Abstract
SRY-box containing gene 17 (Sox17) is a member of the high mobility group (HMG) transcription factor superfamily, which plays critical roles in the regulation of development and stem/precursor cell function, at least partly through repression of Wnt pathway activity. Modulators controlling aberrant Wnt signaling activation are frequently disrupted in human cancers through complementary effects of epigenetic and genetic changes. Our recent global analysis of CpG island hypermethylation and gene expression in colorectal cancer (CRC) cell lines revealed that SOX17 gene silencing is associated with DNA hypermethylation of a CpG island in the promoter region. Here, we report that CpG island methylation-dependent silencing of SOX17 occurs in 100% of CRC cell lines, 86% of colorectal adenomas, 100% of stage I and II CRC, 89% of stage III CRC, 89% of primary esophageal cancer, and 50% of non–small cell lung cancer. Overexpression of SOX17 in HCT116 CRC cells inhibits colony growth and β-catenin/T-cell factor–dependent transcription. Structure-based deletion analysis further shows the presence of a Wnt signaling repression domain in the SOX17 HMG box. Together, our studies suggest that SOX17 is a negative modulator of canonical Wnt signaling, and that SOX17 silencing due to promoter hypermethylation is an early event during tumorigenesis and may contribute to aberrant activation of Wnt signaling in CRC.
Introduction
The Sox gene family was first identified by virtue of its strong homology (>50%) to the high mobility group (HMG) box of the sex-determining gene SRY (1). There are at least 30 members of the Sox family expressed in many different cell types and tissues, and at multiple stages during development (2). Sox genes have been classified into seven groups based on their amino acid sequence and genomic organization, and Sox17, together with Sox7 and Sox18, belongs to Sox group F (2). Sox17 encodes a HMG box transcription factor and has been implicated in oligodendrocyte development (3), vascular development (4), formation of definitive endoderm (5), and embryonic hematopoiesis (6). Sox17 binds to a common Sox target DNA sequence 5′-(A/T)(A/T)CAA(A/T)G-3′ in the minor groove (7) and is known to regulate the transcription of a number of target genes, including Foxa1 and Foxa2 via the physical interaction of its COOH-terminal transcriptional activation domain with β-catenin (8). The importance of Sox17 for embryonic development has been shown by two knockout experiments in mice. Sox17-null embryos exhibit a deficiency of gut definitive endoderm, which leads to embryonic lethality before day E10.5 (9). A recent conditional knockout study reveals that Sox17 is critical for the generation or maintenance of fetal blood stem cells (6).
A growing body of evidence suggests that an important function of Sox17 is to inhibit canonical Wnt pathway signaling. Injection of Sox17β mRNA can effectively suppress the induction of a second axis in Xenopus embryos induced by Wnt activators, but failed to do so when coinjected with mRNAs encoding Wnt targets (10). Sox17 is also indispensable for the specification of cardiac mesoderm in embryonic stem cells by inactivating the canonical Wnt pathway (11). A recent study suggests that mouse Sox17 suppresses canonical Wnt signaling by GSK3β-independent protein degradation of β-catenin and T-cell factor/lymphoid enhancer factor (TCF/LEF) in human SW480 colorectal cancer (CRC) cells (12).
Mutations in the intracellular components of the Wnt/β-catenin pathway, such as APC, Axin2, and β-catenin, are thought to cause constitutive activation of downstream signaling independent of extracellular Wnt ligands in CRC (13). Our previous studies revealed that epigenetic gene silencing of secreted frizzled-related proteins (SFRPs), which encode secreted Wnt antagonists, occurs aberrantly in CRC and enhances constitutive Wnt signaling (14). Furthermore, another two extracellular Wnt inhibitors, Wnt inhibitory factor-1 (WIF-1) and DICKKOPF-1 (DKK-1), are also silenced in cancer cell lines and primary tumors (15, 16).
Here, we show that SOX17 is frequently silenced by promoter hypermethylation in colonic neoplasia and CRC. Reexpression of SOX17 in CRC cells leads to a significant reduction in colony formation, suggesting a potential role as a tumor suppressor. Additionally, we show that overexpression of SOX17 suppresses β-catenin/TCF–regulated transcription in a dose-dependent manner. Deletion analysis in this present study, when combined with previous work of others (10, 12), further suggests that the HMG box of SOX17, but in our hands, not the COOH-terminal transcription activation domain, is essential for this transcriptional repression in colon cancer cells. In view of these and other findings, we conclude that SOX17 gene silencing is an early frequent event associated with aberrant Wnt signaling in CRC, and SOX17 inhibits Wnt signaling through the NH2-terminal HMG box.
Materials and Methods
Cell culture
HCT116, DKO, and SW480 CRC cells were cultured in McCoy’s 5A modified medium; RKO and Caco-2 cells were maintained in MEM; HEK293T cells were maintained in DMEM. All media (Cellgro) were supplemented with 10% fetal bovine serum (HyClone) and antibiotics and grown at 37°C in 5% CO2 atmosphere. For drug treatments, log phase CRC cells were cultured in the above-described medium supplemented with 1 μmol/L 5-aza-2′-deoxycytidine (DAC; Sigma) for 96 h, with replacement of medium and DAC every 24 h.
