without ATP yield no detectable peptides in the immune complex apart from Brd2 and histone H4. We reasoned that this ATP recruitment mechanism defines sufficiently stringent methods for Brd2 complex purification.
In the studies we describe herein, we used proteomics analysis methods, including multicomponent immunoaffinity purification and liquid chromatography tandem mass spectrometry (LC–MS/MS) for low abundance protein identification. We isolated and characterized the components of a Brd2 complex in a model tissue culture system, the hypotriploid human embryonic kidney cell line 293T. We chose these cells to work out our methods and approach as the first stage of our proteomic investigation of Brd2 complexes because of the ease of expression of recombinant Brd2 protein and the high levels of transcription and protein synthesis that can be obtained with this line. Our strategy was to use immunoaffinity purification of recombinant Brd2 protein tagged with the HA epitope at the amino terminus. We have previously established that this modification does not affect transcription or nuclear localization functions,11,22 and that the high affinity of anti-HA antibody (Ab) binding preserves the interaction between the insoluble matrix and the recombinant protein. MS identification and validation by immunoblot of several conserved proteins of known function in this complex supported our hypothesis that the complex is part of a large, chromatin modification machine. We show herein that the complex contains histones, E2F, TAFII250 and representatives of the Swi/Snf chromatin-remodeling complex. Consistent with this scheme, several of the identified proteins are known to have important roles in transcriptional control and growth regulation.
Experimental Section
Adenovirus Constructs and Cell Culture
A cDNA that encoded full length, wild-type Brd2 was cloned into the EcoRI and BamHI sites of the CMV-directed expression plasmid pAC–CMV, to achieve high-level expression of cDNA as previously described.11 The vector encoded an in-frame, amino-terminal, HA epitope tag22 corresponding to the amino acid sequence MAYPYDVPDYASLSLVPSSDRS. Protocols for the preparation and manipulation of Adenovirus 5 to deliver expression constructs are well established.27 293T cells were maintained in Dulbecco’s modified minimal essential medium (Invitrogen/GIBCO-BRL, Grand Island, NY) buffered with sodium bicarbonate and supplemented with 10% fetal bovine serum, glutamine and antibiotics, and were maintained at 75% confluence in a 37 ° C incubator at 5% CO2 and 100% humidity. Cells in 10-cm dishes were infected at 50% confluence with 1 × 1011 plaque-forming units of Adenovirus and harvested after 24 h. Extracts were prepared as described previously,8 based on a canonical, simple method for preparation of soluble extracts that are competent for in vitro transcription initiation from RNA Polymerase II promoters. This method prepares all the general transcription factors, polymerase holoenzyme and TAFs necessary for proper in vitro transcription and was most appropriate for preparation of Brd2 complexes that play a role in transcriptional control. Measurement of optical density ratio at 260 and 280 nm confirmed that the extracts did not contain detectable nucleic acid. A schematic overview of sample preparation is shown in Figure 1.
Figure 1.

Sample preparation/analysis protocol. Schematic flow of work, showing cell extraction, immunoaffinity purification with αHA Ab to isolate recombinant HA-Brd2, elution by pH drop, tryptic digestion of eluted peptides and on-line LC–MS/MS for peptide identification.
Brd2 Immunoaffinity Chromatography and Immunoblots
Methods for αBrd2 rabbit polyclonal immunoaffinity chromatography13 were adapted for 293T extracts. Cytosolic and nuclear extracts8 of Adenovirus-infected cells were pooled for biochemical analyses, using methods adapted from similar experiments in fibroblasts.11
In this first, comprehensive identification of Brd2-associated proteins, we included cytosolic proteins to eliminate the possibility of bias that might arise due to nuclear-cytoplasmic partitioning, a known mechanism of Brd2 regulation.22 Combined extracts (0.5 mg) were diluted (1:2, v:v) with ice-cold Buffer A (50 mM disodium glycerol-2-phosphate, 20 mM Tris-HCl pH 7.4, 2 mM MgCl2, 5 mM disodium ATP, 1 mM Na3VO4, 1 mM DTT, supplemented with the protease inhibitors leu-peptin, pepstatin, aprotinin and phenylmethylsulfonyl fluoride), then applied to an Ab affinity column13 comprised of mouse ascites monoclonal antibody (mAb; 3.5 mg) directed against the HA epitope (Covance, Berkeley, CA) covalently coupled to 0.5 mL agarose (AminoLink; Pierce, Rockford, IL). Unless otherwise specified, all reagents were from Sigma (St. Louis, MO). The columns were washed extensively in the cold with 20 column volumes of ice-cold Buffer A and eluted with a pH drop (100 mM glycine-HCl, pH 2.5), into 1/10 volume of 1.0 M Tris-HCl pH 8.0 to neutralize. Eluted proteins were precipitated on ice with 10% TCA, washed twice with ice-cold 100% acetone, separated with SDS-PAGE and visualized with Coomassie blue stain. For immunoblots, proteins in gels were electroblotted to PVDF membranes and detected with human-reactive, poly-clonal primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) against pan-E2F (H-111, rabbit), Brg-1 (G-7, mouse monoclonal), Baf155 (H-76, rabbit) and CAF-1 (C-16, goat). The antibodies were then detected with horseradish peroxidase-conjugated secondary antibodies (goat anti-mouse IgG or anti-rabbit IgG, Boehringer Mannheim, Indianapolis, IN; or donkey anti-goat IgG, Santa Cruz), as previously described.11 Secondary Ab was visualized by chemiluminescence (New England Nuclear, Boston, MA) and XB-1 blue film (Kodak, Rochester, NY). We found that covalent coupling was necessary to minimize Ab peptides in the subsequent digest of the eluted proteins; otherwise, significant signals observed in the LC–MS/MS analyses were attributable to peptides derived from the Ab (typically immunoglobulin heavy chains). When Ab leached off the column under pH drop elution conditions, peptides derived from Ab overwhelmed the signals of peptides from the Brd2 complex. After we adapted the protocol to include this cross-linking step, the Ab-related signals were still detected, but their abundances had been effectively minimized and informative spectra of Brd2 complex peptides could be obtained. Washing that was less extensive than 20 column volumes of Buffer A increased the complexity of the elution profile of associated proteins, whereas more extensive washing did not significantly reduce the number of associated proteins. This result indicated that the selected degree of washing conferred maximum stringency with minimum loss of signal. For MS analysis, TCA precipitation was avoided and the eluted proteins were dialyzed overnight at 4 °C against 50 mM ammonium bicarbonate buffer containing 5 mM DTT, to remove elution buffer.
