Abstract
Unresectable hepatic colorectal cancer (CRC) metastases are a leading cause of cancer mortality. These tumors, and other epithelial tumors, often express both cyclooxygenase 2 (COX-2) and carcinoembryonic antigen (CEA). Because adenovirus vectors infect liver and lack tumor tropism, they cannot be utilized for systemic therapy of hepatic metastases. We used COX-2 transcriptional restriction, in combination with transductional adenovirus hepatic untargeting and tumor retargeting by a bispecific adapter, sCARhMFE, composed of sCAR (the Coxsackie and Adenovirus Receptor ectodomain) and MFE-23 (a single-chain anti-CEA antibody), to untarget liver following intravenous administration of adenovirus vectors expressing firefly luciferase and to retarget virus to hepatic colorectal tumor xenografts and non-small cell lung tumor xenografts. To improve both liver untargeting and tumor retargeting, we developed sCARfMFE, a trimerized sCARhMFE adapter. Trimerization greatly improves both untargeting of CAR-dependent adenovirus infection and CEA-dependent virus retargeting, in culture and in vivo.
Combining sCARfMFE bispecific adapter transductional liver untargeting and transductional tumor retargeting with COX-2 transcriptional tumor-restricted transgene expression increases systemically-administered adenovirus therapeutic efficacy for hepatic CRC tumors, using Herpes Virus Type 1 thymidine kinase as a therapeutic gene in conjunction with the prodrug ganciclovir. Both transductional untargeting and COX-2 transcriptional restriction also reduce HSV1-tk/GCV hepatic toxicity. In addition, transductional sCARfMFE untargeting reduces the innate immune response to systemic adenovirus administration. Combined transductional liver Ad untargeting, transductional tumor retargeting and transcriptional transgene restriction suggests a means to engineer practical, effective therapeutic agents for hepatic CRC metastases in particular, as well as hepatic metastases of other epithelial cancers.
Keywords: adenovirus, colorectal cancer, COX-2 transcriptional restriction, transductional retargeting
Introduction
Liver is the most common site for colorectal cancer (CRC) metastasis, and a common site of metastasis for lung, breast and other epithelial cancers (1, 2). Unfortunately, the 5-year survival rate for patients with CRC liver metastases is less than 5% (3–5) and current treatments for CRC liver metastases, including surgery, radiation and chemotherapy, are disappointing (6). As a result, it is of paramount importance to develop new therapeutic approaches for hepatic CRC metastases. Gene therapy presents an approach distinct from conventional cancer therapies. Adenovirus serotype 5 (Ad) vectors are currently the most widely used gene delivery vehicle. However, systemically administered Ad preferentially infects the liver (7–9). Consequently, systemically delivered Ad vectors expressing toxic or suicide genes can result in severe hepatic dysfunction and mortality (10). Methods to reduce hepatic infection and to restrict transgene expression to tumors following systemic Ad vector administration are major goals of Ad gene therapy.
Ad tumor targeting can be achieved by “transductional untargeting” of normal cells and tumor cell “retargeting” with bispecific adapter molecules (11). Ad binds to most cells via interaction between the virus fiber “knob” and cellular Coxsackie/Adenovirus Receptors (CAR) (12). However, recent data suggest that Ad liver infection is CAR independent, and is mediated by a hexon/Factor × complex (13). Nevertheless, bispecific adapters that bind to the viral knob dramatically reduce Ad liver infection following intravenous administration (14, 15). Alternatively, “transcriptional” restriction by “tumor-restricted” promoters also enhances Ad vector transgene expression in tumor cells and reduces hepatic Ad transgene expression (16–20).
Many epithelial tumors, including almost all CRC, 70% of non-small cell lung cancers (NSCLC) and 50% of breast cancers are reported to express carcinoembryonic antigen (CEA) (21). In addition, 70% of primary CRC tumors and all hepatic CRC metastases are reported to express elevated cyclooxygenase-2 (COX-2) (22, 23). Here we demonstrate that (i) sCARfMFE (Fig. 3A), a trimeric bispecific adapter in which the CAR ectodomain (sCAR) is linked to a single-chain anti-CEA antibody (MFE-23), can ”untarget” liver infection and enhance CEA+ CRC and non-small cell lung cancer infection; (ii) the COX-2 promoter can restrict Ad vector transgene expression to hepatic COX-2+ tumor xenografts and prevent transgene expression in liver following intravenous injection; and (iii) combining sCARfMFE liver untargeting, sCARfMFE tumor retargeting and COX-2 promoter-restricted expression substantially enhances tumor-specific transgene gene expression and therapy in COX-2+/CEA+ hepatic tumors following systemic administration. Moreover, both hepatic transductional untargeting and COX-2 transcriptional restriction attenuate therapeutic gene/prodrug liver toxicity following systemic Ad administration.
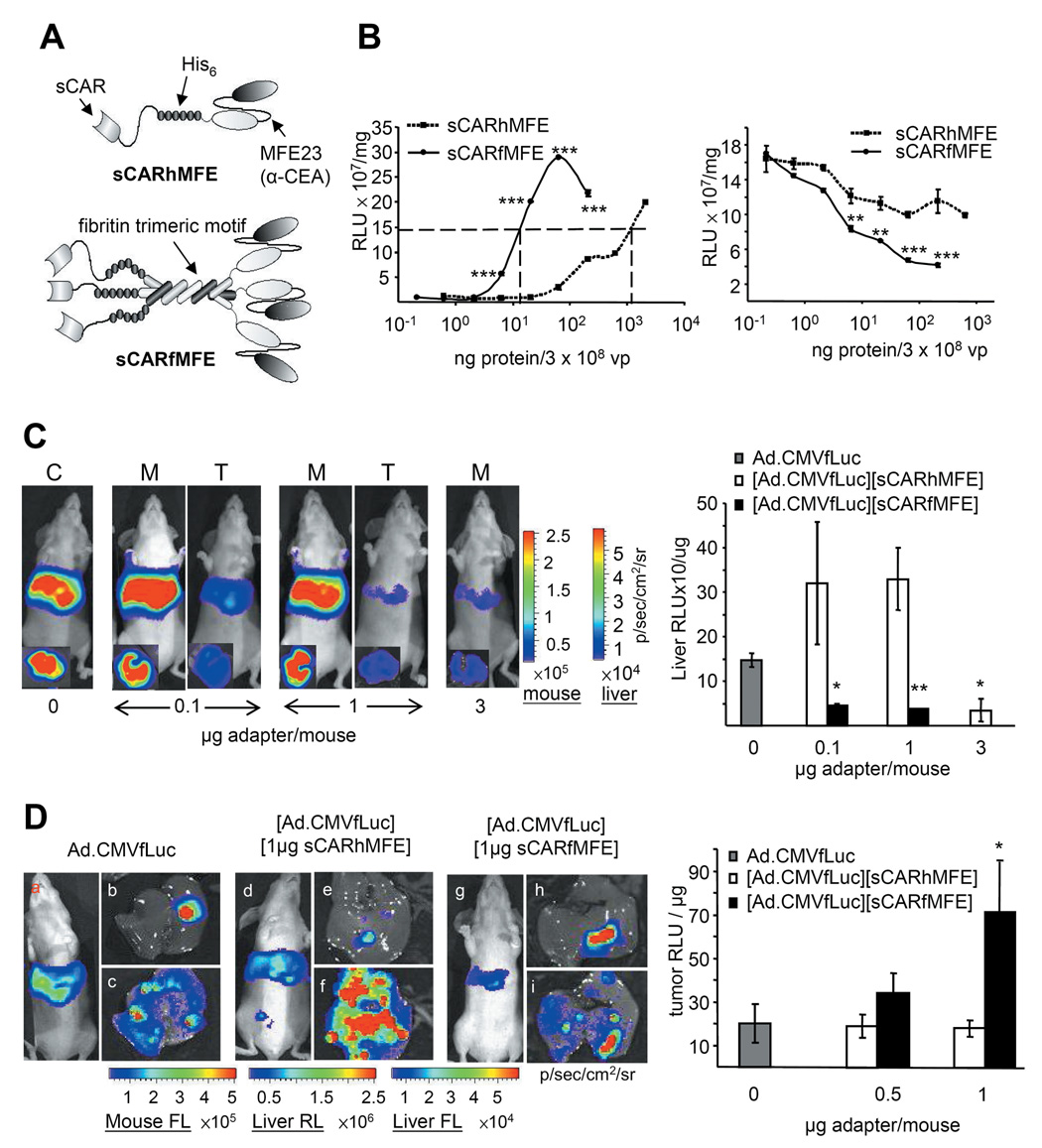
Figure 3. Adenovirus untargeting and retargeting by monomer sCARhFME and trimer sCARfMFE.
(A) sCARhMFE includes the CAR ectodomain (sCAR), a six-histidine tag (His6) and single-chain antibody MFE23. sCARfMFE also includes the fibritin trimerization motif. (B) CEA targeting and CAR untargeting by sCARhMFE and sCARfMFE. To analyze CEA retargeting, LS174T(CEA+) cells were infected with Ad.CMVfLuc (3 × 108 vp/well) pre-incubated with increasing sCARhMFE or sCARfMFE concentrations (left graph). To analyze CAR untargeting by sCARhMFE and sCARfMFE. 293 cells were infected with Ad.CMVfLuc (3 × 108 vp/well) preincubated with increasing sCARhMFE or sCARfMFE. Data are averages ± SEM (n = 3; **p < 0.01, ***p < 0.001 compared to [Ad.CMVfLuc][sCARhMFE]-infected cells). (C) sCARfMFE and sCARhMFE untargeting of Ad liver infection. Ad.CMVfLuc (C, 4.5 × 1010 vp/mouse) (C), [Ad.CMVfLuc][sCARhMFE] (M), and [Ad.CMVfLuc][sCARfMFE] (T) were injected intravenously. Living mice and isolated livers (inset) were imaged for fLuc expression three days after virus injection. After imaging, luciferase activity of liver extracts was quantified. Data are averages ± SEM (n = 3; *p < 0.05, **p < 0.01) compared to Ad.CMVfLuc-injected mice. (D) sCARfMFE and sCARhMFE adenovirus retargeting to CEA+ tumor xenografts. Mice bearing LS174T(rLuc) hepatic tumors were injected intravenously with [Ad.CMVfLuc (3 × 1010 vp/mouse)], [Ad.CMVfLuc (3 × 1010 vp/mouse)][sCARhMFE] (0.5 or 1.0 µg/mouse), or [Ad.CMVfLuc][sCARfMFE] (0.5 or 1.0 ug/mouse). Following fLuc (FL) imaging of the living animals (a,d,g), livers were imaged for tumor-derived rLuc (b,e,h) and adenovirus-directed fLuc (c,f,i). After imaging, fLuc tumor extract activity was quantified. Data are averages ± SEM (n = 3; *p < 0.05) compared to Ad.CMVfLuc-injected mice.