Vector constructs
SOX17 (Genbank accession number NM_022454) was cloned by reverse transcription-PCR (RT-PCR) from cDNA derived from normal colon mucosa. To generate SOX17 expression constructs, the entire encoding region of its cDNA was subcloned in frame into the pcDNA3.1/V5-His B vector (Invitrogen) via KpnI and EcoRI sites. Truncated SOX17 mutants were generated by PCR. All constructs were verified in each case by DNA sequencing.
Gene expression analysis
RNA was isolated with TRIzol reagent (Invitrogen). One microgram RNA was treated with DNase I (Invitrogen) and reverse-transcribed into cDNA by using SuperScript III (Invitrogen) according to the manufacturer’s instructions. SOX3, SOX7, SOX9, SOX17, and SOX18 RT-PCR primers used in this study are as follows: SOX3 forward, 5′-AGACCAGGACCGTGTGAAAC-3′; SOX3 reverse, 5′-GTCGATGAATGGTCG CTTCT-3′; SOX7 forward, 5′-CAAGATGCTGGGAAAGTCGT-3′; SOX7 reverse, 5′-ACTCACCCCTGTCCTCCTTC-3′; SOX9 forward, 5′-GAGGAAGTCGGTGAAGAACG-3′; SOX9 reverse, 5′-AAGTCGATAGGGGGCTGTCT-3′; SOX17 forward, 5′-TTCACGTGTACTACGGCGCGAT-3′; SOX17 reverse, 5′-AGTTGCAGTAATATACCGCGGAGC-3′; SOX18 forward, 5′-TGAACGCCTTCATGGTGTGGGCAAA-3′; SOX18 reverse, 5′-CGGTACTTGTAGTTGGGGTGGTCGC-3′.
Western blots and antibodies
Antibodies used for Western blots were anti-SOX17 (R&D Systems) and anti–β-actin (Sigma).
Methylation-specific PCR and bisulfite sequencing
Genomic DNA from primary colonic, esophageal, and lung tissue samples and from the CRC cell lines was prepared using the proteinase-K method (17). After chloroform/phenol extraction, DNA was precipitated in ethanol and later dissolved in low TE buffer and stored at −20°C. Genomic DNA was bisulfite treated using the EZ DNA methylation Kit (Zymo Research). Methylation-specific PCR (MSP) primers specific for the unmethylated and methylated promoter sequences were designed using MSPPrimer.5 MSP primers are as follows: SOX17-M forward, 5′-CAAAAACGAATCCCGTATCCGACG-3′; SOX17-M reverse, 5′-ACTCACGTACATAATAACGAAAATCCG-3′; SOX17-U forward, 5′-CAAACCAAAAACAAATCCCATATCCAACA-3′; SOX17-U reverse, 5′-GATTTTGTTGTGTTAGTTGTTTGTGTTTG-3′. Each MSP was done by using ~ 100 ng of bisulfite-treated DNA, 25 pmol of each primer, 100 pmol deoxynucleotide triphosphates, 2.5 μL 10× PCR buffer, and 1 unit of JumpStart Red Taq Polymerase (Sigma) in a final reaction volume of 25 μL. Amplifications were done as follows: 95°C × 5 min; 35 cycles × (95°C × 30 s, 60°C × 30 s, 72°C × 30 s); 72°C × 5 min. MSP products were analyzed using 6% PAGE. For bisulfite sequencing, we first did PCR on bisulfite-treated DNA using the primers that amplify both methylated and unmethylated alleles. The following oligonucleotides were used as PCR primers: 5′-ATATGAAGGTGAAGGGCGAGG-3′ (SOX17 BS 5′) and 5′-CTACACACCCCTAATTTTAAAC-3′ (SOX17 BS 3′). PCR products were gel purified and cloned into the vector pCR2.1-TOPO according to the manufacturer’s protocol (Invitrogen). Integrated PCR fragments were verified by EcoRI digestion, and sequenced with the M13 reverse primer by the Johns Hopkins Medical Institutions DNA sequencing facility.
Colony formation assay
HCT116 cells were plated in 10-cm plate culture plate dishes 24 h before transfection. Ten micrograms of empty control vector or SOX17 expression vector were transfected using 50 μL FuGENE6 reagent (Roche Applied Science). Twenty-four hours later, the transfected cells were diluted, replated, and selected in 10-cm plates containing 0.4 mg/mL G418 for 10 d. Staining, visualization, and counting of triplicate wells were done as previously described (14).
Luciferase reporter assays
Reporter gene assays were done as previously described (14). Briefly, 5 × 104 cells were seeded in 24-well tissue culture plates 24 h before transfection. The TOPFLASH or FOPFLASH reporter vectors (gifts from Dr. Bert Vogelstein, Cancer Biology Division, The Sydney Kimmel Comprehensive Cancer Center, The Johns Hopkins University, Baltimore, MD) were transfected at 70 ng/well, and the pRL-TK control vector (Promega) was cotransfected at 7 ng/well as an internal control reporter. For the reporter assays in 293T cells, pCI-neo-β-catenin–expressing wild-type β-catenin (70 ng/well) was used to activate the reporter gene. Increasing amounts of pcDNA3.1-SOX17 wild-type, mutants, or the empty vector were transfected into cells using FuGENE 6 (Roche Applied Science). Forty-eight hours posttransfection, cells were washed and lysed in Passive Lysis Buffer (Promega). We measured luciferase activity in a luminometer (BD Biosciences) and transfection efficiency was normalized using the paired Renilla luciferase activity by using the Dual Luciferase Reporter Assay system (Promega) according to the manufacturer’s instructions.