Tryptic Digestion and LC–MS/MS
Before digestion, the samples were purified and buffer exchanged. Trypsin digestions (enzyme:substrate, 1:100) were carried out in 100 mM ammonium bicarbonate buffer at pH 8, at 37 °C from 5.5 to 18 h. Drying the reaction mixture in a Speedvac concentrator quenched each enzymatic digestion. HPLC electrospray ionization mass spectrometry/tandem mass spectrometry (LC–MS/MS) was performed using a capillary HPLC system (CapLC, Waters, Milford, MA) coupled with a quadrupole orthogonal time-of-flight mass spectrometer (Q-TOF API US, Micromass/Waters) the ion source of which is equipped with NanoLock-Spray and Z-Spray. Peptides were dissolved in 0.2% formic acid. Sample preconcentration and desalting were performed using a peptide trap cartridge in line with the auto-sampler and column. Separation was on a 300 μm × 15 cm Magic C18 (5 μm, 300 Å) capillary column (Michrom BioResources, Auburn, CA). A linear gradient was used to elute peptides into the mass spectrometer at a flow rate of 1 μL min−1: 5% B to 65% B over 55 min (A: 95% H2O, 5% ACN, 0.1% formic acid, 0.001% TFA; B: 5% H2O, 85% ACN, 10% 2-propanol, 0.1% formic acid, 0.001% TFA). Columns were washed and reequilibrated between LC experiments. ESI was carried out at ~2.8 kV, with the ion source temperature at 80 °C and the cone voltage at 22 V. Mass spectra were acquired in the positive-ion mode over the range m/z 400–2000. For accurate mass MS and MS/MS, NanoLockSpray was used with a constant infusion of renin substrate hexadecapeptide or glu1-fibrinopeptide (Sigma) at 1 × 10−6 M in 50% ACN, 0.2% formic acid, at 0.2 μL min−1. Mass accuracy was ~10 ppm and resolution was ca. 1:10 000 (fwhm). Automated-MS/MS parameters were as follows: MS/MS performed on the three most abundant MS precursor ions having intensities >25 counts; MS/MS collision energies were set directly proportional to the mass and inversely proportional to the charge of the precursor (~16 to 40 V). The Ar collision gas pressure was ~2 × 10−5 Torr in the cell. MS/MS spectra were recorded as the summation of ≤4 × 1-second scans over the range m/z 100–2000. Data were processed with MassLynx 4.01 and ProteinLynx Global Server (PLGS) 2.1 (Micromass/Waters). Data from LC–MS/MS experiments were analyzed against user programmed Swiss-Prot (SP)/TREMBL databases. MS/MS data that did not return conclusive protein identifications were BLAST searched28 against the NCBI nonredundant protein and expressed sequence tag databases within PLGS 2.1/2.2 and further searched with the NCBI BLAST server (www.ncbi.nih.gov). All assignments based on MS/MS spectra were validated manually irrespective of the database search scores.
Chromatin Immunoprecipitation Analysis
293T cells were treated essentially the same as Rat1 fibroblasts, as previously described.11 Briefly, asynchronously growing, HA-Brd2 Aden-ovirus-infected 293T cells were exposed to formaldehyde to cross-link protein and DNA. Chromatin was prepared, sheared by sonication and divided into equal samples. Anti-HA mAb (Covance) or anti-His tag mAb (Upstate Biotechnology, Lake Placid, NY) as a negative control and Protein A/G PLUS agarose (Santa Cruz) were used to immunoprecipitate chromatin in the presence of salmon sperm DNA and BSA. Cross-links were reversed with heat, proteins were removed with proteinase K and organic extraction, and DNA of the human cyclin A promoter was amplified with forward primer 5′-ATGTTGG-GCAACTCTGCGCCGGGG-3′ and reverse primer 5′-GGGTC-CAGGTAAACTAATGGCTGAATTAAAAGCCAGGGC-3′, to yield an amplicon of 411 nucleotides that began at the translational start site. Products were separated in ethidium bromide-containing 1% agarose gels and visualized by ultraviolet light.
Histone Acetylation and Cyclin-Dependent Kinase Assays
Total histone-directed acetylation activity in the Brd2 immunoaffinity eluate was measured as previously described.11 Briefly, protein complexes in extracts were bound to HA mAb or nonimmune Ig Ab agarose columns, washed extensively with Buffer A as above, and incubated with 5 μg mixed calf thymus histones and 0.25 μCi [14C]acetyl-CoA (59 mCi/mmol; Amersham Pharmacia Biotech). Histones were then separated from the agarose by centrifugation, precipitated with TCA and separated on Triton X-100/urea/acetic acid gels. Incorporated radioactivity was visualized by autoradiography. Cyclin-dependent kinase (cdk) associated with cyclin A in immuno-affinity eluates was determined under co-immunoprecipitation conditions as previously reported11 with an assay kit (Upstate) and 10 μCi of [γ-32P]ATP (3000 Ci/mmol; New England Nuclear). Serum stimulation of HA-Brd2 Adenovirus-infected 293T cells or control infections defined the time course of cdk induction.