We demonstrate, for the first time, substantial therapeutic index gains achievable by improving and combining strategies for liver untargeting, tumor retargeting and tumor-specific transgene expression. Because ligands for untargeting, ligands for retargeting, promoters for restricted gene expression and cargo genes can be varied independently, these data illustrate a generalized application of gene therapy methods to a variety of disease targets.
Materials and Methods
Cell culture, transfection, immunoblotting, COX-2 promoter activity analysis in cell culture, CEA competition of virus infection and Ki-67 immunohistochemistry are described in Materials and Methods, Supplementary Material.
Virus production
Ad.CMVfLuc, carrying the firefly luciferase (fLuc) transgene under control of the cytomegalovirus (CMV) promoter, and Ad.cox2fLuc, expressing fLuc from the human hCOX-2 promoter, were constructed as described (20), and prepared according to Li et al. (15). Ad.cox2NTP, created for these studies, utilizes the hCOX-2 promoter to drive the NTP transgene encoding fLuc, thymidine kinase and green fluorescent protein, The NTP gene construct was provided by Dr. Owen Witte (University of California at Los Angeles). Virus strains were prepared in 293 cells by double cesium chloride (CsCl) gradient centrifugation (15). Viral particle number was determined by measuring 260 nm (24) absorbance. Viral titers were determined with the Adeno-X Rapid Titer Kit (BD Clontech, Mountain View, CA) (15).
Monomer sCARhMFE and trimer sCARfMFE construction and purification
sCARhMFE was prepared as described previously (25). To construct the plasmid encoding sCARfMFE, we replaced the CD40L cDNA of the pcDNA/CAR/F/m40L plasmid (26) with the cDNA for anti-CEA scFv MFE-23. MFE-23 was amplified from a plasmid obtained from Kerry Chester (University College London). NotI and XhoI restriction sites were added at the 5’ and 3’ ends, along with a TAA stop codon. The pcDNA/CAR/F/m40L plasmid and the MFE-23 PCR product were digested with NotI and XhoI and gel purified. The MFE fragment was ligated into the vector to create cDNA/CAR/F/MFE. 293 cells were transfected with pcDNA/CAR/F/MFE plasmid linearized with PvuI restriction enzyme, using Superfect reagent (Qiagen, Valencia, CA). The medium from sCARfMFE-expressing 293 cells was collected and proteins were precipitated using ammonium sulfate. Precipitate was collected by centrifugation, redissolved in PBS and dialyzed against PBS. Recombinant protein was purified using Ni-NTA Superflow resin (Qiagen, Valencia, CA), followed by dialysis against PBS.
Comparing retargeting and untargeting efficacy of sCARfMFE and sCARhMFE in cell culture
Ad.CMVfLuc (3 × 108 vp) was incubated with sCARfMFE or sCARhMFE, in concentrations described for specific experiments, in 10 µl for 60 minutes at room temperature. sCAR-MFE:adenovirus complexes were diluted to 200 µl or 1000 µl with medium containing 2% FBS, then added to LS174T or 293 cells in 24-well plates (2 × 105 cells/well). Virus-treated cells were incubated at 37 °C for 90 minutes. The virus-containing medium was then aspirated and cells were washed with PBS. After a 40-hour incubation at 37 °C in medium containing 10% FBS, cells were lysed and luciferase activity was measured.
Comparison of hepatic untargeting efficacy for sCARfMFE and sCARhMFE
Viruses were incubated for one hr either with sCARfMFE, sCARhMFE or PBS in a volume of 250 µl. 4.5 × 1010 vp/mouse of Ad.CMVfLuc, [Ad.CMVfLuc][sCARfMFE] or [Ad.CMV.fluc][sCARhMFE] were administered (250 µl) by tail-vein injection into eight week-old (nu/nu) nude mice (Charles River Laboratories, Wilmington, MA). On the third day after virus administration, mice were injected intraperitoneally with D-luciferin (250 µl; ~125 mg/kg body weight) and scanned to image Ad-directed fLuc activity. Immediately after imaging, mice were sacrificed and the livers were removed and imaged for fLuc (Ad-dependent) bioluminescence. After optical imaging, liver extracts were prepared and assayed for fLuc activity.
Preparation of liver xenografts
Eight week-old (nu/nu) nude mice were anesthetized with ketamine/xylazine (100/10 mg/kg). A transverse incision was made across the xyphoid process and extended approximately two cm. LS174TrLuc or H2122 cells (1 × 106/mouse) in 15 µl were injected into the front of the left upper liver lobe, using a 27-gauge needle. The lobe was returned to the abdomen and the incision was closed with sutures and wound clips. Buprenorphine was administered every 12 h for 48 h. Wound clips were removed after seven days. Experiments were performed 10–15 days after surgery.
Systemic Ad administration and measurement of Renilla and firefly luciferase activities
For transcriptional targeting, Ad.CMVfLuc or Ad.cox2fLuc (1 × 109 ifu/mouse) in 100 µl were administered by tail-vein injection. For transductional untargeting/retargeting, 5 × 108 ifu/mouse of AdCMVfLuc, Ad.Cox2fLuc, [Ad.CMVfLuc][sCAR-MFE], or [Ad.cox2fLuc][sCAR-MFE] were administered by tail-vein injection. Before injection, viruses were incubated for 1 h either with 10 µg sCAR-MFE/mouse or with PBS. Injection volumes were 100 µl.
To image tumor burden, coelenterazine (100 µl; ~0.7 mg/kg) was injected via tail-vein and mice were scanned for three min using the Xenogen IVIS Optical Imaging System to monitor tumor-directed rLuc activity. To monitor virus transgene expression, mice were injected intraperitoneally with D-luciferin (250 µl; ~125 mg/kg) and scanned at times indicated to image Ad-directed fLuc activity. After sacrifice, livers were removed and imaged for fLuc (Ad-dependent) bioluminescence. The livers were then immersed in a solution containing coelenterazine (0.2 mg/ml) and imaged again, using one-minute scans to monitor tumor-derived rLuc activity. After optical imaging, tumor and liver extracts were prepared and firefly luciferase activity of the extracts was measured with the Luciferase assay system (Promega, Madison, WI). Protein concentrations were determined with the Bio-Rad Protein Assay (Bio-Rad, Hercules, CA).
Bioluminescent images were analyzed using Living Image software version 2.20 (Xenogen). Regions of interest (ROI) were drawn over tumor and liver areas and total photons were calculated. The ROI sizes in all images for a single experiment were maintained as a consistent area (15).
HSV-tk/GCV Therapy
Mice were injected intravenously with Ad.CMVtk, Ad.cox2NTP, [Ad.CMVtk][sCARfMFE] or [Ad.cox2NTP][sCARfMFE]) at concentrations and times indicated in the experiments. For transductional targeting, Ad.CMVtk or Ad.cox2NTP were pre-incubated with sCARfMFE, at concentrations described for each experiment, in 20 µl for 1 hour prior in injection. Mice were injected intraperitoneally (i.p.) with 100 mg/kg GCV twice daily, according to schedules described in the individual experiments. Immunocompetent C57BL/6 mice were from Charles River Laboratories (Wilmington, MA).
sGOT, sGPT and total bilirubin assays
Mice were anesthetized with ketamine/xylazine (100/10 mg/kg). 1 cm of the tail was removed with a razor and blood was collected in a capillary tube. The sera were subsequently analyzed for sGOT (aspartate aminotransferase; Catachem, Bridgeport, CT), sGPT (alanine aminotransferase; Catachem, Bridgeport, CT) and bilirubin assays (Catachem, Bridgeport, CT). Assays were performed according to manufacturer’s protocols.
TUNEL staining
Livers were immersed in 10% formalin at room temperature overnight, then transferred into 50% ethanol to prepare paraffin sections. Apoptosis was evaluated by TUNEL staining (DeadEnd Colorimetric TUNEL System; Promega, Madison, WI). The assay was performed according to manufacturer’s protocols.
Quantitative PCR
Mouse livers, with or without tumors, were homogenized and genomic DNA was purified, using the DNeasy Tissue kit (Qiagen, Valencia, CA). Human GAPDH DNA, mouse GAPDH DNA and viral genomic DNA were measured by Quantitative PCR. The human-specific GAPDH primer pair is 5’ primer: ACG CTT TCT TTC CTT TCG CGC TCT GCG GGG / 3’ primer: CTA ACG GCT GCC CAT TCA TTT CCT TCC CGG, which can amplify a DNA region in a human GAPDH intron. The mouse-specific GAPDH primer pair is 5’ primer: CTG CGG AAA TGG TGT GATCTTCCC CAA GGG/ 3’ primer: AGG GAG CTC CAT TCA TGT GCT AAA CAG GCC, which can amplify a DNA region in a murine GAPDH intron. The virus-specific primer pair is 5’ primer: TAC CCG AGC CGA TGA CTT AC/ 3’ primer: CAC AGC ATG AAG GCG ATG, which can amplify a DNA section in the HSV1-tk coding region of the viral genome. A two-step amplification reaction (Cycle 1: [95°C for 3 min] × 1; Cycle 2: [95°C for 30 seconds/ 65°C for 45 seconds] × 40) was performed, using an Applied Biosystems 7500 Real-Time PCR System. The human-to-mouse or virus-to-mouse DNA ratios were expressed by Comparative Ct Method (A to B DNA ratio = 2−(Ct of A – Ct of B)) (27). The standard curve to determine tumor burden was determined by Q-PCR of DNAs extracted from mixtures of mouse liver homogenate and LS174T(rLuc) cells. Tumor cells per liver were determined by comparison of PCR data from extracts of tumor-bearing livers with this standard curve.
Cytokine induction in immunocompetent mice
C57BL/6 mice were injected i.v. with 1 × 1010 ifu/mouse Ad.CMVtk or [Ad.CMVtk][sCARfMFE (4 µg/mouse)]. Blood samples were collected prior to virus injection and at 6 and 24 hours after virus injection. Sera were isolated as described above, and the cytokine panel was analyzed with Q-Plex mouse cytokine array assays (Quansys Biosciences, Logan, Utah), according to manufacturer’s protocols.