Results
SOX17 is a candidate for aberrant gene silencing in CRC cells
Our earlier studies described a novel approach to identify genes silenced by DNA hypermethylation in CRC based on a genome-wide microarray expression assay (18). After treatment of HCT116 CRC cells with either DAC, a drug that inhibits DNMT-mediated hypermethylation of promoter CpG islands, or the histone deacetylase inhibitor trichostatin A, we identified SOX17 as a candidate promoter CpG island hypermethylated gene by the criteria previously outlined (18). In this approach, candidate DNA hypermethylated genes are identified by selecting those which have no basal expression on the microarray, have a promoter CpG island, and reside in a zone where gene expression did not respond to trichostatin A (<1.4-fold), but increased >2-fold with DAC treatment (Fig. 1A).
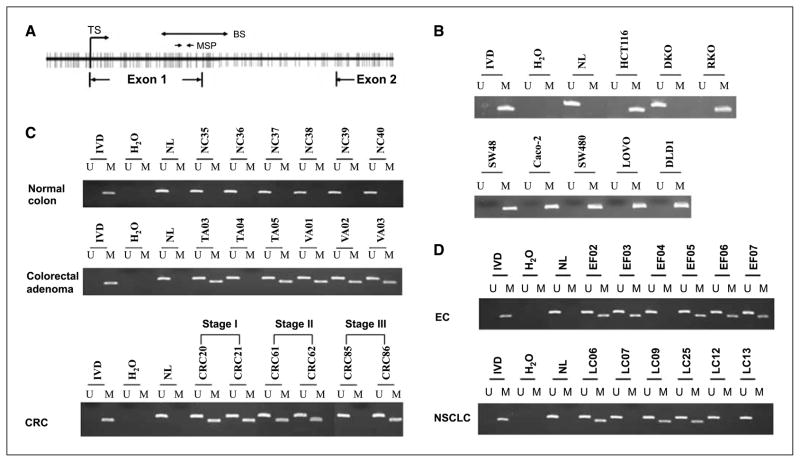
Figure 1.
Identification of SOX17 as a hypermethylated gene in CRC cells. A, the distribution of gene expression changes for HCT116 cells treated with trichostatin A (X axis) or DAC (Y axis) is analyzed and displayed (18). Black dots, individual genes; red dot, SOX17, which is a candidate DNA hypermethylated gene. SOX17 is identified by its position in a zone where gene expression did not respond to trichostatin A (<1.4-fold) but increased >2-fold with DAC treatment. B, RT-PCR analysis for expression of SOX genes in HCT116, DKO, and normal colonic mucosa. Gene symbols are indicated on the left; cell lines are indicated above the data; and NC indicates normal colon. β-Actin is used as a internal control. C, RT-PCR analysis for expression of SOX genes before and after treatment of cells with 1 μmol/L DAC (+) for 96 h. Restoration of SOX17 expression is observed in seven CRC cells. As a control for SOX17 expression, PCR was performed on DNase I–treated RNA without reverse transcription. D, Western blot analysis of SOX17 in different CRC cells. The positive control for SOX17 protein detection uses whole-cell lysate from HCT116 cells transfected with the pcDNA3.1-SOX17 expression vector. Left, of the five cell lines tested, endogenous SOX17 protein was only detected in DKO cells. β-Actin is shown as a gel loading control for the different cell lysates. Right, SOX17 protein is seen after treatment with 1 μmol/L DAC (+) for 96 h in HCT116 and Caco-2 cells.
To validate the expression array results, we first did RT-PCR to examine the expression of SOX17 and the other SOX group F genes, including SOX7 and SOX18 in the CRC cell line HCT116. Two additional SOX genes, SOX3 and SOX9, are also included in this study because they have been shown to inhibit the TCF-mediated Wnt signaling activity (10, 12). In addition, we studied normal human colon cells and a cell line isogenic to HCT116 cells in which two DNA methyltransferases (DNMT1 and DNMT3b) have been genetically disrupted (DKO HCT116 cells). The latter cells have ~95% reduction in the genomic 5-methylcytosine (19). Of the five genes we examined, SOX3 and SOX7 are not expressed in normal colon whereas SOX9, SOX17, and SOX18 are expressed in normal colon and CRC cells. Strikingly, SOX17 is the only gene exhibiting loss of expression in HCT116 cells. Furthermore, this gene is reexpressed in the HCT 116 isogenic DKO cells (Fig. 1B).
To address the role of epigenetic gene silencing, we analyzed SOX17 expression in the absence or presence of the demethylation agent DAC. Using RT-PCR, we found that SOX17 mRNA was undetectable or expressed at extremely low levels in seven CRC cell lines (HCT116, RKO, SW48, Caco-2, SW480, LOVO, and DLD1). After treating these cells with 1 μmol/L DAC for 96 hours, SOX17 was reexpressed in all of the seven cell lines (Fig. 1C). As a control for SOX17 expression, RT-PCR without reverse transcription showed absence of SOX17 bands (Fig. 1C). Consistent with the RT-PCR results, Western blots established that endogenous SOX17 protein is detectable in DKO cells, but not in the HCT116, SW480, Caco-2, and RKO cells (Fig. 1D). These data are consistent with a previous expression analysis in which SW480 cells lacked SOX17 expression (20). In addition, expression of SOX17 protein was restored in HCT116 and Caco-2 cells with DAC treatment (Fig. 1D). Together, these data strongly suggest that SOX17 expression is down-regulated in CRC cells in association with abnormal promoter region DNA methylation.
