Abstract
The genome of Lactococcus lactis encodes a single long chain 3-ketoacyl-acyl carrier protein synthase. This is in contrast to its close relative, Enterococcus faecalis, and to Escherichia coli, both of which have two such enzymes. In E. faecalis and E. coli one of the two long chain synthases (FabO and FabB, respectively) has a role in unsaturated fatty acid synthesis that cannot be satisfied by FabF, the other long chain synthase. Since L. lactis has only a single long chain 3-ketoacyl-acyl carrier protein synthase (annotated as FabF), it seemed likely that this enzyme must function both in unsaturated fatty acid synthesis and in elongation of short chain acyl carrier protein substrates to the C18 fatty acids found in the cellular phospholipids. We report that this is the case. Expression of L. lactis FabF can functionally replace both FabB and FabF in E. coli, although it does not restore thermal regulation of phospholipid fatty acid composition to E. coli fabF mutant strains. The lack of thermal regulation was predictable because wild type L. lactis was found not to show any significant change in fatty acid composition with growth temperature. We also report that overproduction of L. lactis FabF allows growth of an L. lactis mutant strain that lacks the FabH short chain 3-ketoacyl-acyl carrier protein synthase. The strain tested was a derivative (called the ΔfabH bypass strain) of the original fabH deletion strain that had acquired the ability to grow when supplemented with octanoate. Upon introduction of a FabF overexpression plasmid into this strain, growth proceeded normally in the absence of fatty acid supplementation. Moreover, this strain had a normal rate of fatty acid synthesis and a normal fatty acid composition. Both the ΔfabH bypass strain that overproduced FabF and the wild type incorporated much less exogenous octanoate into long chain phospholipid fatty acids than did the ΔfabH bypass strain. Incorporation of octanoate and decanoate labeled with deuterium showed that these acids were incorporated intact as the distal methyl and methylene groups of the long chain fatty acids.
Introduction
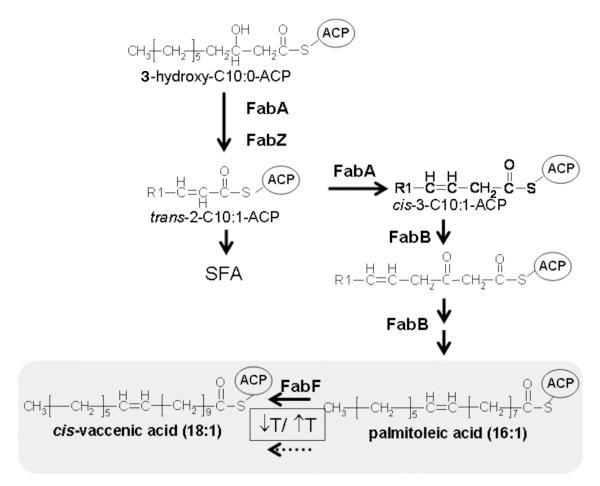
Although Lactococcus lactis synthesizes a mixture of phospholipid fatty acid moieties similar to that made by Escherichia coli, the genome encodes only a single long chain 3-ketoacyl-acyl carrier protein (ACP) synthase (KAS) (Bolotin et al. 2001) whereas E. coli encodes two such enzymes, both of which are required for a normal cellular fatty acid composition (de Mendoza and Cronan 1983; Campbell and Cronan 2001). In E. coli the two long chain KAS enzymes (along with the FabA dehydratase/isomerase) are required for synthesis of the normal complement of UFAs, although either enzyme will support saturated fatty acid synthesis (de Mendoza and Cronan 1983; Campbell and Cronan 2001). KAS I (encoded by fabB) is believed to be required for elongation of the 10-carbon cis-unsaturated intermediate formed in the first step of the UFA biosynthetic pathway (Fig. 1) whereas KAS II (encoded by fabF) is required for elongation of palmitoleate, the C16 monounsaturated acid, to cis-vaccenate, the C18 monounsaturated acid. Thus fabB mutants are unsaturated fatty acid auxotrophs whereas fabF mutants lack cis-vaccenate and are unable to regulate the ratio of unsaturated to saturated fatty acids in the cellular lipids (de Mendoza and Cronan 1983; Campbell and Cronan 2001).