PET/CT scan
PET/CT scans were performed with a microPET FOCUS 220 PET scanner (Siemens Preclinical Solutions, Knoxville, TN) and a MicroCAT II CT scanner (Siemens Preclinical Solutions, Malvern, PA). The mice were injected i.v. with liver contrast agent, Fenestra LC (300 µl/25 mg of body weight) 3h prior to the CT scans and were injected i.p. with ~200 µCi/mouse of 18F-FDG 1h prior to the microPET scans. The mice were anesthetized with 2% isoflurane and kept at 34°C during probe uptake. Mice were scanned in a chamber that minimizes positioning errors (28). MicroPET imaging was started 60 min after 18F-FDG injection. Image acquisition time was 10 min for PET scans, followed by 7 min CT scans, using routine image acquisition variables (28). Images were reconstructed using a 3-dimensional filtered back-projection reconstruction algorithm (29) and using a maximum a posteriori reconstruction protocol (30) for presentation of images. The images were analyzed and displayed with AMIDE software (31).
Statistical analysis
All experiments were performed at least in triplicate. Data are presented as means +/− SEM. The statistical significance (p values) in mean values of two-sample comparison (Fig. 1, 2, 3C, 3D, 4, 5B lower graph, 5C and 6) was determined with Student's t-test. The statistical significance in the comparison of multiple samples was examined with Bonferroni's post-hoc test (Fig. 3B, and 5B upper graph) after two-way ANOVA.
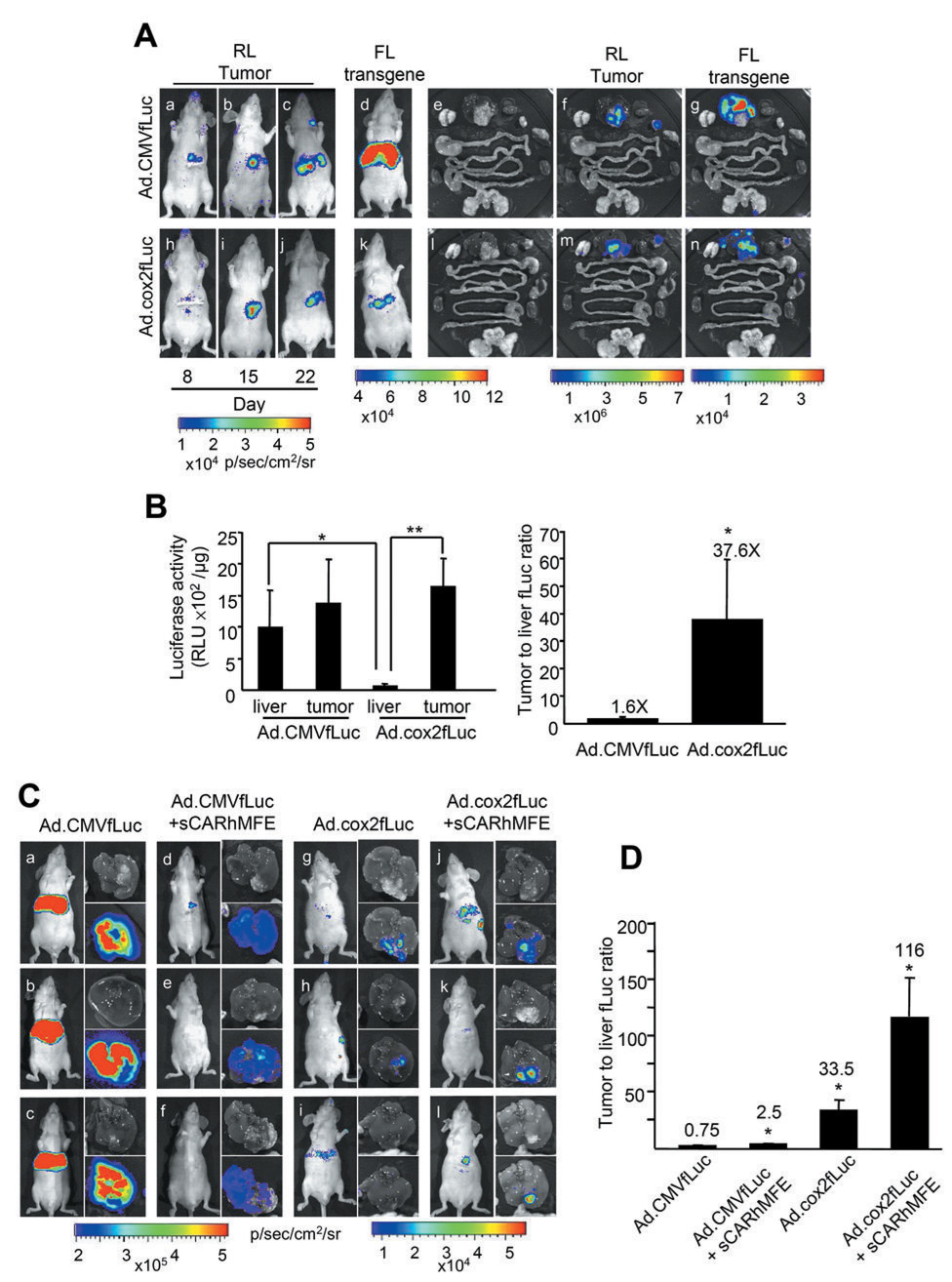
Figure 1. Combined COX-2 promoter transcriptional restriction and sCARhMFE transductional liver untargeting/tumor retargeting of Adenovirus-directed transgene expression in hepatic COX2+CEA+ CRC xenografts.
(A) LS174T(rLuc) tumor burden was monitored by rLuc bioluminescence (RL) in living animals (a–c and h–j) at 8, 15 and 22 days after tumor implantation. Ad.CMVfLuc or Ad.cox2fLuc (1×109 ifu/mouse) were injected intravenously into LS174T(Luc)-bearing mice on day 18 after surgery. Ad-directed transgene expression was monitored in living animals by fLuc bioluminescence (FL) (d and k) on day 22. Following FL imaging, mice were sacrificed and tissues were removed and imaged for tumor-derived rLuc activity (f and m) and for Ad-directed fLuc transgene expression (g and n). Photographs of tissues; e and l. (B) Quantitation of Ad-directed fLuc transgene expression in liver and tumor. Extracts were prepared from dissected tumors and from tumor-free liver regions. fLuc activity, reflecting virally directed gene expression, and tumor:liver fLuc activity ratios were quantitated with conventional luciferase and protein assays. Data are averages ± SEM (n = 3; *p < 0.05, **p < 0.01). (C) Ad.CMVfLuc (5 × 108 ifu/mouse, a–c), [Ad.CMVfLuc][sCARhMFE (10 µ;g/mouse)] (d–f), Ad.cox2fLuc (g–i) and [Ad.cox2fLuc][sCARhMFE] (j–l) were injected intravenously into LS174T(rLuc)-bearing mice. fLuc-derived bioluminescence was monitored. Photographs of the livers show the locations of tumors; bioluminescent overlays identify liver and tumor tissues where viral fLuc transgene expression occurs. (D) Quantitation of Ad-directed fLuc transgene expression from livers shown in panel C. Extracts were prepared from isolated tumors and tumor-free liver regions. Tumor:liver fLuc activity ratios, quantitated with conventional luciferase and protein assays, are shown. Data are averages ± SEM (n = 3; *p < 0.05, compared to mice receiving Ad.CMVfLuc).
Figure 2. Combined sCAR-MFE transductional hepatic untargeting, sCAR-MFE transductional tumor retargeting and COX-2 promoter transcriptional restriction of Ad infection in hepatic COX-2+CEA+ NSCLC tumor xenografts.
(A) Ad.CMVfLuc (5×108 ifu/mouse), Ad.cox2fLuc and [Ad.cox2fLuc][sCAR-MFE (10 µg/mouse)] were injected intravenously into nude mice bearing H2122 NSCLC liver xenografts. fLuc-derived bioluminescence was monitored in living mice and from isolated livers five days after virus administration. Photographs of the livers show the locations of tumors; bioluminescent overlays identify liver and tumor tissues where viral fLuc transgene expression occurs. (B) Quantitation of Ad-directed fLuc transgene expression. After bioluminescent imaging, extracts were prepared from the isolated tumors and from tumor-free liver regions. Tumor:liver fLuc activity ratios, quantitated with conventional luciferase and protein assays, are shown. Data are averages ± SEM (n = 3; *p < 0.05, **p < 0.01 compared to mice injected with Ad.CMVfLuc only).
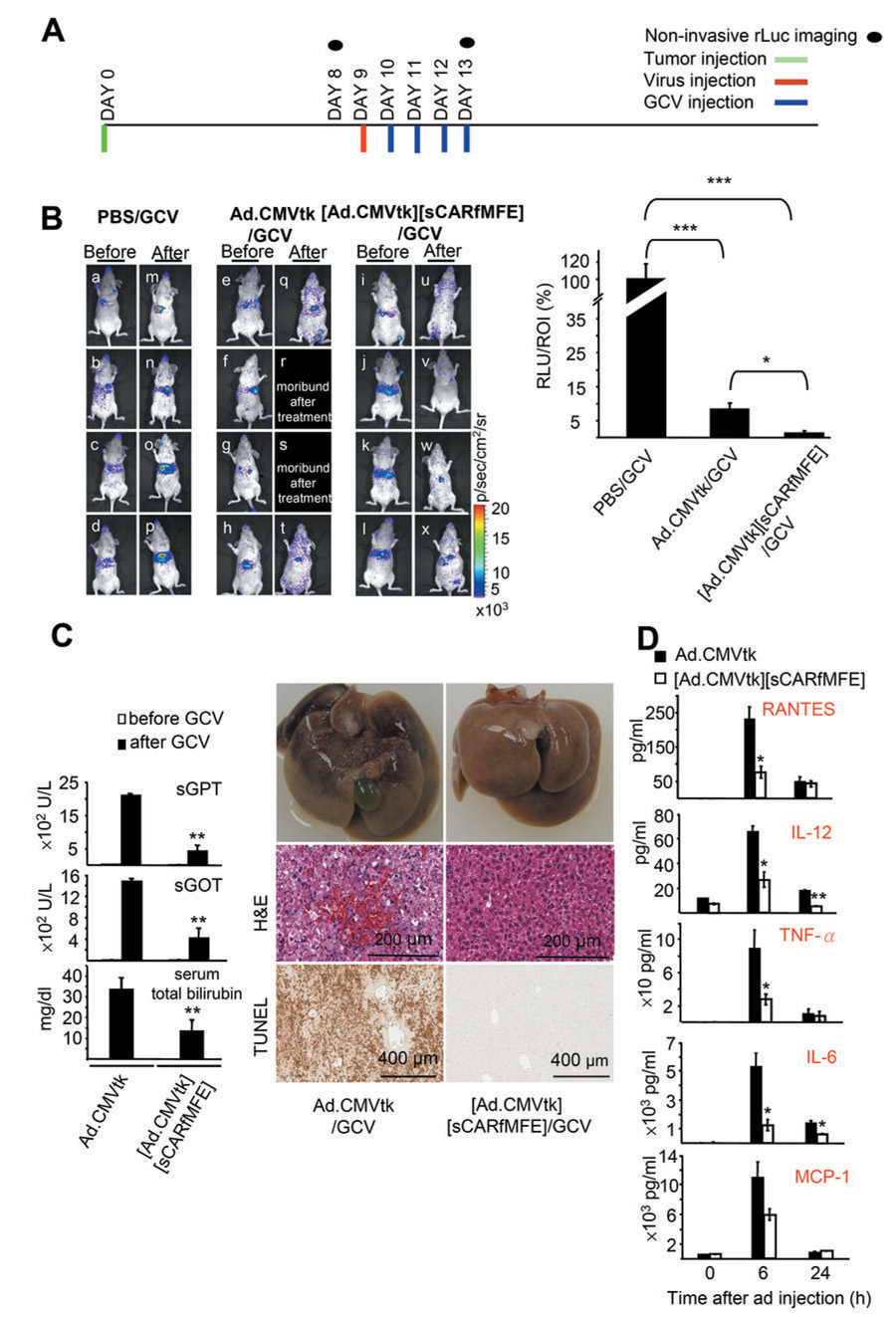
Figure 4. sCARfMFE-targeted Ad.CMVtk/GCV therapy of CEA+ hepatic CRC tumors.