Silencing of SOX17 is associated with its promoter CpG island hypermethylation
Genomic DNA sequence analysis of SOX17 5′ regulatory regions shows that there is a CpG island encompassing its transcription start site (CpG frequency >68%; Fig. 2A). To analyze the methylation status of the SOX17 promoter–associated CpG island, we first designed a set of primers, in a region downstream to the transcription start site, to screen human CRC cell lines by MSP (18). As predicted by our RT-PCR and Western blot results, hypermethylation of SOX17 promoter was detected in seven of the studied colon cancer cell lines (Fig. 2B). In contrast, there is no methylation detected in isogenic DKO cells (Fig. 2B). We obtained the same results with a second set of MSP primers encompassing the transcription start site (supplementary Fig. 1). We then analyzed methylation status in a series of human colon specimens from unselected patients to determine whether aberrant methylation of the SOX17 gene in human CRC cell lines reflects an epigenetic process of colon cancer initiation and progression in humans. Normal colon (n = 20), tubular adenomas (n = 15), villous adenomas (n = 22), and stage I (n = 35), stage II (n = 36), and stage III (n = 28) CRCs were included in this study. Strikingly, SOX17 hypermethylation was found in 12 of 15 (80%) colorectal tubular adenomas and 20 of 22 (91%) villous adenomas (Fig. 2C). Overall, the frequency of SOX17 methylation in adenomas was 86% (32 of 37). Similarly, extensive methylation at the same loci was seen in primary CRCs: 35 of 35 (100%) in stage I CRC, 36 of 36 (100%) in stage II CRC, and 25 of 28 (89%) in stage III CRC (Fig. 2C). Notably, promoter methylation has not been detected in 20 samples of normal colon. These data strongly suggest that hypermethylation of SOX17 is established at a very early stage in the initiation of colorectal carcinogenesis.
Figure 2.
Methylation of the SOX17 CpG island in cell lines and primary tumors. A, schematic of the SOX17 CpG island spanning the 5′ upstream region, exon1, intron 1, and exon 2. Bold vertical lines, individual CpG sites. TS, transcriptional start site in exon 1. Two small solid arrows, the location of primers used in the MSP assay. Double-headed arrow, the bisulfite genomic sequencing region (marked BS). B, aberrant methylation of SOX17 in human CRC cell lines. A visible PCR product in lanes marked U indicates the presence of unmethylated alleles; a visible PCR product in lanes marked M indicates the presence of methylated alleles. Note that in DKO cells, SOX17 is unmethylated, whereas in HCT116, RKO, SW48, Caco-2, SW480, LOVO, and DLD1 cells, the gene region queried is completely methylated. C, representative results of MSP analysis for SOX17 hypermethylation in human normal colon, adenomas, and stage I to III CRC. SOX17 is not methylated in normal colon but is frequently methylated in adenomas and in different stages of CRC. D, representative results of SOX17 methylation analysis in human esophageal squamous cancer (EC) and non–small cell lung cancer (NSCLC).
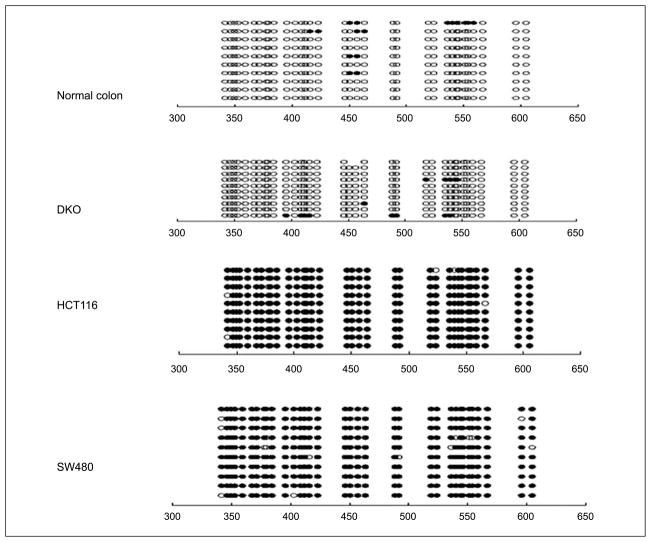
Hypermethylation of the SOX17 promoter region was not limited to colorectal neoplasms. We observed SOX17 methylation in 17 of 19 (89%) esophageal squamous cancers and 6 of 12 (50%) non–small cell lung cancer samples (Fig. 2D). To confirm all of the above data and especially the findings for normal versus tumor DNA, we did sodium bisulfite sequencing on selected samples. We observed that the SOX17 promoter CpG island is not hypermethylated in normal colon and DKO cells, whereas HCT116 and SW480 cells are completely hypermethylated (Fig. 3).
Figure 3.
Bisulfite genomic DNA sequencing results of SOX17 in normal colon, DKO, HCT116, and SW480 cells. ○, unmethylated CpG sites; ●, methylated CpG sites. Locations of CpG sites are given relative to the transcription start site.