Results
We have previously reported immunoaffinity purification studies of the Brd2 multiprotein complex in different cell types.13 The use of advanced MS and proteomics methods to describe the Brd2 complex components is a logical extension of our previously published work, which has linked the function of this bromodomain complex to transcriptional control of the cyclin A locus and thereby to cell cycle control.1,9–11 First, we confirmed recombinant protein overexpression in a CMV-directed Adenovirus 5 infection system and found agreement with our previous studies.11 Adenoviral expression of recombinant Brd2 protein increased intracellular levels by about 7-fold, as quantitated by densitometry (Figure 2A). Immuno-affinity chromatography was used to isolate Brd2-associated proteins. We used Coomassie blue stain to estimate the number of proteins that were involved in this Brd2 complex and observed five strongly staining proteins in the eluate of (Figure 2B), along with several minor species (arrows). Immunoprecipitation of large protein complexes for MS analysis has been validated in several laboratories and in practice does not pull down significant amounts of noninteracting proteins.29 No proteins were visualized in the eluate of a control column comprised of nonimmune rabbit IgG, with extract chromatographed under the same conditions (Figure 2C).
Figure 2.

Brd2 expression and immunoaffinity chromatography. (A) αHA immunoblot of an Adenovirus 5-delivered, CMV-directed, expression vector for HA-Brd2 to 293T cells. (B) Coomassie stain of proteins eluted from immunoaffinity columns that were synthesized with anti-HA mAb or (C) nonimmune polyclonal IgG Ab. Extracts were applied to the columns as described and unbound proteins were collected; columns were then washed extensively (20 column volumes with binding buffer) and the bound proteins were eluted by pH drop. Abundant eluted species are noted (arrows). Note lack of detectable protein in control column eluate.
MS Analysis
MS/proteomics methods were used to identify proteins within the Brd2 complex. MALDI time-of-flight mass spectra indicated that the detected proteins had molecular masses corresponding to the apparent molecular weights derived in a separate experiment from gel migration distance after SDS-PAGE. These MALDI mass spectra also confirmed that the complex was relatively pure. MALDI MS of the tryptic digest of the complex indicated that there were several associated proteins (results not shown). We then used LC–MS/MS to accomplish the actual characterization of the proteins found within the complex by analyzing the complex in its entirety. The peptides were characterized by on-line LC with automated tandem mass spectrometry (MS/MS). Figure 3A–C illustrates the identification of one of the proteins obtained in one LC–MS/MS experiment. The peptides from all Brd2-associated proteins were separated on-line by LC and sequentially introduced into the mass spectrometer; the sum of signals from the peptides constituted the TIC (Figure 3A). The single ion chromatogram (SIC) of m/z 590.7, corresponding to the [M+2H]2+ ion of the peptide eluting at 25 min, is marked (Figure 3A) and the MS recorded at the signal apex is shown in Figure 3B. Automated collision-induced dissociation (CID) of this precursor peptide produced the MS/MS spectrum shown in Figure 3C. The MS and MS/MS data were then formatted for query against a protein database. Endogenous histone H4 was conclusively identified from the MS/MS data of this and seventeen other peptide signals observed in the LC–MS/MS experiment (Table 1). The amino acid sequence is consistent with this spectrum and accounts for all the major fragment ions. In control experiments, LC–MS/MS data obtained on eluates from preimmune Ab control columns gave only background noise and no conclusive identifications of proteins except trypsin (used for digestion) and immunoglobulins (used for affinity purification). This result is consistent with the expectation that endogenous histone H4 should associate with Brd2 through its bromodomains.15
Figure 3.
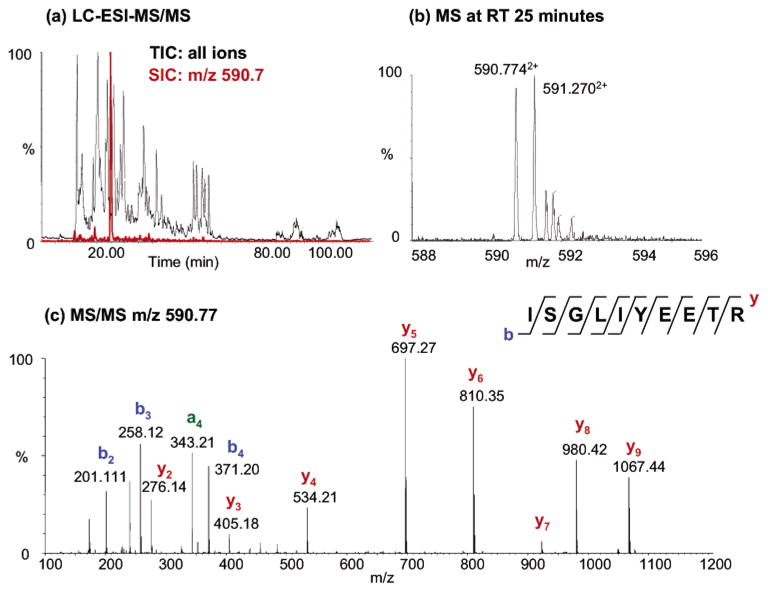
LC–MS/MS of tryptic peptides from the immunoaffinity purified protein complex: an example of the identification of histone H4. (a) Total ion chromatogram (TIC) for LC–ESI–MS/MS of tryptic peptides; shown here is the single ion chromatogram (SIC) for m/z 590.7. (b) MS spectrum of the peptide eluting at RT ≈ 25 min, [M+2H]2+ m/z 590.774. (c) LC–MS/MS spectrum of [M+2H]2+ m/z 590.77 corresponding to the peptide ISGLIYEETR assigned to histone H4. Assignments use Roepstorff and Fohlman nomenclature,49 as modified by Biemann.50
Table 1.