(A) Experimental time line. Days of tumor (1 × 106 cells/mouse), Ad.CMVtk (2 × 109 ifu/mouse), [Ad.CMV.tk][sCARfMFE], and GCV (100 mg/kg i.p. twice/day) injection, and bioluminescent imaging are indicated. (B) LS174T(rLuc) tumor burden was monitored by rLuc bioluminescence before (Day 8, a–l) and after (Day 13, m–x) therapy. For quantitation of tumor-derived Renilla luciferase activity from the livers, in vitro luciferase activities of whole liver extracts measured by conventional luciferase assay were normalized by the average liver ROI values acquired before therapy for all twelve untreated mice. Data are averages ± SEM (n = 4; *p < 0.05, ***p < 0.001) (C) sCARfMFE untargeting decreases liver damage from HSV1-tk/GCV treatment. C57BL/6 mice were injected with Ad.CMVtk or [Ad.CMVtk][sCARfMFE)], followed with GCV injection (100 mg/kg twice/day) for two days. sGPT, sGOT and total bilirubin were measured from sera collected before and after Ad/GCV treatment. Data are averages ± SEM (n = 4; **p < 0.01). Liver sections from these mice were stained with hematoxylin and eosion (H&E) and were also used for TUNEL assays. The livers from Ad.CMVtK/GCV treated mice are hemorrhagic and damaged, in contrast to the livers of mice treated with [Ad.CMVtk][sCARfMFE)]/GCV. (D) Ad-induced cytokine release is decreased by sCARfMFE untargeting. C57BL/6 mice were injected with Ad.CMVtk or [Ad.CMVtk][sCARfMFE]. The RANTES, IL-12, TNF-α, IL-6 and MCP-1 levels were measured from sera collected before and after Ad injection. Data are averages +/− SEM (n = 4, *p < 0.05, **p < 0.01).
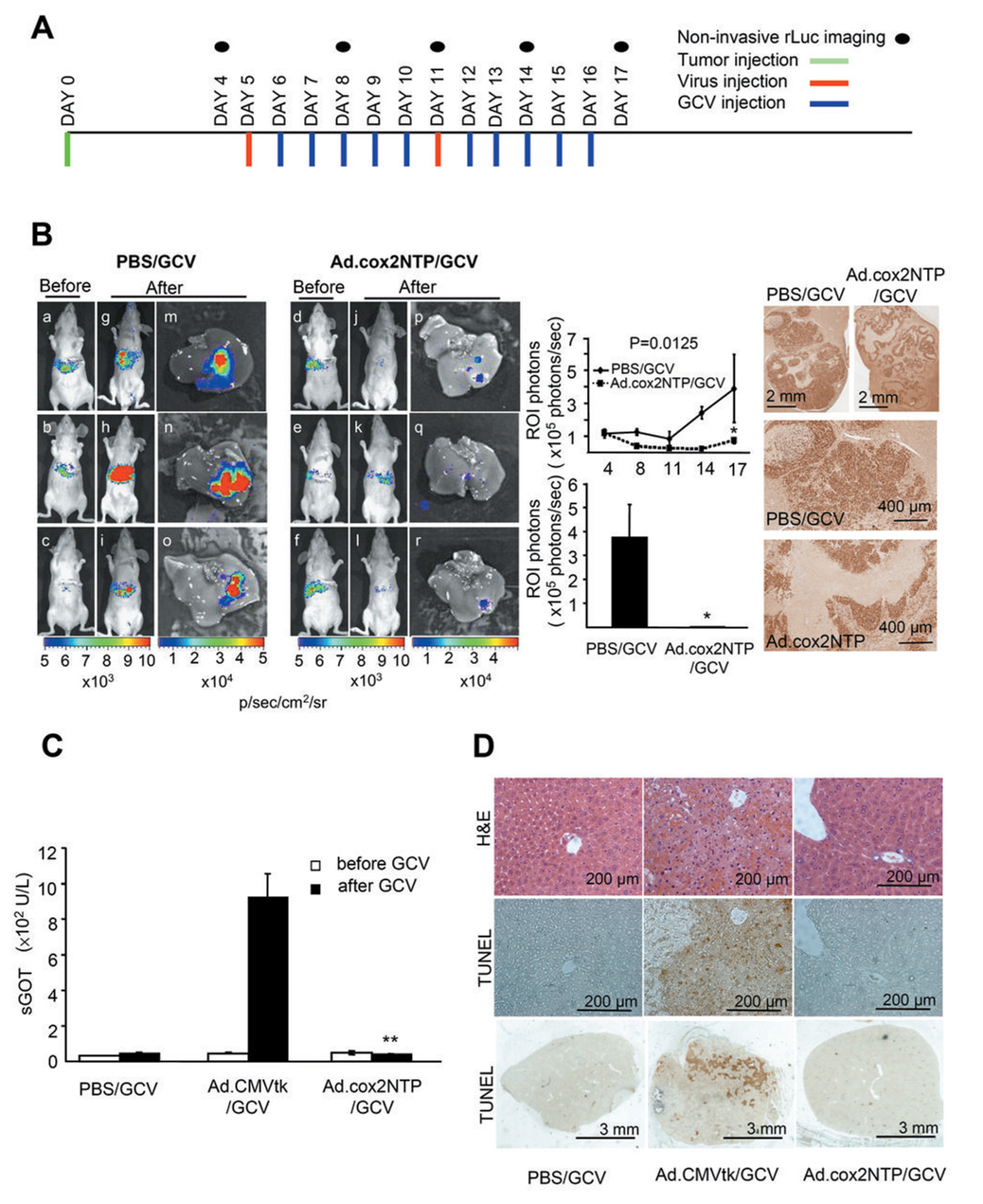
Figure 5. COX-2 promoter-directed HSV1-tk/GCV therapy of LS174T hepatic tumors by Ad.cox2NTP.
(A) Experimental time line. LS174T(rLuc) tumor (1 × 106 cells/mouse), Ad.cox2NTP injection (2 × 109 ifu/mouse), GCV (100 mg/kg i.p. twice/day) injection, and bioluminescent imaging are indicated. (B) LS174T(rLuc) tumor burden was monitored by rLuc bioluminescence before (day 4, a–f) and after (day 17, g–l) therapy. Following d17 rLuc imaging, mice were sacrificed. Livers were removed and imaged for tumor-derived rLuc activity (m–r). Luciferase activity was quantitated by ROI analysis of bioluminescence in the living mice in the upper graph and for livers isolated at day 17 in the lower graph. Data are averages ± SEM (n = 3; *p < 0.05). To measure cell proliferation in the hepatic CRC tumors, formalin-fixed, paraffin embedded sections of the tumor bearing livers were stained with antibody to Ki-67. Note nuclear staining of tumor cells. (C) COX-2 transcriptional restriction reduces liver damage from HSV1TK/GCV treatment following systemic Ad.cox2NTP administration; serum enzymes. Mice were injected with PBS, Ad.CMVtk or Ad.cox2NTP, followed by GCV injection (100 mg/kg twice/day) for four days. Sera were collected before and after GCV treatment and serum GOT (sGOT) levels were measured. Data are averages ± SEM (n = 3; **p < 0.01). (D). COX-2 transcriptional restriction reduces liver damage from HSV1-tk/GCV following systemic Ad.cox2NTP administration; liver histology. Livers were removed after GCV treatment. Formalin fixed, paraffin embedded liver sections were stained with hematoxylin and eosion (H&E) and also used for TUNEL assay.
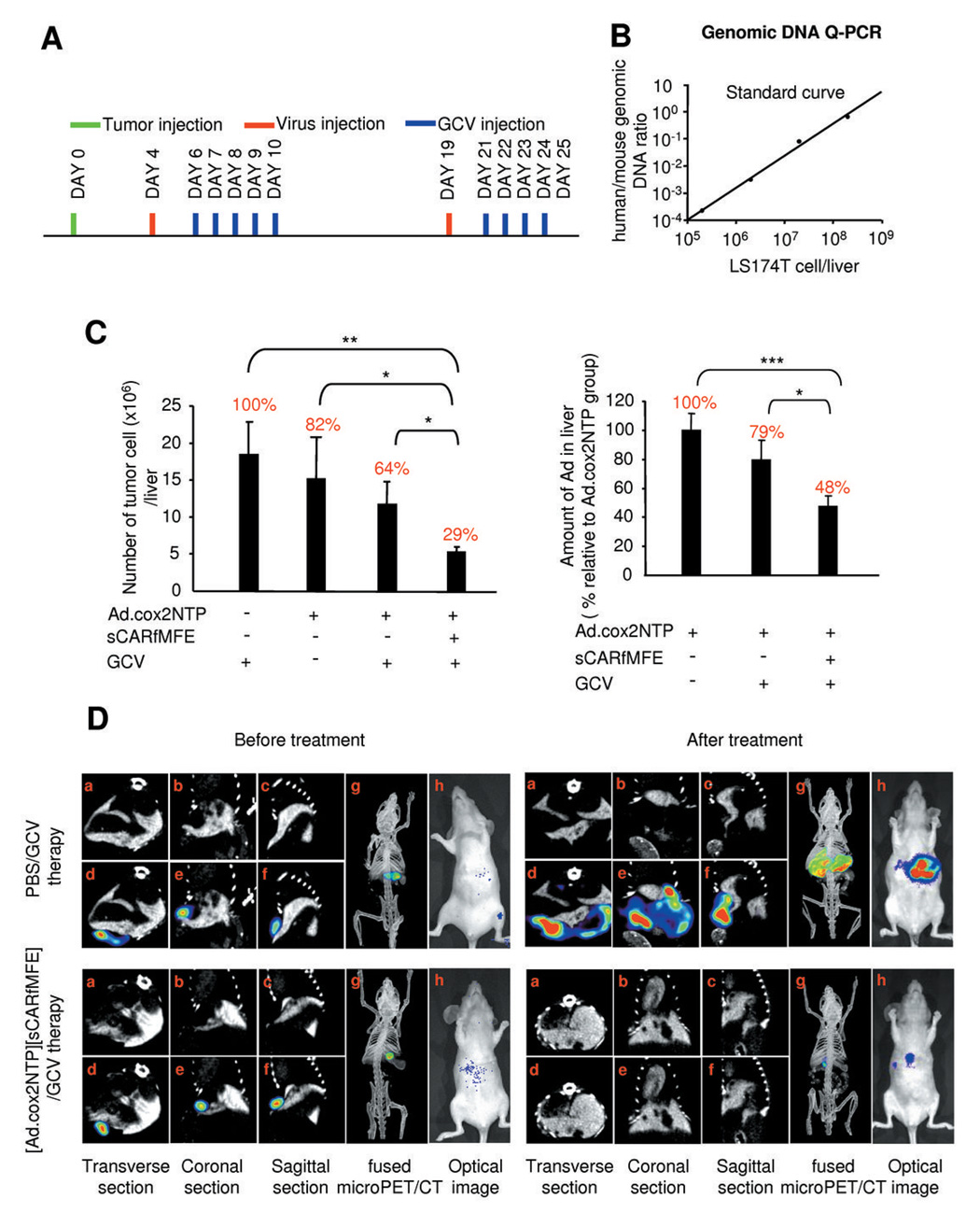
Figure 6. Combined Ad.cox2NTP transcriptional restriction and sCARfMFE transductional untargeting/retargeting therapy of LS174T(rLuc) CRC hepatic tumors.