Fig. 1.
The anaerobic pathway of unsaturated fatty acid biosynthesis in E. coli. The 3-hydroxylacyl-ACP intermediate is dehydrated by either FabA or FabZ. FabA catalyzes the first committed step in UFA synthesis, the isomerization of the double bond of the 10-carbon intermediate. 3-Ketoacyl-ACP synthase I (KASI or FabB) is required for condensation of malonyl-ACP with the cis-3-C10:1-ACP. Further elongation of the unsaturated fatty acyl-ACP intermediates can be carried out by either FabB or FabF (KAS II) to produce a 16-carbon monounsaturated fatty acid, palmitoleate. Elongation of 16:1 to 18:1 is catalyzed by KAS II. However, at high temperatures (42 °C), KAS II is inactive and palmitoleate accumulates. The major UFA species are shaded. The R1 group is CH3-(CH2)5.
The close relative of L. lactis, E. faecalis also has a fatty acid composition similar to that of E. coli, but contains two genes that encode long chain KAS proteins, one of which can replace fabB function in E. coli whereas the other cannot (Wang and Cronan 2004). These comparisons suggested the possibility that the single long chain KAS of L. lactis (annotated as a FabF homologue) might have the functions of both FabB and FabF proteins. We have tested this possibility by complementation of E. coli strains lacking functional FabB and FabF proteins. We also tested if, upon overproduction, the L. lactis FabF protein could functionally replace FabH (KAS III), an enzyme required for initiation of fatty acid synthesis. We report that indeed, expression of L. lactis FabF remedies each of these enzymatic defects.
MATERIALS AND METHODS
Plasmids, Bacterial Strains and Growth
All E. coli K-12 and L. lactis subspecies lactis IL1403 strains as well as the plasmids used in this study are given in Table 1. E. coli strains were typically grown in Luria-Bertani medium (Lai and Cronan 2003). L. lactis strains were grown in M17 medium (Difco) supplemented with 0.5% glucose. The fabH bypass strain, CL114, was grown in M17 supplemented with either octanoate or oleate at a concentration of 100 μM. Antibiotics were used in E. coli at the following final concentrations (in μg/mL): sodium ampicillin, 100; and erythromycin sulfate, 200. In L. lactis the erythromycin concentrations were 5 μg/ml for plasmid borne determinants. All fatty acid supplements were solubilized in Tergitol NP-40 and used at a concentration of 100 μM unless stated otherwise. Arabinose was added at a final concentration of 0.05%.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid |
Relevant Characteristicsa | Reference |
|---|---|---|
| E. coli strains | ||
| CY244 | fabB(Ts)fabF1 | (de Mendoza et al. 1983) |
| GRT23 | ΔfabB::cml Δcfa::kan | G. Tulik (University of Illinois) |
| L. lactis strains | ||
| IL1403 | Wild type | (Bolotin et al. 2001) |
| CL114 | ΔfabH bypass | (Lai and Cronan 2003) |
| Plasmids | ||
| pORI23 | EmR; shuttle-expression vector | (Que et al. 2000) |
| pBAD24 | AmpR, expression vector | (Guzman et al. 1995) |
| pRK51 | EmR, pORI23 carrying L. lactis fabF | This work |
| pRK52 | AmpR, pBAD24 carrying L. lactis fabF | This work |
The abbreviations are; cml, chloramphenicol resistance; kan, kanamycin resistance. EmR, erythromycin resistance and AmpR, ampicillin resistance
The cyclopropane fatty acid, cis-9,10-methylenehexadecanoic acid, was purchased from Larodan Fine Chemicals (Malmo, Sweden). Deuterated fatty acids were purchased from Cambridge Isotope laboratories Inc. (Andover, MA). Radiolabeled substrates were obtained from American Radiochemicals. Unless otherwise indicated, all other chemicals were purchased from Sigma.