Proteins Identified in the Brd2 Complex
| protein | accession | MW | no. peptides matcheda by MS/MS |
|---|---|---|---|
| Chromatin-related | kDa | ||
| Histone H2B | P02278 | 13.8 | 12 |
| Histone H2A | 121992 | 15.1 | 2 |
| Histone H3 | 122064 | 11.3 | 3 |
| Histone H4 | P02304 | 11.2 | 18 |
| Snf2β; Brg-1 (brahma) | P51532 | 184.6 | 2 |
| Swi/Snf p155, Brg-1 associated (Baf155) | AAC50693 | 122.6 | 4 |
| Swi/Snf-related chromatin regulator p270 | O14497 | 205.9 | 2 |
| Histone deacetylase(HDAC) 11 | Q96DB2 | 39.2 | 4 |
| Chromatin assembly factor(CAF) 1, subunit B | Q13112 | 61.5 | 2 |
| Nucleosome assembly protein (NAP) 1-like 3 | Q99457 | 57.5 | 4 |
| Nucleosome assembly protein 1, centrosome- associated | NP_009117 | 281.1 | 2 |
| DNA-dependent protein kinase | P78527 | 468.8 | 4 |
| Transcription-associated factors and co-activators | |||
| TATA Box Binding Protein (TBP) | O29874 | 20.1 | 3 |
| TAFII170 (Snf2 family) | O14981 | 206.8 | 1 |
| TAFII250 | P21675 | 212.7 | 2 |
| TAFII55 | NP_005633 | 40.3 | 2 |
| RNA Polymerase β | Q889X7 | 154.6 | 3 |
| E2F-1 | JC4929 | 47.0 | 2 |
| DP-1 (E2F binding partner) | Q22703 | 67.9 | 2 |
| Zn finger protein 157 (Krüppel- type C2H2 Zn finger) | P51786 | 58.2 | 2 |
| CREB Binding Protein (CBP) | NP_004371 | 265.3 | 1 |
| p300 | NP_001420 | 264.2 | 1 |
| Homeobox protein Meis1 | O00470 | 45.8 | 3 |
| Med6 | CAG46498 | 28.4 | 2 |
| Cell cycle regulators | |||
| NimA-related mitotic regulator Nek9 | Q8TD19 | 107.1 | 3 |
| Proliferation antigen KI-67 | P46013 | 358.5 | 4 |
| Brd2 | NP_005095 | 88.1 | 3 |
| Cyclin A2 | P20248 | 48.5 | 2 |
| Controlsb | |||
| Ig κ chain | P01837 | 11.8 | 7 |
| Ig μ chain | 4959482 | 13.5 | 5 |
| Staphylococcus protein Ac | P02976 | 57.3 | 11 |
| Serum albuminc | P02768 | 69.3 | 7 |
Peptides were matched based on positive identifications within PLGS 2.1/2.2, and ladder scores >10 defined a cutoff for initial identification. Conclusive matches were confirmed manually for all spectra; please see text for details.
Positive controls that were expected and confirmed in all column eluates, including mock eluates of columns without protein extracts applied.
Nonreactive component of column purification matrix or common constituent of cell growth medium.
Additional MS Results
Each LC–MS/MS experiment typically produced 300+ MS/MS spectra that were subsequently queried against protein databases for identification (Table 1). Stringent manual confirmation of each identified protein was performed against the original MS/MS data. Ambiguous results were re-interrogated via BLAST homology queries to ensure accuracy of results and eliminate potential false positives. The majority of proteins were identified based on matching two or more MS/MS spectra. A very small number of proteins were assigned with only one positive MS/MS match. These were included if the protein was expected to be present or if an additional protein of the same class was also identified, i.e., TAF. We assigned several proteins expected to be in the complex, including Brd2, E2F-1, its binding partner DP-1 and MED6 (a component of Brd2-containing Mediator complexes).13,23,24 These results give further confidence to the identifications; Brd2 association with E2F proteins13 and acetylated histones15 has been verified by other investigators.24 We identified histones H2A, H2B, H3, and H4, as expected from the association between bromodomains and nucleosomes, although we could assign 12 peptides to histone H2B and 18 to histone H4, but only 2 to histone H2A and 3 to histone H3. The quantity of histones is reflected by the number of MS/MS signals and may explain the histone acetylation assay results shown below. In addition, we identified the chromatin and nucleosome assembly factors CAF-1 and NAP1, consistent with the posited role for bromodomain-containing proteins in binding nucleosomes and regulating chromatin. With respect to candidate HATs in the complex, we detected TAFII250, which acetylates H3 and H4; and CBP/p300, which acetylate H2A, H2B, H3, and H4.30 We also identified known proteins that potentially have important negative regulatory functions: HDAC 11 and TAFII55, a negative regulator of TAFII250 HAT activity and Swi/Snf proteins (Brg-1), which also have positive regulatory functions.1,21 The complex also contained elements of the basal transcription machinery, such as TBP, TAFII170 and RNA Polymerase (the latter of which has previously been reported24 to associate with Brd2), and participants in transcription activation and regulators of proliferation and the cell cycle that are also likely to play a role in development and differentiation.
Verification of MS Identifications
We confirmed the MS findings for the most important chromatin-related proteins of functional interest that are reported in the Table: these were detected with immunoblot of eluted complex that had been purified by immunoaffinity chromatography conducted under the same conditions. Immunoblot (Figure 4A) revealed that E2F proteins, which were barely detectable in column input, were highly enriched in the eluate of the specific αHA mAb column (αA eluate), but completely absent in the eluate of the nonimmune IgG control column (IgG agarose eluate), consistent with our earlier report.13 This result lent additional confidence to the level of stringency we employed in subsequent experiments with MS methods to identify comprehensively the other components of the complex. Likewise, we used the same methodology to detect Brg-1, Baf155, and CAF-1 in the eluates from Brd2-specific immunoaffinity columns, but not in eluates of nonspecific IgG control columns (Figure 4B).
Figure 4.

Brd2 nuclear complex contains E2F proteins and chromatin modifying factors. To verify the MS-based assignments shown in Table 1, the presence of several important proteins in the Brd2 complex was further investigated by immunoblot. (A) Pan-E2F Ab immunoblot of fractions from αHA-Brd2 immunoaffinity column (αHA), comprising 1/10 of input and wash fractions of columns, and all of eluted fraction. As a control, nonimmune rabbit IgG agarose immunoaffinity column chromatography was performed under the same conditions (IgG). Immunodetected E2F protein is indicated (arrow). (B) Additional immunoblots, carried out in the same manner as (A) detected chromatin modifying proteins Brg-1, Baf155, and CAF-1 in eluate of Brd2-specific immunoaffinity column but not in eluate of IgG control column.