Restoration of SOX17 expression suppresses tumor cell growth
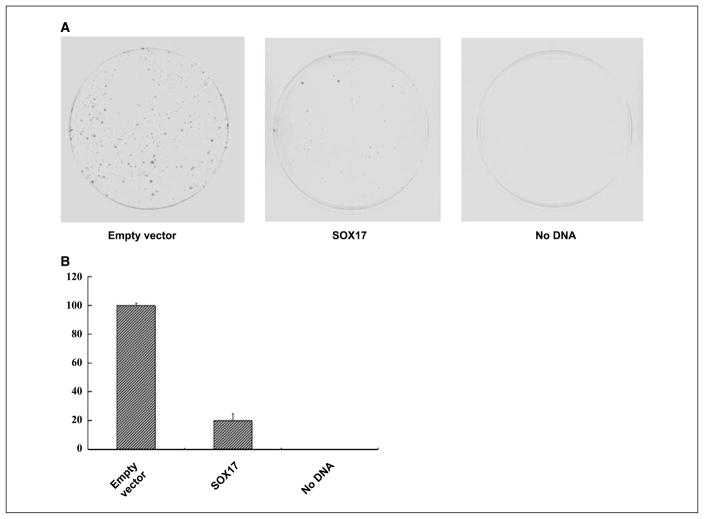
To address the functional significance of SOX17 gene silencing, we tested its ability to suppress tumor growth in HCT116 cells, in which endogenous expression is silenced by DNA hypermethylation. Colony formation assays were carried out following transient transfection of a vector with the cDNA for wild-type SOX17 (pcDNA3.1-SOX17) or the corresponding empty vector into HCT116 cells. The cells were selected in the presence of 400 μg/mL G418, and the number of colonies was counted 10 days later. As shown in Fig. 4A and B, reexpression of SOX17 markedly decreased colony formation, which is consistent with a similar assay done by overexpressing mouse Sox17 in SW480 cells (12). Furthermore, seven surviving clones transfected with SOX17 expression plasmids were randomly selected for subculture. Western blots indicate that there is no detectable SOX17 protein among these colonies (data not shown). These results reveal that SOX17 can inhibit colony growth in CRC cells.
Figure 4.
Suppression of cancer cell growth by SOX17. A, expression vectors encoding wild-type SOX17 or empty control vectors were transfected into HCT116 cells, which were then selected for G418 resistance. After 10 d, the cells were fixed with 10% formaldehyde and stained with Giemsa. B, quantitative analysis of surviving colonies after G418 selection. Each experiment was repeated three times and the average number of colonies is indicated with error bars on the histogram.
SOX17 inhibits β-catenin/TCF–driven transcription
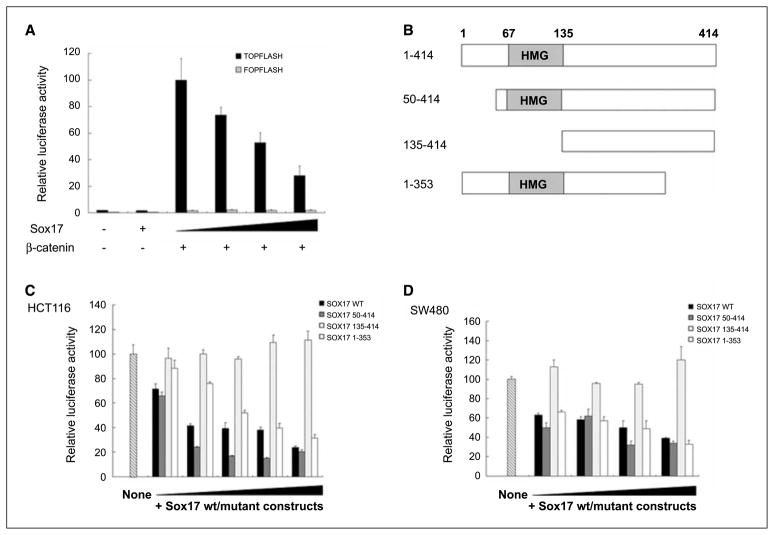
Previous studies have shown that several members of the Sox gene family can antagonize the canonical Wnt signaling pathway (10, 12, 21, 22). For Sox17, the mechanisms identified, to date, includes the following: (a) Xenopus Sox17β protein competes with TCF/LEF for β-catenin binding (10), and (b) mouse Sox17 induces degradation of both β-catenin and TCF/LEF in SW480 cells (12). We tested the ability of SOX17 to repress Wnt signaling in the context of our findings. To examine whether SOX17 can inhibit wild-type β-catenin–mediated Wnt activation, HEK293T cells were transfected with the TOPFLASH or FOPFLASH reporter constructs, concomitantly with a wild-type β-catenin expression vector and increasing amounts of pcDNA3.1-SOX17. The TOPFLASH transfection plasmid contains three consensus TCF binding sites fused to a minimal FOS promoter and firefly luciferase gene. The FOPFLASH plasmid is identical except that the TCF binding site sequences are mutated to serve as a negative control (23). Wild-type β-catenin expression induced a 60-fold activation of TOPFLASH activity, which can be repressed by overexpression of SOX17 in a dose-dependent manner (Fig. 5A). In contrast, SOX17 did not affect transcription from the plasmid FOPFLASH (Fig. 5A). We also examined whether SOX17 could inhibit β-catenin/TCF activity in CRC cells that have a high basal transcriptional activity driven by this protein complex as measured by the TOPFLASH assay (23, 24). In keeping with our observation in HEK293T cells, inhibition of TOPFLASH reporter activity by SOX17 was seen in HCT116 cells (Fig. 5C), which contain the high basal activity driven by an endogenously mutated β-catenin gene (24).