Plasmid construction
The annotated fabF gene was amplified from L. lactis genomic DNA (Bolotin et al. 2001) with primer sets pOR-fabF-L (GGATCC AATTCCATATGACAAATAGA- note: double underline indicate engineered ribosomal binding site) and pOR-fabF-R (GTTCATCTGCAGTTACTCTCCAGTCCA), or BAD-fabF-L (TAAGAGGAGGATCCTATGACAAATAGA) and BAD-fabF-R (GTTCATTCTAGATTACTCTCCAGTCCA) using PfuTurbo polymerase (Stratagene), and cloned into pCR2.1 (Invitrogen) to give plasmids pRK49 and pRK50, respectively. Both plasmids were sequenced to verify fidelity (Keck Genomics Center, University of Illinois). pRK49 was digested with the restriction enzymes BamHI and PstI and ligated into the Lactococcus expression-shuttle vector pOri23 (Que et al. 2000) cut with the same enzymes to produce pRK51. Plasmid pRK50 was digested with the restriction enzymes EcoR1 and XbaI and the fabF fragment was ligated into E. coli expression vector pBAD24 cut with the same enzymes to produce the plasmid pRK52.
AATTCCATATGACAAATAGA- note: double underline indicate engineered ribosomal binding site) and pOR-fabF-R (GTTCATCTGCAGTTACTCTCCAGTCCA), or BAD-fabF-L (TAAGAGGAGGATCCTATGACAAATAGA) and BAD-fabF-R (GTTCATTCTAGATTACTCTCCAGTCCA) using PfuTurbo polymerase (Stratagene), and cloned into pCR2.1 (Invitrogen) to give plasmids pRK49 and pRK50, respectively. Both plasmids were sequenced to verify fidelity (Keck Genomics Center, University of Illinois). pRK49 was digested with the restriction enzymes BamHI and PstI and ligated into the Lactococcus expression-shuttle vector pOri23 (Que et al. 2000) cut with the same enzymes to produce pRK51. Plasmid pRK50 was digested with the restriction enzymes EcoR1 and XbaI and the fabF fragment was ligated into E. coli expression vector pBAD24 cut with the same enzymes to produce the plasmid pRK52.
Preparation of L. lactis electrocompetent cells and electroporation parameters
L. lactis cultures were prepared for electroporation essentially according to McIntyre and Harland (1989), with some modifications. Standing cultures were grown overnight at 30 °C in SGM17 medium which consisted of M17 broth (Terzaghi and Sandine 1975) supplemented with 0.3 M sucrose, 0.5% (w/v) glucose. The overnight culture was diluted 50-fold in 100 mL of SGM17 medium supplemented with either 0.5% or 0.2% glycine in wild type strain IL1403 or strain CL114, respectively. The cultures were grown at 30°C at a slow shaking speed (150 rpm) for 3 to 5 hours until an OD600~0.7. The cells were harvested and washed three times in 25 mL of ice-cold 0.5 M sucrose containing 10% glycerol, then resuspended in 2 mL of the sucrose-glycerol solution and centrifuged for 3 min at 12,000 xg at 4 °C. The pellet was then resuspended in 0.5 mL of the above buffer.
Purified plasmid DNA (200-1000 ng) was added to 50 μL of freshly prepared electrocompetent cells, and the samples were transferred to a 0.2 mm gap cuvette (Biorad). A voltage was of 2.25 kV was applied from a Biorad Gene Pulser. The time constant varied from 4.7-5.3 ms. Following electroporation, cells were allowed to recover for 2 h at 30 °C in 1 mL of the SGM17 medium with additionally supplemented with 2 mM CaCl2, 20 mM MgCl2, 50 ng/ml erythromycin, and 100 μM oleic acid. Following recovery, the cells were recovered by centrifugation and then plated on SGM17 agar plates supplemented with 100 μM oleic acid and erythromycin at either 3 or 5 μg/mL as given above.