Functional Analysis of Brd2 Complex
We have previously published studies in fibroblasts, in which we demonstrated that Brd2 is physically associated with the cyclin A promoter.11 We have also shown that complexes contain histone H4-directed HAT activity and that Adenovirus expression of Brd2 accelerates the cell cycle, whereas Adenovirus controls do not. Evidence from Tg mice, transfection or Adenovirus delivery to tissue culture models suggests that Brd2 transactivates the cyclin A locus, and that overexpression increases cyclin A transcription, destabilizing the cell cycle by a general, conserved mechanism. Biochemically, this forward pressure on the cell cycle is characterized by increased levels of cyclin A-associated cyclin-dependent kinase activity (principally cdc2 and cdk2). We confirmed that the Brd2 complex had similar functions in the 293T system. Chromatin immunoprecipitation of the cyclin A promoter confirmed that the Brd2 protein was physically present on cyclin A promoter chromatin (Figure 5A). The carboxyl terminal portion of Brd2 mediates Brd2 association with the multiprotein complex,13 establishing a potential anchor point to DNA through E2F DNA binding. Bromodomains, which associate with nucleosomal histones,5,6,15–17,19 and, in the case of Brd2 and Brd4, selectively with acetylated nucleosomes,15,16 could also provide this anchor point.11 The Brd2 complex contains HAT activities against all core nucleosomal histones (Figure 5B). The activity of cyclin/cdk complexes drives cell cycle progression and attenuation of cdk2 activity arrests cells.31 We have also previously reported that Brd2-dependent elevation of cyclin A protein levels accelerates cell cycle progression in fibroblasts,11 through increased cyclin A-associated cdk2 activity, a well-established and conserved mechanism for S phase progression, which recalls the mechanism of the S phase-specific DNA ‘replitase’ complex that Pardee and others defined in pioneering work with Chinese hamster embryo fibroblast cells.32 We determined cyclin A-associated histone H1 kinase activity here and confirmed that, in 293T cells, greater cyclin A/cdk activity was mobilized in cells infected with Brd2-expressing Adenovirus than in cells infected with control Adenovirus (Figure 5C), consistent with our earlier observations,11 and demonstrating conserved functions of the Brd2 multiprotein complex.
Figure 5.

Functional analysis of Brd2 complex. (A) Asynchronous 293T cells infected with HA-Brd2-expressing Adenovirus. Chromatin was immunoprecipitated with anti-HA primary mAb and secondary Ab coupled to agarose; the cyclin A promoter was amplified by PCR. Input chromatin was amplified and compared to HA or anti-(His)6 Ab control. PCR products and primers for the human cyclin A promoter are identified with arrows. (B) HAT activity associated with recombinant proteins was co-immuno-precipitated with anti-HA mAb. Left panel shows histone standards, separated by Triton X-100/urea/acetic acid PAGE,11,30 either individually or in an equimolar mixture (mix) and stained with Coomassie Blue. Right panel shows autoradiogram of incorporation of [14C]acetate from [14C]acetyl CoA into histone mix. All core nucleosomal histones were acetylated, whereas the linker histone H1 was not. (C) Cell extracts were assayed for cyclin A-associated cyclin-dependent kinase activity in the presence of [γ-32P]ATP, as determined by the histone H1 kinase activity in cyclin A co-immunoprecipitated complexes.11 Hours after serum stimulation of Adenovirus-infected cells are indicated; 100% of cells were infected.27 Adenoviruses either encoded Brd2 or an “empty vector” control. Standard deviation (n = 3) is shown.
Discussion
In this report, we confirm and extend our previous partial characterizations of the Brd2 multiprotein complex,8,11,13 with MS and immunoaffinity analysis of its components. We have reported that the Brd2 complex in fibroblasts contains E2F proteins and histone H4-directed HAT activity. These results supports a model of dual control of cyclin A transcription through (a) direct transcription factor recruitment and (b) histone acetylation/deacetylation.11,13,31,33 Furthermore, chromatin immunoprecipitation has established, in synchronized cells, that Brd2 is associated with the cyclin A promoter at both G1 and S phase of the cell cycle,11 suggesting that Brd2 functions as a scaffold that mediates access of transcriptional control proteins to chromatin.20 These observations imply that the Brd2 complex, at least when physically associated with cyclin A promoter chromatin, has dynamic, time-ordered functions of transcriptional repression, then activation, then repression again, as the cell progresses through the stages of the cell cycle. It is reasonable to hypothesize, therefore, that the complex also has dynamic composition, with shifting alliances between transcriptionally repressing and activating sub-complexes.1 In this report, we studied asynchronously cycling 293T cells as an established model system in order first to develop our methods and then to obtain a time-averaged, “global” picture of Brd2 complex composition.
Brd2 complexes are likely to provide a functional link between mitogenic signal transduction pathways and mechanisms of cell cycle progression, particularly cyclin A transcriptional control.11 In a Tg mouse model wherein Brd2 constitutive expression is B cell-restricted, Brd2 upregulates cyclin A transcription, destabilizes the cell cycle and leads to B cell malignancy.10 Furthermore, constitutive expression of BRD2 in the lymphoid compartment increases cyclin A transcription, “priming” pre-malignant, Tg B cells for over-proliferation. Thus, the identities of Brd2-associated proteins in the constitutively expressed state are likely to be informative of lymphoid carcinogenesis. In the case of Adenovirus delivery to 293T cells, we have controlled for Brd2 expression with “empty vector” Adenovirus constructs, which gives us confidence that Brd2 overexpression is sufficient to increase cyclin A transcription, although any interpretation of cell cycle “destabilization” in 293T cells must be made with caution because of the already highly transformed nature of these cells. Nevertheless, having worked out our MS methods in this system, we are poised to explore Brd2 complex composition and function in normal systems with endogenous Brd2 levels, such as activated human primary B cells; in nonmalignant Tg B cells and Tg B cell lymphomas;10 and in an important control case, murine brd2(−/−) null B cells, which we are currently generating with Cre-Lox technology. We expect these experiments will illuminate the role of Brd2 complexes in the control mechanisms of normal B cell mitogenesis and deregulated proliferation in malignancy.