Figure 5.
SOX17 inhibits Wnt stimulated transcription. A, SOX17 inhibits wild-type β-catenin–activated transcription. HEK293T cells were transfected with 70 ng of TOPFLASH or FOPFLASH plasmids, 7 ng of pRL-TK, and increasing amounts of pcDNA3.1-SOX17 or empty vector control (0, 30, 50, and 100 ng; black triangle, increasing dose), and stimulated for 48 h with cotransfection of 70 ng wild-type β-catenin expression vectors. The results are normalized to those for empty control vectors and are expressed as a relative ratio of firefly luciferase to Renilla luciferase. Bars, +1 SD. B, deletion analysis of SOX17. Individual SOX17 deletion mutants are depicted. The HMG box of SOX17 is shown in schematic form. C, SOX17 inhibits endogenous TCF/β-catenin–mediated transcription through the NH2-terminal HMG box. HCT116 cells were transfected with 70 ng of TOPFLASH, 7 ng of pRL-TK, and increasing amounts of pcDNA3.1-SOX17 or the different deletion mutants (SOX17 constructs 50–414, 135–414, and 1–353) or empty vectors (2, 10, 30, 50, and 100 ng; black triangle, increasing dose). Transfection of TOPFLASH reporter vectors reveals that HCT116 cells have high levels of endogenous β-catenin/TCF transcription activity (lane marked “None,” and set at 100% for normalization of data in all other lanes). Columns, mean of three independent experiments; bars, SD. D, a similar suppression effect mediated by the SOX17 HMG box is observed in SW480 cells. The SW480 cells were transfected with 10, 30, 50, or 100 ng SOX17 expression constructs, respectively. Data are expressed relative to the high basal activity in nontransfected cells as in C.
To better understand the mechanisms by which human SOX17 represses β-catenin/TCF activity, segments of SOX17 that are necessary for inhibition were mapped by structure-based deletion analysis (Fig. 5B). An NH2-terminal deletion (yielding SOX17 fragment 50–414) did not affect the inhibitory function of the protein at all (Fig. 5C). The deletion of the HMG domain (yielding SOX17 fragment 135–414) led to a complete loss of repression of TCF/β-catenin activity (Fig. 5C), which is consistent with the result of reporter gene assays acquired in studies of the Xenopus Sox17β protein (10). In contrast to a recent study that suggests that the COOH terminus of mouse Sox17 is also required for the repressive ability on β-catenin/TCF activity, our COOH-terminal deletion construct (human SOX17 fragment 1–353) exhibited a similar repression effect as wild-type SOX17 expression vector. Interestingly, alignment of mouse and human SOX17 protein sequences reveals that they share the identical HMG box located in the NH2 terminus (residue 67–135), and that the COOH-terminal fragments are strongly homologous. Previous studies suggest that Xenopus Sox17β and mouse Sox17 have a β-catenin interaction domain at their COOH termini (8, 10). Because the deletion construct (SOX17 1–353) does not contain this domain, our results suggest that the repressive function of SOX17 does not depend on β-catenin binding. The inhibitory capability of the SOX17 HMG domain on TOPFLASH reporter transcription was also seen in SW480 cells (Fig. 5D), which harbor a mutant APC gene (24). These data collectively show that SOX17 can inhibit canonical Wnt signaling triggered by mutations in either APC or β-catenin, and that the HMG domain is required for this effect.
Discussion
It is now well established that loss of proper gene function by gene silencing contributes heavily to CRC initiation and progression. In this regard, several important Wnt signaling inhibitors such as SFRPs, WIF-1, and DKK-1 have been previously reported to be frequently hypermethylated in primary colorectal tumors (14–16). Our results suggest that epigenetic silencing of SOX17 is another important step in activation or amplification of aberrant Wnt signaling in CRC. This conclusion is supported by our observations that (a) methylation of SOX17 is strongly associated with the loss of gene expression in seven CRC cell lines; (b) SOX17 is unmethylated in normal colon but methylated with high frequency in a variety of human primary tumors including CRC; (c) methylation of SOX17 occurs frequently in premalignant colonic neoplasms; (d) restoration of SOX17 function reduces colony formation in colon cancer cells; and (e) SOX17 can efficiently suppress both wild-type and mutant β-catenin–mediated transcriptional activity. These findings suggest that SOX17 hypermethylation may play an important role in the early steps of cancer formation.