Analysis and Extraction of Phospholipids
For analysis of phospholipid contents cultures were typically grown to mid-log phase, washed three times in unsupplemented growth medium and resuspended to an OD~0.1 in 5 mL of growth media supplemented with various fatty acid substrates (100 μM) and typically allowed to grow overnight. Phospholipids were typically extracted from overnight cultures grown in either LB (E. coli) or GM17 (L. lactis), unless stated otherwise. Total membrane lipids were extracted according to Lai and Cronan (2003) using the method of Bligh and Dyer (1959). The extracted lipids were dried under nitrogen and concentrated 50-fold in chloroform:methanol (1:2), and the mass spectra were generated via collision-induced dissociation electrospray mass spectrometry on a VG Quattro spectrometer at a cone voltage of 90 V. This cone voltage resulted in complete degradation of the phospholipids into the head groups and fatty acid components, and mass ranges from 100 to 500 m/z were typically collected. Fatty acids were identified based on their molecular masses (Sweetman et al. 1996).
Fatty acid methyl ester extraction and argentation thin layer chromatography
For the purposes of analysis of fatty acid methyl esters (FAME) by thin layer chromatography, 5 mL cultures were grown overnight as described above in the presence of either [1-14C] sodium acetate (5 μCi) or various [14C]-labeled fatty acid substrates. Samples were washed in cold growth medium 3 times prior to phospholipid extraction. Extracted phospholipids were transesterified to their methyl esters using the base-catalyzed method of Christie (2003)as described by Lai and Cronan (2003). The methyl esters were separated by silver nitrate chromatography on Analtech Silica Gel GHL plates impregnated with 20% AgNO3. Plates were developed twice in 100% toluene at −20°C. Radiolabeled methyl esters were visualized by autoradiography or by phosphorimager (Amersham Biosciences). Note that transesterification will not convert free fatty acids to their methyl esters and thus only phospholipid-bound acids were analyzed.
RESULTS
Functions of L. lactis FabF deduced from expression of the protein in E. coli fabB and fabF mutant strains
To test the ability of L. lactis to functionally replace the long chain KAS enzymes of E. coli we used two mutant strains, strain GRT23 which carries a fabB null mutation, and strain CY244, which carries a fabF null mutation plus a temperature-sensitive fabB mutation (Ulrich et al. 1983). As expected from prior data strain GRT23 is nonviable in the absence of long chain unsaturated fatty acids (Fig. 2A) whereas strain CY244 is nonviable at 42°C due to the lack of both KAS I and KAS II activities resulting in the failure of UFA supplementation to support growth (Fig. 2B). The lack of growth results because the double mutant strain is unable to elongate any nascent fatty acyl chains produced by FabH to the chain lengths required for synthesis of essential membrane lipids (Garwin et al. 1980; Ulrich et al. 1983; Jackowski et al. 1989). Therefore, even in the presence of UFA supplementation these strains fail to grow because they are unable to synthesize the saturated fatty acid chains required for phospholipids and the essential lipid A component of the outer membrane.
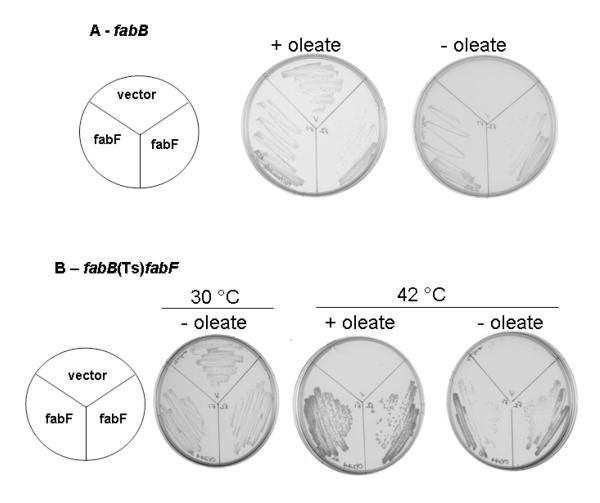
Fig. 2.