(A) Experimental time line for LS174T(rLuc) tumor, Ad.cox2NTP (5 × 108 ifu/mouse), [Ad.cox2NTP][sCARfMFE (500 ng)], and GCV (100 mg/kg i.p. twice/day) injection. (B) Standard curve to determine LS174T genomes per mouse liver. (C) LS174T(rLuc) tumor burden and hepatic viral burden, day 25. Genomic Q-PCR analyses for human and mouse GAPDH were performed and the standard curve in panel B was used to determine the number of tumor cells present in each liver. Results are percentages of tumor GAPDH DNA/liver GAPDH DNA for mice treated with PBS/GCV and are presented as averages ± SEM (n=8 for PBS/GCV and Ad.cox2NTP groups, n=9 for Ad.cox2NTP/GCV and [Ad.cox2NTP][sCARfMFE]/GCV groups; *p < 0.05, **p < 0.01). Viral genomes in liver extracts from the tumor-bearing mice were measured by Q-PCR with Ad.cox2NTP specific primers, and were normalized by mouse GAPDH genomic DNA values. Results are percentages of Ad DNA in livers of mice treated with Ad.cox2NTP. Data are averages ± SEM (n=8 for Ad.cox2NTP group, n=9 for Ad.cox2NTP/GCV and [Ad.cox2NTP][sCARfMFE]/GCV groups; *p < 0.05, ***p < 0.001). (D) PET/CT scanning to monitor tumor burden following combined transcriptional restriction and transductional untargeting/retargeting Ad therapy. LS174T(rLuc) tumor burden in mice treated either with PBS + GCV (upper panels, before and after treatment) or with [Ad.cox2NTP][sCARfMFE] + GCV (lower panels, before and after treatment), monitored by 18F-FDG-PET/contrast CT (a–g) and by rLuc bioluminescence (h) in living animals before (left panels) and after (right panels) therapy. (a–c: contrast CT images; d–g: PET/contrast CT overlaid images).
Results
The COX-2 promoter transcriptionally restricts adenovirus transgene expression to hepatic CRC metastases
COX-2 and CEA protein expression (Supplementary Fig. S1) and hepatic tumor-forming efficacy (not shown) were determined for several human CRC and NSCLC cell lines. LS174T (COX-2+, CEA+), was chosen as a hepatic CRC metastasis model. To evaluate the COX-2 promoter transcriptional restriction efficacy for liver CRC metastases, Ad.CMVfLuc (firefly luciferase) and Ad.cox2fLuc were administrated intravenously to mice bearing hepatic LS174T(rLuc) (expressing Renilla luciferase) CRC xenografts. For mice receiving Ad.CMVfLuc, Ad-mediated fLuc bioluminescence (Fig. 1A, d and g) is not co-localized with tumor-encoded rLuc bioluminescence (Fig. 1A, c and f). In contrast, for mice receiving Ad.cox2fLuc, fLuc bioluminescence (Fig. 1A, k and n) co-localizes with rLuc bioluminescence (Fig. 1A, j and m). Ad.cox2fLuc-mediated expression is restricted in liver, but is strong in the COX-2+ tumor. Following systemic Ad.CMVfLuc infection, the hepatic tumor:liver luciferase ratio is 1.6:1 (Fig. 1B). In contrast, the Ad.cox2fLuc tumor-liver luciferase ratio is 37.6:1, illustrating the targeting gains using this tumor-restricted promoter.
sCARhMFE hepatic untargeting, sCARhMFE tumor retargeting, and COX-2 promoter transcriptional restriction combine to enhance Ad transgene expression in hepatic CEA+/COX-2+ CRC tumor xenografts
To achieve maximal transgene hepatic tumor targeting, we combined sCARhMFE hepatic untargeting (15), sCARhMFE tumor retargeting (15), and COX-2 promoter-restricted expression to target CEA+/COX-2+ liver metastases. Ad.CMVfLuc, [Ad.CMVfLuc][sCARhMFE], Ad.cox2fLuc and [Ad.cox2fLuc][sCARhMFE] were administered intravenously to LS174T(rLuc)-bearing mice (Fig. 1C). Mice receiving [Ad.CMVfLuc][sCARhMFE] have reduced hepatic fLuc bioluminescence, when compared to Ad.CMVfLuc-treated tumor-bearing mice (Fig. 1C, d–f versus a–c). The tumor:liver fLuc ratio for Ad.CMVfLuc infection is increased from 0.75:1 to 2.5:1 by sCARhMFE-mediated adenovirus hepatic untargeting and tumor retargeting (Fig. 1D). Ad.cox2fLuc-treated mice exhibit substantially reduced hepatic bioluminescence (Fig. 1C, g–i); the fLuc tumor:liver ratio resulting from COX-2 transcriptional restriction is 33.5:1 (Fig. 1D). [Ad.cox2fLuc][sCARhMFE]-treated mice, combining sCARhMFE transductional untargeting/retargeting and COX-2 promoter-dependent transcriptional restriction, demonstrate both low hepatic bioluminescence and strong tumor bioluminescence (Fig. 1C, j–l). The combination brings the tumor:liver ratio following systemic Ad infection from 0.75:1 to 116:1 (Fig. 1D).
sCARhMFE hepatic untargeting, sCARhMFE CEA-mediated tumor retargeting, and COX-2 promoter transcriptional restriction combine to enhance Ad transgene expression in hepatic CEA+/COX-2+ NSCLC tumor xenografts
A number of other epithelial tumors (e.g., breast, lung), in addition to CRC tumors, often express CEA and elevated COX-2. The combined efficacy of sCARhMFE transductional hepatic untargeting, sCARhMFE transductional tumor retargeting, and COX-2 promoter transcriptional restriction is also evaluated in a CEA+COX-2+ NSCLC liver metastasis model. Ad.CMVfLuc, Ad.cox2fLuc and [Ad.cox2fLuc][sCARhMFE] were injected intravenously into mice carrying H2122 human CEA-positive, COX-2-positive NSCLC hepatic tumors. Once again, mice infected with Ad.CMVfLuc (Fig. 2A, a–c) have much greater hepatic bioluminescence levels than do mice infected with Ad.cox2fLuc (Fig. 2A, d–f). The tumor:liver luciferase activity ratio in extracts is increased from 0.4:1 to 2.2:1 by COX-2 promoter transcriptional restriction (Fig. 2B). Mice infected with [Ad.Cox2-fLuc][sCARhMFE] have even lower hepatic bioluminescence and increased tumor bioluminescence (Fig. 2A, g–i). The tumor:liver luciferase ratio is increased from 2.2:1 for Ad.cox2fLuc to 11.1:1 when Ad.cox2fLuc virus is untargeted from liver and retargeted to CEA+ H2122 xenografts by sCARhMFE (Fig. 2B). Combining sCARhMFE transductional hepatic untargeting, sCARhMFE transductional tumor retargeting and COX-2 promoter restricted transgene expression enhances the tumor:liver luciferase ratio from 0.4:1 to 11.1:1, an increase in tumor-restricted transgene expression of 28 fold for this NSCLC hepatic xenograft model (Fig. 2B)
Trimeric sCARfMFE is more effective than monomeric sCARhMFE for endogenous receptor untargeting and for CEA retargeting, both in culture and in vivo
Before moving to therapeutic studies, we increased the efficacy of sCAR- mediated adapter untargeting and MFE-mediated adapter retargeting by trimerizing the bispecific adapter. The CAR-binding adenovirus knob domain is a trimer (32, 33); consequently, trimerization should increase sCARhMFE binding to the Ad knob (34, 35). To trimerize sCARhMFE, the phage T4 fibritin protein trimerization sequence, which promotes efficient trimerization of sCAR adapters (34), was inserted to produce sCARfMFE, a trimeric adapter for CEA-mediated retargeting (Fig. 3A).
Prior incubation with ~11 ng of trimeric sCARfMFE increased Ad.CMVfLuc infectivity to 50% of maximum sCARfMFE retargeting for CEA+ LS174T cells (Fig. 3B, left). In contrast, to reach the same retargeting level ~1100 ng of monomeric sCARhMFE is required; trimeric sCARfMFE retargeting efficacy is ~100-fold greater than monomeric sCARhMFE for CEA+ cells, using these culture conditions.