Our current findings for SOX17 make yet another addition of epigenetic inactivation events to the growing list of DNA hypermethylated Wnt antagonist genes in colon and other cancers. The question arises as to why so many potentially redundant steps for Wnt pathway activation would simultaneously be present in a single tumor. The answer may be 2-fold, as we and others have postulated (14, 25, 26). First, that each of these individual gene epigenetic inactivation steps alone may be less potent than single mutations for driving the Wnt pathway and that they are required to summate to yield the full epigenetic drive for tumorigenesis. Second, this summation may be additionally necessary to give Wnt pathway mutations their full effect to help drive abnormal activation of the Wnt pathway. In this regard, silenced Wnt antagonist genes can be divided into three broad classes, each contributing to individual steps in amplifying the effects of increasing nuclear β-catenin function as the final readout for the active Wnt pathway. The first class, including the secreted SFRPs, WIF-1, and DKK, acts at the level of the cell membrane to prevent ligand-receptor interactions. The inactivation of this class, as we and others have shown for SFRPs (14, 27, 28), can up-regulate the Wnt pathway at the cell membrane and this leads to increased cellular levels of β-catenin. When these increases meet a crippled cytoplasmic degradation complex for this protein, such as in colon cancer cells with APC mutations, or when this leads to increased levels of mutant β-catenin, which can evade the complex, then more β-catenin reaches the nucleus to transcriptionally drive Wnt pathway target genes (23, 24). The second class comprises certain members of the cytoplasmic degradation complex for β-catenin and the example here is the APC gene. Thus, APC promoter hypermethylation is an alternative mechanism to mutations for inactivation of this key gene in colon cancer development (29) and can, especially in the setting of inactivation of the above membrane Wnt antagonists, result in Wnt nuclear activation. Finally, we now show epigenetic inactivation of another class of Wnt antagonist. Certain nuclear proteins, including SOX17, inhibit Wnt signaling at the level of the nuclear complex between β-catenin and TCF. Inactivation of SOX17 is then added to abnormal activation of the pathway by stabilizing and/or facilitating this key Wnt-driven transcription complex. In summary, we hypothesize that simultaneous epigenetic down-regulation of SFRPs, WIF-1, DKK, and SOX17, especially in the setting of key pathway mutations, act in a complementary manner for the constitutive activation of Wnt signaling, which can drive tumor initiation and progression.
Our detection of SOX17 hypermethylation now adds this gene to the repertoire of DNA hypermethylated genes that may potentially be used as new molecular markers for cancer detection and/or risk. The high frequency of SOX17 methylation in 50% of non–small cell lung cancers and nearly 90% of esophageal squamous cancers further supports this possibility and is consistent with the known role of aberrant activation of Wnt signaling during tumorigenesis for multiple cancer types (30, 31). Given that APC inactivation or β-catenin mutations rarely occurs in these tumor types other than CRC (32, 33), our findings suggest that SOX17 maybe have wide-ranging importance for tumor suppression.
Our present results and those from other groups all indicate that negative regulation of β-catenin/TCF transcription activity is an important function of Sox17 (10, 12). However, how the full range of mechanisms underlying SOX17 repression of Wnt activity in CRC and other cancers must still be determined. Three Sox proteins in Xenopus embryos (Sox17α, Sox17β, and Sox3) have been reported to inhibit Wnt signaling by directly competing with TCF/LEF for β-catenin binding (10). Most recently, another model based on studies mainly in SW480 colon cancer cells suggests that mouse Sox17 antagonizes Wnt signaling by physically associating with TCF/LEF (via the NH2-terminal HMG) and β-catenin (via the COOH terminus) to promote the protein degradation of both TCF/LEF and β-catenin (12). In contrast to the two models discussed above, our structure-based deletion analysis data have clearly shown that only the NH2-terminal HMG box of SOX17 is required for inhibition of TCF/β-catenin transcriptional activity, whereas the COOH-terminal β-catenin binding domain may not be heavily required for this inhibition. The discrepancy between our data and other studies may be due to different Sox17 genes from different species used in the experimental systems.
Although the HMG box of Sox family proteins is a conserved motif for minor-groove DNA recognition, accumulating studies suggest that it is also an important domain involved in protein-protein interaction. For instance, HBP1 can down-regulate Wnt signaling by blocking the DNA-binding ability of TCF4 protein via its HMG box (34). The HMG domain of SOX8 and SOX10 can interact with numerous transcription factors through the COOH-terminal part of the HMG box, including homeodomain proteins Meox1 and Pax6 and the zinc-finger protein Hivep1 (35). Importantly, structural biology studies show that SOX2 and Oct1 bind to adjacent sites on DNA, and interact with each other through the COOH-terminal region of the HMG box in SOX2 and the POU domain in Oct1 to regulate transcription synergistically (36). Furthermore, mouse Sox17 can physically interact with TCF/LEF family members, including TCF3, TCF4, and LEF1, and this interaction is mediated by their respective HMG domains (12). In light of all of the above results, we speculate that SOX17 could physically interact with TCF/LEF or yet to be identified protein(s) via its HMG box, which can affect TCF/LEF DNA binding–dependent transcription, thereby repressing Wnt signaling target transcription in vivo. Thus, an important aspect of future studies is to investigate the DNA elements on endogenous SOX17 target gene promoters through which SOX17 can repress transcription. Understanding the molecular mechanisms underlying tumor growth control by SOX17 may improve our ability to provide diagnostic, prognostic, and treatment paradigms for cancer patients.
Acknowledgments
Grant support: National Institute of Environmental Health Sciences grant ES011858 and National Cancer Institute grant CA043318 (S.B. Baylin), and the American Surgical Association Fellowship Award and the Richard Ross Clinician Scientist Award (N. Ahuja).