Complementation by L. lactis fabF of E. coli strains carrying either a fabB deletion (panel A) or mutations in both long chain KAS encoding genes (panel B). Panel A. The fabB strain GRT23 carrying either pBAD24 or pRK52 (pBAD24 carrying L lactis fabF) was grown on LB plates at 37°C for 16 h in the presence and absence of 100 μM oleate. Panel B. The E. coli fabB(Ts) fabF strain CY244 carrying pRK52 was grown for 16 h on LB plates supplemented with 0.02% arabinose at either 30°C or 42°C in the presence or absence of oleate (0.1 mM).
Strains GRT23 and CY244 were transformed with plasmid pRK52 which encoded the wild type L. lactis fabF under the control of the pBAD promoter and ribosome binding site of the E. coli expression vector, pBAD24. As expected, the fabB mutant strain (GRT23) carrying the empty pBAD24 vector was unable to grow on plates without exogenously supplied unsaturated fatty acids (Fig. 2A). However, cultures of strain GRT23 carrying the FabF plasmid pRK52 grew overnight on plates in the absence of the fatty acid supplement (Fig. 2A) thereby demonstrating that expression of L. lactis FabF could replace the function of KAS I.
Given the fact that there is only a single recognizable long chain KAS-encoding ORF in L. lactis it seemed probable that L. lactis FabF could replace the functions of both E. coli long chain KAS proteins. This was tested by introducing pRK52 into the E. coli fabB(Ts) fabF strain CY244. As expected the strain carrying the empty pBAD24 vector was nonviable at the nonpermissive growth temperature (42°C) even in the presence of an exogenous unsaturated fatty acid (Fig. 2B). In contrast, cultures of CY244 containing the L. lactis fabF plasmid grew at 42°C in the presence or absence of the UFA supplement (Fig. 2B). However, it should be noted that cultures containing oleic acid grew significantly better than those lacking the fatty acid (Fig. 2B) indicating that in vivo complementation of the fabB lesion by L. lactis FabF was incomplete when E. coli FabF was absent. This weak complementation indicates that L. lactis FabF was not present in significant functional excess.
The function of L. lactis FabF in E. coli KAS mutants was also assayed by measuring de novo fatty acid synthesis by [1-14C]acetate incorporation (Fig. 3) and by analyses of the fatty acid compositions of the membrane phospholipid fatty acids (Table 2). The fabB strain, GRT23, was grown at 37°C in medium supplemented with the cyclopropane fatty acid, cis-9,10-methylenehexadecanoate. The presence of the exogenously added cyclopropane fatty acid satisfied the unsaturated fatty acid auxotrophy of this strain, but because the acid was unlabeled and had a mass significantly different from those of the UFAs, its presence did not interfere with either of the assays for UFA production (the cfa mutation of this strain prevented synthesis of cyclopropane fatty acids by the host). As expected cultures of strain GRT23 carrying the empty pBAD24 vector had no UFA synthetic ability detected by either assay (Fig. 3A, Table 2). In contrast, in cultures carrying the L. lactis fabF plasmid, pRK52, more than 50% of the total membrane phospholipid acyl chains were UFA (Table 2). Similarly, argentation TLC analysis of radiolabeled fatty acids synthesis of both palmitoleate and cis-vaccenate in the absence of induction and greater synthesis of the longer UFA in the presence of inducer (Fig. 3A).
Fig. 3.