The presence of components of the SWI/SNF nucleosome remodeling complex in association with Brd2 is consistent with the long-established mechanism whereby histone acetylation and nucleosome remodeling work together in transcriptional activation.1,2,30,33 The SWI/SNF complex is the best-characterized nucleosome remodeling complex2,4,21 and is composed of 11 subunits, two of which (Baf155 and Brg-1) we have so far identified. Brg-1 and the related protein brahma (Brm) are catalytic components of SWI/SNF complex. They do not bind to specific DNA sequences, but they form separate, related SWI/SNF complexes that are recruited to promoters and that are essential for growth and development.34 Apart from histones and histone modifying enzymes, SWI/SNF components are also known to interact with E2F proteins,35 a property that underscores the functional links between Brd2 and proliferation, as well as with the general transcription machinery, TAFs, Mediator and RNA Polymerase II.36,37 Definition of a Brd2-dependent structural assembly that requires ATP to associate offers in vitro support for the idea that these sub-complexes actually do physically associate in a single “supercomplex”, whereas earlier discussion has tended to use transitive logic: E2F proteins are associated with basal transcription machinery at promoter DNA and SWI/SNF is associated with promoter chromatin, therefore E2Fs must be associated with SWI/SNF and TAFs at promoter chromatin.
We summarize our findings of the aggregate Brd2 complex in the scheme presented in Figure 6. We have used anti-HA- Brd2 immunoaffinity chromatography to isolate components of the DNA/chromatin-directed subcomplexes shown. The major subcomplexes represented include: (a) the core promoter and TATA binding factor (TBP)-associated factors (TAFs) TBP, TAFII250, TAFII55, TAFII170, and RNA Polymerase II; (b) activated transcription factors E2F and DP-1; (c) Mediator/scaffold proteins Brd2 and MED6; (d) chromatin/histone modification enzymes HDAC11, CBP, and p300; and (e) SWI/SNF remodeling complex components Brg-1/Snf2β, p270, and Baf155. Interestingly, all of these complexes are represented, but none in its entirety. For example, several of the RNA Polymerase holoenzyme components and general transcription factors have not been detected. Also not detected were some of the TAFs, such as TAFII130 and TAFII150; many of the Mediator elements, such as MED1–5, 7–31, CDK8, or CycC; and some of the Swi/Snf proteins, such as Brm/Snf2α and Swi2/Snf2. Neither did we detect representative elements of other nucleosome remodeling machines such as RSC, ISWI, SAGA, or ADA. The presence of the basal transcription factor TAFII250 (and its inhibitor38 TAFII55) is significant because the double bromodomain structure of Brd2 is very closely related to TAFII250.7 Histone acetylation of cyclin A promoter chromatin, probably in part through HAT activity that is intrinsic or associated with TAFII250, is essential for cyclin A transcription and cell cycle progression.14 Indeed, TAFII250 was originally named CCG1, for Cell Cycle Gene 1.7 On the basis of these similarities, we have suggested11 that certain Brd2 functions may be redundant with TAFII250.
Figure 6.

Aggregate model of Brd2 complex. Schematic diagram showing summary of proteins associated with Brd2 after immunoaffinity purification, including general transcription factors (TBP), TAFs, and RNA Polymerase II; Mediator components, transcription factor E2F, histone modification machinery of HAT (CBP) and HDACs, and nucleosome remodeling machinery such as SWI/SNF complex. The “x” indicates that more proteins may be involved and that this scheme is not intended to be comprehensive. The model suggests an integrated mechanism for the transcriptional control of the mammalian cyclin A locus.
The presence in a supposed transcriptional activation complex of HDAC11, a protein with transcriptional repression activity, was initially a paradox, but upon consideration of the dual nature of Swi/Snf complexes,1 which include the repressor Brg-1, its detection here is not so surprising. Specifically, the transcriptional control of E2F-regulated cell cycle genes is essential to proper progression through each stage of the cell cycle. The cyclin A locus, for example, is transcriptionally repressed in G1 phase, then activated in S phase, then repressed again as the cell exits S phase, a mechanism that clearly requires coordinated regulation,11,31,33,35 as we have discussed elsewhere.1 The dual nature of chromatin remodeling was first observed in yeast, wherein Swi/Snf complexes, which were initially associated with transcriptional activation,39 were later linked to repression as well: swi/snf mutations transcriptionally activate more yeast genes than they repress.40
Swi/Snf complexes can perform different functions, depending on the promoter context, and sometimes work in collaboration with HAT/HDAC exchange.33 Ectopic expression of Brg-1 is known to repress E2F1 and cyclin A promoters through interaction with the retinoblastoma tumor suppressor protein (RB) to enhance RB-mediated transcription repression during G1 phase.31,41,42 These considerations led to a hypothesis that the Brd2 complex is present at the cyclin A promoter not only during S phase, when it acts as part of a transcriptional activating machine, but during G1 phase as well, when transcriptional repression is required. We have recently confirmed this hypothesis in fibroblasts.11 The complex analyzed above was purified from asynchronous, cycling cells, and is therefore a time-average of complex composition, with every stage of the cell cycle represented. Our results do not imply that all the elements illustrated (Figure 6) are simultaneously present in association with each Brd2 molecule. Cell cycle-specific changes in complex composition may alter the functions of the complex. We suspect that this mechanism underlies cyclin A transcriptional deregulation in Tg mice.10