To determine whether sCARfMFE trimer binds more effectively than sCARhMFE monomer to the viral knob, we examined the ability of the monomer and the trimer to block Ad infectivity of CAR+CEA− 293 cells. [Ad.CMVfLuc][sCARhMFE] infection of 293 cells is reduced to 40% of AdCMVfLuc infection at ~60 ng of monomeric sCARhMFE, and cannot be further reduced by increased sCARhMFE (Fig. 3B, right). In contrast, trimeric sCARfMFE reduces Ad.CMVfLuc infection by 40% at approximately one-tenth the concentration (~6 ng) and – at higher concentrations – can block ~80% of AdCMVfLuc infection; trimeric sCARfMFE untargeting efficacy is at least 10-fold greater than monomeric sCARhMFE untargeting efficacy for CAR+ cells, using these culture conditions‥
At concentrations where monomeric sCARhMFE is ineffective, trimeric sCARfMFE both untargets adenovirus liver infection in non-tumor bearing mice (Fig. 3C) and retargets adenovirus infection to CEA+ LS174T hepatic tumors (Fig. 3D). These imaging data suggest that a combination of hepatic untargeting/tumor retargeting with trimeric sCARfMFE and COX-2 promoter transcriptional restriction should be an effective combination for CEA+/COX-2+ CRC metastasis eradication, using a therapeutic transgene. We next examined Herpes Virus Type I thymidine kinase/ganciclovir (HSV1-tk/GCV) mediated tumor killing and hepatic toxicity, using sCARfMFE transductional retargeting alone, COX-2 transcriptional restriction alone, and a combination of the two therapies.
sCARfMFE adapter untargeting and tumor retargeting enhances Ad.CMVtk/GCV therapy of CEA+ CRC hepatic metastases and reduces both HSV1-tk/GCV liver toxicity and the immediate immune response to Ad infection
To examine transductional untargeting/retargeting directed therapy, mice bearing LS174T(rLuc) CRC tumors were injected intravenously with PBS, Ad.CMVtk (an adenovirus in which the Herpes Virus type 1 thymidine kinase gene is driven by the early CMV promoter (36, 37)) or [Ad.CMVtk][sCARfMFE], followed by ganciclovir (GCV). Times of tumor grafting, virus administration, GCV injection and optical imaging are shown in Fig. 4A. In tumor-bearing mice treated with only with GCV (PBS/GCV), tumor-derived rLuc bioluminescence increased (Fig. 4B; compare a–d and m–p), indicating tumor growth. In Ad.CMVtk/GCV-treated mice, although rLuc bioluminescence was reduced when compared to PBS/GCV-treated controls (~83%, Fig. 4B, right graph); half the mice became moribund (Fig. 4B, compare e–h and q–t). rLuc bioluminescence decreased even more extensively, relative to controls (~94%), in mice receiving [Ad.CMVtk][sCARfMFE]/GCV (Fig. 4B, compare i–l and u–x); moreover, all [Ad.CMVtk][sCARfMFE]/GCV-treated mice survived with no signs of distress. sCARfMFE liver untargeting and tumor retargeting improves Ad.CMVtk/GCV therapy and reduces morbidity.
Since sCARfMFE untargets Ad liver infection, hepatic HSV1-tk/GCV toxicity should be attenuated. Ad.CMVtk and [Ad.CMVtk][sCARfMFE] were injected intravenously into immunocompetent C57BL/6 mice, followed by GCV. All Ad.CMVtk/GCV-treated mice became moribund, exhibiting impaired mobility and unkempt appearance. In contrast, [Ad.CMVtk][sCARfMFE]/GCV-treated mice displayed no obvious adverse effects. Serum GPT, GOT and bilirubin levels were significantly reduced in [Ad.CMVtk][sCARfMFE]/GCV-treated mice when compared to levels for Ad.CMVtk/GCV-treated mice (Fig. 4C, graph). Abnormal gross morphology, histology and positive TUNEL staining for Ad.CMVtk/GCV-treated mice indicate severe hepatic necrosis (Fig. 4C, left photos). In contrast livers of [Ad.CMVtk][sCARfMFE]/GCV-treated mice appear normal (Fig. 4C, right photos). sCARfMFE-mediated hepatic untargeting protects the liver from Ad.CMV-mediated HSV1-tk/GCV toxicity.
Adenovirus-induced cytokine release can cause severe host damage (38) and even kill virus recipients (39). Since sCARfMFE can untarget Ad from liver (14, 15), we hypothesized untargeting might decrease Ad-induced cytokine release. IL-6, IL-12, TNF-α, RANTES, and MCP-1 levels were strongly induced at 6h in Ad.CMVtk-treated immunocompetent mice (Fig. 4D). In contrast, cytokine levels at 6h after injection were substantially attenuated in [Ad.CMVtk][sCARfMFE]-treated mice; sCARfMFE untargeting reduced cytokine release following systemic adenovirus administration.
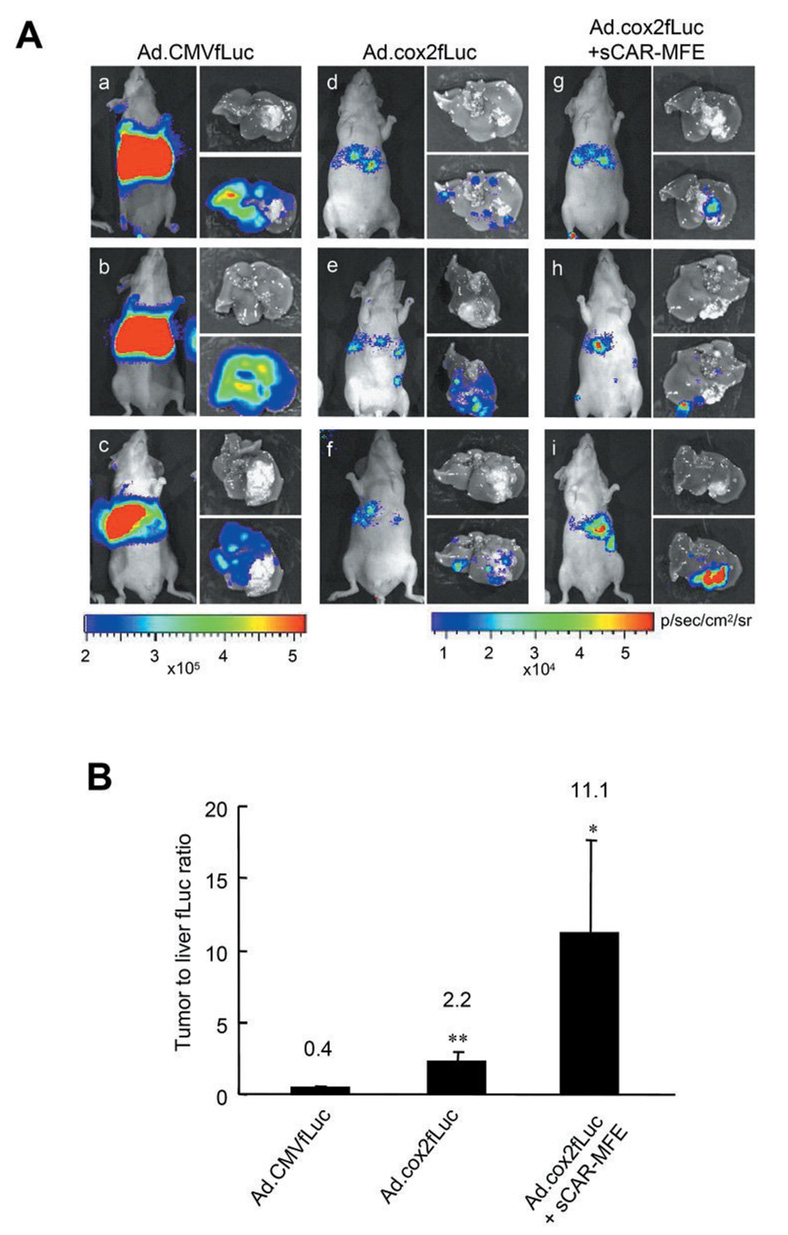
Ad.cox2NTP transcriptionally-restricted HSV1-tk/GCV therapy is effective for COX-2+ CRC hepatic metastases, with reduced liver toxicity
To utilize COX-2 promoter transcriptional restriction for tumor elimination, we created a new Ad gene delivery vector. Ad.cox2NTP expresses GFP, fLuc and a truncated HSV1-tk protein (Supplementary Fig. S2A). Ad.cox2NTP-directed transgene expression following systemic administration, like Ad.cox2fLuc transgene expression, is restricted in liver but is strong in LS174T hepatic xenografts (Supplementary Fig. S2B).
To evaluate COX-2 transcriptionally restricted HSV1-tk/GCV therapy of COX-2+ CRC tumors, mice bearing LS174T(rLuc) tumors were injected with PBS or with Ad.cox2NTP, followed by GCV. Times of hepatic tumor grafting, virus injection, GCV injection and optical imaging are shown in Fig. 5A. In mice treated only with GCV (PBS/GCV), LS174T(rLuc) tumor-derived rLuc bioluminescence increased (Fig. 5B, compare a–c with g–i). In contrast, in Ad.cox2NTP/GCV-treated mice, rLuc bioluminescence decreased during therapy (Fig 5B, compare d–f with j–l). Fig 5B, upper graph shows ROIs for repeated imaging of the two groups. At the conclusion of the experiment, the tumor burden in control PBS/GCV-treated mice was 170 fold greater than the tumor burden in Ad.cox2NTP/GCV-treated mice (Fig. 5B, lower graph). While extensive tumor cell proliferation, measured by Ki-67 staining, occurred in tumor nodules of control PBS/GCV treated mice, the livers of tumor-bearing Ad.cox2NTP/GCV-treated mice have empty cysts with reduced Ki-67 immunostaining (Fig. 5B, right photo).
To evaluate HSV1-tk/GCV hepatic toxicity resulting from COX-2 promoter-restricted expression, mice injected intravenously with PBS, Ad.CMVtk, or Ad.cox2NTP then received GCV. There is no significant sGOT increase in PBS/GCV- or Ad.cox2NTP/GCV-treated mice. In contrast, sGOT levels are greatly increased in Ad.CMVtk/GCV-treated mice (Fig. 5C). Abnormal liver histology and positive TUNEL staining indicate hepatic toxicity in Ad.CMVtk/GCV-treated mice, but not in PBS/GCV- or Ad.cox2NTP/GCV-treated mice (Fig. 5D).
Combined transcriptional (COX-2 promoter) and transductional (sCARfMFE) adenovirus therapy of CEA+/COX-2+ CRC hepatic metastases enhances tumor elimination and reduces liver infection
Combining transductional hepatic untargeting/tumor retargeting with COX-2 transcriptional restriction enhances adenovirus transgene tumor:liver targeting for both a CRC metastatic model and for a NSCLC metastatic model (Fig. 1 and 2). We anticipated, therefore, that combined transductional retargeting/transcriptional restriction should also enhance Ad-mediated HSV1-tk/GCV tumor therapy.