We thank T. Chan, J. Licchesi, E. Greene, Y. Cai, L. Liang, J. Ohm, Y. Huang, M. Dhir, and M. Riojas for helpful discussions and technical advice; K. Bender for manuscript preparation and submission; and B. Vogelstein for providing the TOPFLASH and FOPFLASH reporter constructs.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Gubbay J, Collignon J, Koopman P, et al. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–50. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 2.Wegner M. From head to toes: the multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–20. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sohn J, Natale J, Chew LJ, et al. Identification of Sox17 as a transcription factor that regulates oligodendrocyte development. J Neurosci. 2006;26:9722–35. doi: 10.1523/JNEUROSCI.1716-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsui T, Kanai-Azuma M, Hara K, et al. Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci. 2006;119:3513–26. doi: 10.1242/jcs.03081. [DOI] [PubMed] [Google Scholar]
- 5.Park KS, Wells JM, Zorn AM, Wert SE, Whitsett JA. Sox17 influences the differentiation of respiratory epithelial cells. Dev Biol. 2006;294:192–202. doi: 10.1016/j.ydbio.2006.02.038. [DOI] [PubMed] [Google Scholar]
- 6.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130:470–83. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanai Y, Kanai-Azuma M, Noce T, et al. Identification of two Sox17 messenger RNA isoforms, with and without the high mobility group box region, and their differential expression in mouse spermatogenesis. J Cell Biol. 1996;133:667–81. doi: 10.1083/jcb.133.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinner D, Rankin S, Lee M, Zorn AM. Sox17 and β-catenin cooperate to regulate the transcription of endodermal genes. Development. 2004;131:3069–80. doi: 10.1242/dev.01176. [DOI] [PubMed] [Google Scholar]
- 9.Kanai-Azuma M, Kanai Y, Gad JM, et al. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–79. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 10.Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 α/β and XSox3 physically interact with β-catenin. Mol Cell. 1999;4:487–98. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Asakura M, Inoue H, et al. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:3859–64. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinner D, Kordich JJ, Spence JR, et al. Sox17 and Sox4 differentially regulate {β}-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27:7802–15. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancho E, Batlle E, Clevers H. Signaling pathways in intestinal development and cancer. Annu Rev Cell Dev Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–22. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 15.Taniguchi H, Yamamoto H, Hirata T, et al. Frequent epigenetic inactivation of Wnt inhibitory factor-1 in human gastrointestinal cancers. Oncogene. 2005;24:7946–52. doi: 10.1038/sj.onc.1208910. [DOI] [PubMed] [Google Scholar]
- 16.Aguilera O, Fraga MF, Ballestar E, et al. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene. 2006;25:4116–21. doi: 10.1038/sj.onc.1209439. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Goodman SN, Galizia G, et al. Counting alleles to predict recurrence of early-stage colorectal cancers. Lancet. 2002;359:219–25. doi: 10.1016/S0140-6736(02)07448-2. [DOI] [PubMed] [Google Scholar]
- 18.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhee I, Bachman KE, Park BH, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 20.Katoh M. Molecular cloning and characterization of human SOX17. Int J Mol Med. 2002;9:153–7. [PubMed] [Google Scholar]
- 21.Takash W, Canizares J, Bonneaud N, et al. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–83. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melichar HJ, Narayan K, Der SD, et al. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–3. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 23.Korinek V, Barker N, Morin PJ, et al. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–7. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 24.Morin PJ, Sparks AB, Korinek V, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 25.Taketo MM. Shutting down Wnt signal-activated cancer. Nat Genet. 2004;36:320–2. doi: 10.1038/ng0404-320. [DOI] [PubMed] [Google Scholar]
- 26.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 27.Zou H, Molina JR, Harrington JJ, et al. Aberrant methylation of secreted frizzled-related protein genes in esophageal adenocarcinoma and Barrett’s esophagus. Int J Cancer. 2005;116:584–91. doi: 10.1002/ijc.21045. [DOI] [PubMed] [Google Scholar]
- 28.Qi J, Zhu YQ, Luo J, Tao WH. Hypermethylation and expression regulation of secreted frizzled-related protein genes in colorectal tumor. World J Gastroenterol. 2006;12:7113–7. doi: 10.3748/wjg.v12.i44.7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–71. [PubMed] [Google Scholar]
- 30.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Powell SM, Papadopoulos N, Kinzler KW, Smolinski KN, Meltzer SJ. APC gene mutations in the mutation cluster region are rare in esophageal cancers. Gastroenterology. 1994;107:1759–63. doi: 10.1016/0016-5085(94)90818-4. [DOI] [PubMed] [Google Scholar]
- 33.Choi YW, Heath EI, Heitmiller R, Forastiere AA, Wu TT. Mutations in β-catenin and APC genes are uncommon in esophageal and esophagogastric junction adenocarcinomas. Mod Pathol. 2000;13:1055–9. doi: 10.1038/modpathol.3880194. [DOI] [PubMed] [Google Scholar]
- 34.Sampson EM, Haque ZK, Ku MC, et al. Negative regulation of the Wnt-β-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–11. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–44. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams DC, Jr, Cai M, Clore GM. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2. Hoxb1-DNA ternary transcription factor complex. J Biol Chem. 2004;279:1449–57. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]