We demonstrate herein the first biochemical purification of endogenous histones in association with a double bromo-domain protein. Previous reports employed nonphysiological methods, such as glutathione-S-transferase pull-downs6 or yeast two-hybrid analysis.17 The presence on mitotic chromosomes of the double bromodomain proteins Brd215 and Brd412,16 probably reflects a high association constant, exploitable in this kind of affinity purification. However, this constant may be too low in single bromodomain protein–histone interactions to permit biochemical isolation. In further stages of these investigations, we will use the methods employed here to map specific amino acid modifications, such as acetylation of histones in the complex, which may also influence complex recruitment.19,39 Comprehensive analysis of lysine acetylation requires MS methods because modifications are spread throughout each histone molecule, and are not exclusively localized to the amino terminal tails, for which commercial Ab reagents exist.43 Of particular interest is our identification of components of the mammalian Swi/Snf complex in association with Brd2. Previous MS analyses of the yeast Swi/Snf complex44,45 or Mediator23 identified a partially overlapping set of proteins; several chromatin remodeling complexes contain the DNA-dependent ATPases Snf2/Swi2p, Sth1p, Brg-1, Brm, and Iswi. We speculate that this Brd2-associated set of Swi/Snf components defines a specific Brd2-dependent chromatin remodeling complex. Intriguingly, MS and immunoblot studies of the human trithorax “supercomplex”46 also identify SWI/SNF components (Brm and Baf155), HDACs, TAFII250, and TBP in association with methylation of lysine 4 of histone H3 and acetylation of histone H4. Human trithorax, named ALL-1 for Acute Lymphocytic Leukemia or MLL for Mixed Lineage Leukemia, is also a bromodomain protein and a homologue of Drosophila trithorax. Its disruption by chromosomal translocation at 11q23 underlies many of these human leukemias. Because fsh genetically activates trithorax during fly development,47 we originally proposed8 that the relationship between Brd2 and ALL-1/MLL is conserved in humans. Thus, these complexes may serve important and specific functions in proliferation and cancer.
We will use isotopic labeling methods and two-dimensional SDS-PAGE with Ab immunoblot to define the relative stoichiometry of Brd2 complex components in further stage-specific experiments. Alternatively, the Brd2 complex may share subunits of nucleosome remodeling complexes but not the complexes in their entirety, inasmuch as many complexes share the histone-like TAFs. These subunits might be important to target Brd2 to sequence specific activators, after which Brd2 might act as a docking platform for other coactivators. We expect that chromatin immunoprecipitation of the MS-identified components in synchronized cells at different stages of the cell cycle or under different regimens of mitogenic stimulation will help resolve these questions for the cyclin A promoter. There is sufficient evidence to hypothesize that increased levels of the Brd2 scaffold are oncogenic in any normal cell because of increased cyclin A transcriptional activation. Finally, proteomics and bioinformatics approaches will allow us to define the characteristic molecular events of the Brd2-driven Tg lymphoma and to define potential biomarkers for diagnosis and therapeutic intervention, such have been validated recently for cancer-specific expression of endothelial cell proteomes.48 Taken as a whole, this research illuminates the regulation of transcription through the coordinated action of large complexes that recruit specific transcription factor proteins, nucleosome remodeling machines, and histone modification enzymes to promoter chromatin.
Acknowledgments
This project was supported by Grant No. RSG0507201 from the American Cancer Society (G.V.D.) and NIH Grant Nos. CA75107, CA102889 (G.V.D.); CA84193 (D.V.F.); P41-RR10888, S10-RR15942, and NHLBI Contract No. N01-HV-28178 (C.E.C.). We thank members of the Cancer Research Center at Boston University School of Medicine and G. Schnitzler of Tufts University School of Medicine for helpful criticism.
Abbreviations
- Ab
antibody
- ACN
acetonitrile
- CBP
CREB Binding Protein
- CID
collision-induced dissociation
- CMV
cytomegalovirus
- DTT
dithiothreitol
- ESI
electrospray ionization
- ESI–MS/MS
electrospray ionization mass spectrometry/tandem mass spectrometry
- HAT
histone acetylase
- HDAC
histone deacetylase
- HPLC
high performance liquid chromatography
- IgG
immunoglobulin G
- LC
liquid chromatography
- LC–MS/MS
liquid chromatography mass spectrometry/tandem mass spectrometry
- mAb
monoclonal antibody
- MALDI
matrix-assisted laser desorption/ionization
- MS
mass spectrometry
- PCR
polymerase chain reaction
- PTM
post-translational modification
- PVDF
polyvinyldifluoride
- Q-TOF
quadrupole orthogonal time-of-flight
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SIC
single ion chromatogram
- TAF
TATA Box-Associated Factor
- TCA
trichloroacetic acid
- TFA
trifluoroacetic acid
- Tg
transgenic, TIC, total ion chromatogram
References
- 1.Denis GV. Duality in bromodomain-containing protein complexes. Front Biosci. 2001;6:D849–852. doi: 10.2741/denis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn PJ, Peterson CL. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front Biosci. 2001;6:D1019–1023. doi: 10.2741/horn. [DOI] [PubMed] [Google Scholar]
- 3.Tackett AJ, Dilworth DJ, Davey MJ, O’Donnell M, Aitchison JD, Rout MP, Chait BT. Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol. 2005;169:35–47. doi: 10.1083/jcb.200502104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamkun JW, Deuring R, Scott MP, Kissenger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma – a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SWI2/SNF2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 5.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 6.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 7.Beck S, Hanson I, Kelly A, Pappin DJC, Trowsdale J. A homologue of the Drosophila female sterile homeotic (fsh) gene in the class II region of the human MHC. DNA Seq. 1992;2:203–210. doi: 10.3109/10425179209020804. [DOI] [PubMed] [Google Scholar]
- 8.Denis GV, Green MR. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 1996;10:261–271. doi: 10.1101/gad.10.3.261. [DOI] [PubMed] [Google Scholar]
- 9.Florence B, Faller DV. You betcha: a novel family of transcriptional regulators. Front Biosci. 2001;6:D1008–1018. doi: 10.2741/florence. [DOI] [PubMed] [Google Scholar]