Systemic Ad.cox2NTP/GCV treatment can eliminate CRC hepatic tumors, if a large virus dose and an appropriate amount of GCV are administered (Fig. 5B). To determine whether the sCARfMFE untargeting/retargeting agent can enhance the efficacy of limiting Ad.cox2NTP/GCV administration, thus reducing the virus titer needed for effective therapy, we determined a maximal Ad.cox2NTP level that – in combination with GCV – cannot significantly reduce tumor burden when compared with virus/GCV treatment alone (data not shown). We then determined whether sCARfMFE untargeting/retargeting can enhance tumor killing by this limiting virus treatment. The protocol for comparison of [Ad.cox2NTP][sCARfMFE]/GCV versus Ad.cox2NTP/GCV is shown in Fig. 6A. To more accurately measure tumor burden, we utilized a genomic DNA Q-PCR assay to determine the numbers of LS174T (human) and mouse GAPDH genes in whole [liver + LSI74T tumor] DNA isolates (Fig. 6B).
Two injections of 5 × 108 ifu/mouse Ad.cox2NTP virus + GCV therapy (determined by a previous titration assay) did not result in statistically significant reduction in hepatic CRC tumor cell growth (Fig. 6C, left); there is no significant difference in tumor burden among GCV, Ad.cox2NTP or Ad.cox2NTP/GCV-treated mice. In contrast, the tumor burden for [Ad.cox2NTP][sCARfMFE]/GCV-treated mice is significantly reduced, relative to these three treatments (Fig. 6C, left).
sCARfMFE retargeting should also reduce Adenovirus infection of liver cells in tumor-bearing mice. Using a genomic DNA PCR assay, we determined the number of adenovirus genomes present per liver in these mice. Livers from [Ad.cox2NTP][sCARfMFE]/GCV-treated mice had only half the Ad DNA present in livers from Ad.cox2NTP-treated mice (Fig. 6C, right). sCARfMFE transductional untargeting/retargeting increases Ad.cox2NTP/GCV therapeutic efficacy for CEA+/COX-2+ hepatic tumors and reduces hepatic adenovirus infection.
Efficacy of combined transcriptional (COX-2 promoter) and transductional (sCARfMFE) adenovirus therapy of CEA+/COX-2+ CRC hepatic metastases can be monitored by conventional clinical imaging technology
To reflect more clinically relevant imaging criteria, the efficacy of combined sCARfMFE adapter CEA retargeting and COX-2 promoter-restricted HSV1-tk/GCV therapy was monitored by 18F-FDG-PET/contrast microCT imaging. 18F-FDG PET scans, like bioluminescent rLuc imaging, demonstrated hepatic CRC tumor growth in PBS/GCV-treated mice and reduced tumor burden in [Ad.cox2NTP][sCARfMFE]/GCV-treated mice (Fig. 6D).
Discussion
Unresectable hepatic CRC metastases are one of the leading causes of cancer mortality. Adenovirus gene-delivery vectors provide substantial promise as therapeutic agents, and have proven effective in cases where local delivery is feasible (40). Unfortunately, in the clinic it is often not possible to directly inject all metastases. Here we demonstrate that systemic adenovirus tumor targeting and therapy can be achieved by combined use of (i) transductional liver Ad untargeting with a trimeric bispecific adapter, to reduce complications of liver infection, (ii) transductional tumor retargeting with this bispecific adapter to increase efficacy and specificity of hepatic tumor transgene delivery and (iii) transcriptional restriction with a tumor-restricted promoter. This demonstration of enhanced adenovirus transgene tumor expression with therapeutic efficacy and reduced liver transgene expression and hepatic toxicity suggests a means to engineer practical, effective therapeutic agents, both for hepatic CRC metastases in particular and for hepatic metastases of other epithelial cancers.
Adenovirus gene transfer efficacy for most tumors, following systemic administration, has been limited. One reason is that many tumors lose CAR expression (41–43). Moreover, over 90% of systemically administered Ad is taken up by the liver (44, 45). We previously demonstrated transductional Ad liver untargeting and CEA+ hepatic tumor reporter gene retargeting following systemic Ad administration, using monomeric sCARhMFE adapter (15). In this report we demonstrate that combining transductional liver untargeting/tumor retargeting with COX-2 mediated transcriptional restriction increases hepatic tumor:liver targeting following systemic virus administration (Fig. 1 and 2). We have now substantially improved the adapter, increasing its binding both to the virus knob and to CEA on target cells, by a trimerization protocol (Fig. 3). We demonstrate here that sCARfMFE trimerization results in (i) greater adapter affinity for the viral knob, (ii) increased untargeting of CAR-mediated infection in cultured cells, (iii) increased hepatic untargeting in vivo and (iv) increased retargeting efficacy for CEA+ tumor cells, both in culture and in vivo.
Although adenovirus-bispecific adapter complexes are prepared at high concentrations, the preparations are diluted substantially when injected intravenously. To determine whether association between the trimeric fiber knob and the trimeric sCARfMFE adapter renders the [Ad][sCARfMFE] complex less likely than the adenovirus-monomeric adapter [Ad][sCARhMFE] complex to dissociate following intravenous infection, we examined the effects of [Ad.CMVfLuc][sCARhMFE] and [Ad.CMVfLuc][sCARfMFE] dilution on Ad retargeting to cultured CEA+ cells (Supplementary Fig. S3). Dilution decreased [Ad.CMVfLuc][sCARhMFE] CEA retargeting efficacy much more extensively than did dilution of [Ad.CMVfLuc][sCARfMFE].
Competition by soluble CEA (sCEA), often found in sera of colorectal cancer patients (46), might also interfere with anti-CEA based Ad tumor retargeting for CEA+ tumors. To determine whether the [Ad][trimeric sCARfMFE] complex is more resistant than the [Ad][monomeric sCARhMFE] complex to sCEA competition for binding to CEA+ cells, infection of CEA+ cells by [Ad][adapter] complexes was competed with recombinant sCEA. Monomer sCARhMFE-mediated CEA retargeting is nearly eliminated by sCEA; only 6% of [Ad.CMVfLuc][sCARhMFE] binding is not blocked (Supplementary Fig. S4). In contrast, nearly half the trimer sCARfMFE Ad retargeting ability cannot be competed by sCEA (Supplementary Fig. S4), suggesting that the trimer binds to cell membrane CEA molecules much more avidly, presumably by multivalent “capping” of CEA molecules that are mobile in the cell membrane. Thus circulating sCEA is less likely to present a problem in retargeting adenovirus to CEA-shedding tumors when trimer sCARfMFE adapter, rather than monomer sCARhMFE adapter, is used.
These data support the potential use of trimerized adapters that untarget adenovirus liver infection and retarget the virus to hepatic tumors as a promising therapeutic strategy for hepatic metastases. In addition to reducing hepatic infection, enhancing CEA-dependent tumor targeting, minimizing dilution effects, and avoiding circulating CEA inhibition of CEA-dependent tumor targeting, sCARfMFE hepatic untargeting also decreases liver toxicity following systemic Ad vector administration both by reducing hepatic therapeutic gene expression and by reducing innate immune responses to Ad particles (Fig. 4).
Transgene expression can also be regulated by transcriptional restriction, using targeted, cell-specific promoters (47). Even if Ad vectors infect hepatocytes, virally-delivered transgenes cannot be expressed if they are under the control of a promoter not expressed in liver cells. The COX-2 gene is ectopically activated in many epithelial cancers (22, 48, 49) but is not expressed strongly in liver (50). Here we used COX-2 promoter restriction to target reporter gene expression to CRC and NSCLC liver metastasis models (Fig. 1 and 2) and demonstrated the efficacy of COX-2 promoter restriction in Ad-mediated HSV1-tk/GCV CRC liver metastasis therapy (Fig. 5). COX-2 promoter transcriptional restriction constrains Ad therapeutic transgene expression, following systemic Ad administration, to COX-2+ CRC liver metastases. The result is both to kill the cancer cells using a therapeutic transgene and, by preventing liver transgene expression, to reduce liver toxicity. These data demonstrate that transcriptional restriction can reduce liver toxicity by preventing hepatic therapeutic gene expression during systemic Ad gene therapy.
Imaging data suggest that combining the two technologies should prove more effective at therapeutic gene delivery than either transductional Ad untargeting/retargeting or transcriptional transgene restriction alone (Fig. 1 and 2). As anticipated, combined transductional untargeting/retargeting and transcriptional restriction enhances therapeutic efficacy (Fig. 6C, left). sCARfMFE transductional liver untargeting and CEA retargeting increased Ad.cox2NTP-mediated HSV1-tk/GCV killing of LS174T hepatic metastases; moreover the increased CRC hepatic tumor cell killing resulting from sCARfMFE transductional retargeting is accompanied by a decreased number of virus particles present in liver (Fig. 6C, right).
Combining transductional untargeting/retargeting and tumor-restricted transcriptional expression offers substantial flexibility. Recombinant proteins can be created using alternative adenovirus untargeting components (e.g. sCAR, anti-knob Fab) and retargeting agents (e.g., receptor ligands, antibodies, lectins). Promoters for restricted expression can be chosen that utilize either tissue/cell specificity or tumor cell specificity. By varying retargeting moieties and transcriptionally restricted promoters, one can “tailor” untargeting/retargeting and transcriptional restriction combinations for specific tumors or for other cells. By utilizing alternative transgenes, different purposes can be achieved; cargo genes whose products can be imaged by bioluminescence, fluorescence, magnetic resonance imaging, positron emission tomography, single photon emission tomography, etc can be targeted. Alternatively, therapeutic genes that target tumor cells, tumor neovasculature, inflammatory cells and other cells that participate in tumor progression can be incorporated into these vectors and retargeted to alternative cells and/or tissues with appropriate bispecific adapters.
Supplementary Material
Acknowledgements
We thank the members of the Herschman and Curiel laboratories for helpful discussions, and David Stout and the members of the UCLA Small Animal Imaging Shared Resource for technical advice and experimental assistance.
Financial support: These studies were supported by National Cancer Institute grants R01 CA84572 and P50 CA86306 (H. R. Herschman) 5R01 CA113454 and 1R01CA121187 (D.R. Curiel)
References
- 1.Wagner JS, Adson MA, Van Heerden JA, Adson MH, Ilstrup DM. The natural history of hepatic metastases from colorectal cancer. A comparison with resective treatment. Ann Surg. 1984;199:502–508. doi: 10.1097/00000658-198405000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert HA, Kagan AR. Metastases: incidence, detection, and evaluation without histologic confirmation. Fundamental Aspects of Metastasis: North Holland Publishing Co; 1976. pp. 385–405. [Google Scholar]
- 3.Macdonald JS. Adjuvant therapy of colon cancer. CA Cancer J Clin. 1999;49:202–219. doi: 10.3322/canjclin.49.4.202. [DOI] [PubMed] [Google Scholar]
- 4.Fong Y, Cohen AM, Fortner JG, et al. Liver resection for colorectal metastases. J Clin Oncol. 1997;15:938–946. doi: 10.1200/JCO.1997.15.3.938. [DOI] [PubMed] [Google Scholar]
- 5.Tuttle TM, Curley SA, Roh MS. Repeat hepatic resection as effective treatment of recurrent colorectal liver metastases. Ann Surg Oncol. 1997;4:125–130. doi: 10.1007/BF02303794. [DOI] [PubMed] [Google Scholar]
- 6.Feliberti EC, Wagman LD. Radiofrequency ablation of liver metastases from colorectal carcinoma. Cancer Control. 2006;13:48–51. doi: 10.1177/107327480601300107. [DOI] [PubMed] [Google Scholar]
- 7.Zinn KR, Douglas JT, Smyth CA, et al. Imaging and tissue biodistribution of 99mTc-labeled adenovirus knob (serotype 5) Gene Ther. 1998;5:798–808. doi: 10.1038/sj.gt.3300659. [DOI] [PubMed] [Google Scholar]
- 8.Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci U S A. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Einfeld DA, Schroeder R, Roelvink PW, et al. Reducing the native tropism of adenovirus vectors requires removal of both CAR and integrin interactions. J Virol. 2001;75:11284–11291. doi: 10.1128/JVI.75.23.11284-11291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Eb MM, Cramer SJ, Vergouwe Y, et al. Severe hepatic dysfunction after adenovirus-mediated transfer of the herpes simplex virus thymidine kinase gene and ganciclovir administration. Gene Ther. 1998;5:451–458. doi: 10.1038/sj.gt.3300637. [DOI] [PubMed] [Google Scholar]
- 11.Glasgow JN, Everts M, Curiel DT. Transductional targeting of adenovirus vectors for gene therapy. Cancer Gene Ther. 2006;13:830–844. doi: 10.1038/sj.cgt.7700928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waddington SN, McVey JH, Bhella D, et al. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell. 2008;132:397–409. doi: 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Liang Q, Dmitriev I, Kashentseva E, Curiel DT, Herschman HR. Noninvasive of adenovirus tumor retargeting in living subjects by a soluble adenovirus receptor-epidermal growth factor (sCAR-EGF) fusion protein. Mol Imaging Biol. 2004;6:385–394. doi: 10.1016/j.mibio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Li HJ, Everts M, Pereboeva L, et al. Adenovirus Tumor Targeting and Hepatic Untargeting by a Coxsackie/Adenovirus Receptor Ectodomain Anti-Carcinoembryonic Antigen Bispecific Adapter. Cancer Res. 2007;67:5354–5361. doi: 10.1158/0008-5472.CAN-06-4679. [DOI] [PubMed] [Google Scholar]
- 16.Glasgow JN, Bauerschmitz GJ, Curiel DT, Hemminki A. Transductional and transcriptional targeting of adenovirus for clinical applications. Curr Gene Ther. 2004;4:1–14. doi: 10.2174/1566523044577997. [DOI] [PubMed] [Google Scholar]
- 17.Mathis JM, Stoff-Khalili MA, Curiel DT. Oncolytic adenoviruses - selective retargeting to tumor cells. Oncogene. 2005;24:7775–7791. doi: 10.1038/sj.onc.1209044. [DOI] [PubMed] [Google Scholar]
- 18.Stoff-Khalili MA, Stoff A, Rivera AA, et al. Preclinical evaluation of transcriptional targeting strategies for carcinoma of the breast in a tissue slice model system. Breast Cancer Res. 2005;7:R1141–R1152. doi: 10.1186/bcr1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto M, Alemany R, Adachi Y, Grizzle WE, Curiel DT. Characterization of the cyclooxygenase-2 promoter in an adenoviral vector and its application for the mitigation of toxicity in suicide gene therapy of gastrointestinal cancers. Mol Ther. 2001;3:385–394. doi: 10.1006/mthe.2001.0275. [DOI] [PubMed] [Google Scholar]
- 20.Liang Q, Yamamoto M, Curiel DT, Herschman HR. Noninvasive imaging of transcriptionally restricted transgene expression following intratumoral injection of an adenovirus in which the COX-2 promoter drives a reporter gene. Mol Imaging Biol. 2004;6:395–404. doi: 10.1016/j.mibio.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest. 2005;23:338–351. doi: 10.1081/cnv-58878. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Sun XF. Overexpression of cyclooxygenase-2 correlates with advanced stages of colorectal cancer. Am J Gastroenterol. 2002;97:1037–1041. doi: 10.1111/j.1572-0241.2002.05625.x. [DOI] [PubMed] [Google Scholar]
- 23.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10:8465–8471. doi: 10.1158/1078-0432.CCR-04-0653. [DOI] [PubMed] [Google Scholar]
- 24.Maizel JV, Jr, White DO, Scharff MD. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 25.Everts M, Kim-Park SA, Preuss MA, et al. Selective induction of tumor-associated antigens in murine pulmonary vasculature using double-targeted adenoviral vectors. Gene Ther. 2005;12:1042–1048. doi: 10.1038/sj.gt.3302491. [DOI] [PubMed] [Google Scholar]
- 26.Pereboev AV, Nagle JM, Shakhmatov MA, et al. Enhanced gene transfer to mouse dendritic cells using adenoviral vectors coated with a novel adapter molecule. Mol Ther. 2004;9:712–720. doi: 10.1016/j.ymthe.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Applied Biosystems. Relative Quantitation Of Gene Expression: ABI PRISM 7700 Sequence Detection System: User Bulletin #2: Rev B. Foster City: Applied Biosystems; 1997. pp. 11–15. [Google Scholar]
- 28.Chow PL, Stout DB, Komisopoulou E, Chatziioannou AF. A method of image registration for small animal, multi-modality imaging. Phys Med Biol. 2006;51:379–390. doi: 10.1088/0031-9155/51/2/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinahan PE RJ. Analytic 3D image reconstruction using all detected events. IEEE Trans Nucl Sci. 1989;36:964–968. [Google Scholar]
- 30.Qi J, Leahy RM, Cherry SR, Chatziioannou A, Farquhar TH. High-resolution 3D Bayesian image reconstruction using the microPET small-animal scanner. Phys Med Biol. 1998;43:1001–1013. doi: 10.1088/0031-9155/43/4/027. [DOI] [PubMed] [Google Scholar]
- 31.Loening AM, Gambhir SS. AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging. 2003;2:131–137. doi: 10.1162/15353500200303133. [DOI] [PubMed] [Google Scholar]
- 32.Xia D, Henry LJ, Gerard RD, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 A resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]
- 33.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 34.Kashentseva EA, Seki T, Curiel DT, Dmitriev IP. Adenovirus targeting to c-erbB-2 oncoprotein by single-chain antibody fused to trimeric form of adenovirus receptor ectodomain. Cancer Res. 2002;62:609–616. [PubMed] [Google Scholar]
- 35.Kim J, Smith T, Idamakanti N, et al. Targeting adenoviral vectors by using the extracellular domain of the coxsackie-adenovirus receptor: improved potency via trimerization. J Virol. 2002;76:1892–1903. doi: 10.1128/JVI.76.4.1892-1903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smythe WR, Hwang HC, Amin KM, et al. Use of recombinant adenovirus to transfer the herpes simplex virus thymidine kinase (HSVtk) gene to thoracic neoplasms: an effective in vitro drug sensitization system. Cancer Res. 1994;54:2055–2059. [PubMed] [Google Scholar]
- 37.Qian C, Bilbao R, Bruna O, Prieto J. Induction of sensitivity to ganciclovir in human hepatocellular carcinoma cells by adenovirus-mediated gene transfer of herpes simplex virus thymidine kinase. Hepatology. 1995;22:118–123. [PubMed] [Google Scholar]
- 38.Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 39.Raper SE, Chirmule N, Lee FS, et al. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol Genet Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 40.Wildner O, Blaese RM, Morris JC. Therapy of colon cancer with oncolytic adenovirus is enhanced by the addition of herpes simplex virus-thymidine kinase. Cancer Res. 1999;59:410–413. [PubMed] [Google Scholar]
- 41.Cripe TP, Dunphy EJ, Holub AD, et al. Fiber knob modifications overcome low, heterogeneous expression of the coxsackievirus-adenovirus receptor that limits adenovirus gene transfer and oncolysis for human rhabdomyosarcoma cells. Cancer Res. 2001;61:2953–2960. [PubMed] [Google Scholar]
- 42.Li Y, Pong RC, Bergelson JM, et al. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res. 1999;59:325–330. [PubMed] [Google Scholar]
- 43.Kanerva A, Mikheeva GV, Krasnykh V, et al. Targeting adenovirus to the serotype 3 receptor increases gene transfer efficiency to ovarian cancer cells. Clin Cancer Res. 2002;8:275–280. [PubMed] [Google Scholar]
- 44.Huard J, Lochmuller H, Acsadi G, Jani A, Massie B, Karpati G. The route of administration is a major determinant of the transduction efficiency of rat tissues by adenoviral recombinants. Gene Ther. 1995;2:107–115. [PubMed] [Google Scholar]
- 45.Li Q, Kay MA, Finegold M, Stratford-Perricaudet LD, Woo SL. Assessment of recombinant adenoviral vectors for hepatic gene therapy. Hum Gene Ther. 1993;4:403–409. doi: 10.1089/hum.1993.4.4-403. [DOI] [PubMed] [Google Scholar]
- 46.Kutun S, Celik A, Hatiboglu C, Ulucanlar H, Cetin A. Carcinoembryonic antigen to detect hepatic metastases of colorectal cancers. Surg Today. 2003;33:590–594. doi: 10.1007/s00595-003-2546-y. [DOI] [PubMed] [Google Scholar]
- 47.Saukkonen K, Hemminki A. Tissue-specific promoters for cancer gene therapy. Expert Opin Biol Ther. 2004;4:683–696. doi: 10.1517/14712598.4.5.683. [DOI] [PubMed] [Google Scholar]
- 48.DuBois RN, Smalley WE. Cyclooxygenase, NSAIDs, and colorectal cancer. J Gastroenterol. 1996;31:898–906. doi: 10.1007/BF02358623. [DOI] [PubMed] [Google Scholar]
- 49.Fujita T, Matsui M, Takaku K, et al. Size- and invasion-dependent increase in cyclooxygenase 2 levels in human colorectal carcinomas. Cancer Res. 1998;58:4823–4826. [PubMed] [Google Scholar]
- 50.Ishikawa TO, Jain NK, Taketo MM, Herschman HR. Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene. Mol Imaging Biol. 2006;8:171–187. doi: 10.1007/s11307-006-0034-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.