- 10.Greenwald R, Tumang JR, Sinha A, Currier N, Cardiff RD, Rothstein TL, Faller DV, Denis GV. Eμ-BRD2 transgenic mice develop B cell lymphoma and leukemia. Blood. 2004;103:1475–1484. doi: 10.1182/blood-2003-06-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinha A, Faller DV, Denis GV. Bromodomain analysis of Brd2-dependent transcriptional activation ofcyclin A. Biochem J. 2005;387:257–269. doi: 10.1042/BJ20041793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Diff. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 14.Dunphy EL, Johnson T, Auerbach SS, Wang EH. Requirement for TAF(II)250 acetyltransferase activity in cell cycle progression. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanno T, Kano Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromo-domain proteins visualized in living cells. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 16.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Nat’l Acad Sci US A. 2003;100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pamblanco M, Poveda A, Sendra R, Rodriguez-Navarro S, Perez-Ortin JE, Tordera V. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 18.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 19.de la Cruz X, Lois S, Sanchez-Molina S, Martinez-Balbas MA. Do protein motifs read the histone code? Bioessays. 2005;27:164–175. doi: 10.1002/bies.20176. [DOI] [PubMed] [Google Scholar]
- 20.Denis GV. Bromodomain motifs and “scaffolding”? Front Biosci. 2001;6:D1065–1068. doi: 10.2741/a668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson CL, Tamkun JW. The SWI–SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 22.Guo N, Faller DV, Denis GV. Activation-induced nuclear translocation of RING3. J Cell Sci. 2000;113:3085–3091. doi: 10.1242/jcs.113.17.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang YW, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway JW, Conaway RC, Kornberg RD. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Nat’l Acad Sci US A. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowley TE, Kaine EM, Yoshida M, Nandi A, Wolgemuth DJ. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol Endocrinol. 2002;16:1727–1737. doi: 10.1210/me.2001-0353. [DOI] [PubMed] [Google Scholar]
- 25.Freitas MA, Sklenar AR, Parthun MR. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J Cell Biochem. 2004;92:691–700. doi: 10.1002/jcb.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 27.Becker TC, Noel RJ, Coats WS, Gomez-Foix AM, Alam T, Gerard RD, Newgard CB. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43(Pt A):161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI–BLAST: a new generation of protein database search programs. Nucl Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen LRH, Strupat K, Hillenkamp F. Analysis of quaternary protein ensembles by matrix assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 1997;8:1046–1052. [Google Scholar]
- 30.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Dürr P, Henglein B. Cell cycle regulation of the cyclin A gene is mediated by a variant E2F site. Proc Nat’l Acad Sci US A. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prem veer Reddy G, Pardee AB. Multienzyme complex for metabolic channeling in mammalian DNA replication. Proc Nat’l Acad Sci US A. 1980;77:3312–3316. doi: 10.1073/pnas.77.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siddiqui H, Solomon DA, Gunawardena RW, Wang Y, Knudsen ES. Histone deacetylation of RB-responsive promoters: requisite for specific gene repression but dispensable for cell cycle inhibition. Mol Cell Biol. 2003;23:7719–7731. doi: 10.1128/MCB.23.21.7719-7731.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barlev NA, Emelyanov AV, Castagnino P, Zegerman P, Bannister AJ, Sepulveda MA, Robert F, Tora L, Kouzarides T, Birshtein BK, Berger SL. A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol Cell Biol. 2003;23:6944–6957. doi: 10.1128/MCB.23.19.6944-6957.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Nat’l Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase II holoenzyme contains SWI/SNF transcriptional activators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 37.Sharma VM, Li B, Reese JC. SWI/SNF-dependent chromatin remodeling of RNR3 requires TAF(II)s and the general transcription machinery. Genes Dev. 2003;17:502–515. doi: 10.1101/gad.1039503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gegonne A, Weissman JD, Singer DS. TAFII55 binding to TAFII250 inhibits its acetyltransferase activity. Proc Nat’l Acad Sci US A. 2001;98:12432–12437. doi: 10.1073/pnas.211444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns LG, Peterson CL. The yeast SWI–SNF complex facilitates binding of a transcriptional activator to nucleosomal sites in vivo. Mol Cell Biol. 1997;17:4811–4819. doi: 10.1128/mcb.17.8.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 41.Stiegler P, De Luca A, Bagella L, Giordano A. The COOH-terminal region of pRb2/p130 binds to histone deacetylase 1 (HDAC1), enhancing transcriptional repression of the E2F-dependent cyclin A promoter. Cancer Res. 1998;58:5049–5052. [PubMed] [Google Scholar]
- 42.Knudsen KE, Fribourg AF, Strobeck MW, Blanchard JM, Knudsen ES. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J Biol Chem. 1999;274:27632–27641. doi: 10.1074/jbc.274.39.27632. [DOI] [PubMed] [Google Scholar]
- 43.Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, Boeke JD. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 45.Peterson CL, Zhao Y, Chait BT. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 47.Mozer BA, Dawid IB. Cloning and molecular characterization of the trithorax locus of Drosophila melanogaster. Proc Nat’l Acad Sci US A. 1989;86:3738–3742. doi: 10.1073/pnas.86.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oh P, Li Y, Durr E, Krasinska KM, Carber LA, Testa JE, Schnitzer JE. Subtractive proteomics mapping of the endothelial surface in lung and solid tumours for tissue-specific therapy. Nature. 2004;429:629–635. doi: 10.1038/nature02580. [DOI] [PubMed] [Google Scholar]
- 49.Roepstorff P, Fohlman J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 50.Biemann K. Mass spectrometry of peptides and proteins. Annu Rev Biochem. 1992;61:977–1010. doi: 10.1146/annurev.bi.61.070192.004553. [DOI] [PubMed] [Google Scholar]