Separation of methyl ester of phospholipid acyl chains by argentation TLC in fabB (panel A) or fabB(Ts) fabF double mutant (panel B) strains carrying a plasmid that expressed L. lactis fabF. The cultures were grown overnight at either 37°C (plate A) or 42°C (plate B) in LB medium supplemented with 5 μCi [1-14C]acetate. Cultures received 0.02% arabinose and 100 μM cyclopropane fatty acid (cis-9,10-methylenehexadecanoic acid) as shown. The fabF designation denotes the presence (pRK52) or absence (pBAD24) of the L. lactis fabF gene; ara denotes arabinose and FA denotes oleic acid.
Table 2.
Complementation of E. coli strains carrying mutations in fabB or in both fabB and fabF by expression of L. lactis FabF. The strains were grown overnight at 30°C or 42°C in the presence of 0.02% arabinose.
| Relevant | Growth Temperature °C |
Fatty acid distribution (percent of total by mass) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Phenotype | Plasmid | C14:0 | C16:1 | C16:0 | C18:1 | C18:0 | 16:1/18:1 | SFA/UFA | |
| GRT23a | fabB | vector | 37 | 34.8 | ND | 65.2 | ND | ND | n/a | n/a |
| GRT23a | fabB | pRK52 | 37 | 6.2 | 7.7 | 41.3 | 44.8 | ND | 0.2 | 0.9 |
| CY244 | fabF fabB(Ts) | vector | 30 | 10.9 | 38.8 | 42.3 | 7.8 | <0.1 | 5.0 | 1.1 |
| CY244 | fabF fabB(Ts) | pRK52 | 30 | 5.9 | 13.9 | 2.0 | 74.3 | 3.9 | 0.2 | 0.1 |
| CY244b | fabF fabB(Ts) | vector | 42 | 48.4 | 24.9 | 19.0 | 5.4 | 2.2 | 4.6 | 2.3 |
| CY244 | fabF fabB(Ts) | pRK52 | 42 | 11.1 | 29.5 | 25.4 | 49.7 | 2.6 | 0.6 | 0.5 |
These cultures were supplemented with 0.02% CFA.
This culture ceased growth due to inhibition of fatty acid synthetic ability.
The fatty acid compositions of cultures of the double mutant fabB(Ts) fabF strain carrying the empty pBAD24 vector grown at 30°C showed accumulation of the 16-carbon monounsaturated acid, palmitoleate and a correspondingly high 16:1/18:1 ratio (Table 2). This was expected since KAS I (FabB) was functional at the permissive growth temperature of 30 °C, whereas KAS II (FabF), which catalyzes the elongation of 16:1 to 18:1, was nonfunctional in this strain. In contrast, when cultures were transferred to the non-permissive temperature of 42°C, there was a 2.1-fold increase in the SFA/UFA ratio and, more significantly, an accumulation of short chain fatty acyl chains due to loss of elongation activity. Thus, the double mutant exhibited a severely reduced de novo fatty acid synthetic capacity regardless of the addition of fatty acid supplements (Fig. 3B). When the L. lactis FabF plasmid pRK52 was present in the fabB(Ts) fabF strain the ratio of 16:1/18:1 was 25-fold lower relative to cells carrying the vector plasmid thereby showing that expression of L. lactis FabF replaced E. coli KAS II function (Table 2). Furthermore, at the nonpermissible growth temperature of 42°C, cultures expressing the L. lactis FabF had relatively low 16:1 to 18:1 ratios as well as SFA to UFA ratios. In agreement with the mass spectral data, the level of [1-14C]acetate incorporation into fatty acids was increased in 42°C grown cultures of the fabB(Ts) fabF mutant when the strain carried pRK52 demonstrating an increase in fatty acid synthetic ability (Fig. 3). These results show that at the nonpermissive growth temperature expression of L. lactis FabF replaced the functions of both KAS I and KAS II in E. coli and also that endogenous long chain fatty acid synthesis suppressed elongation of exogenously supplied octanoate.
Expression of L. lactis FabF fails to restore thermal control of fatty acid composition to E. coli FabF mutants
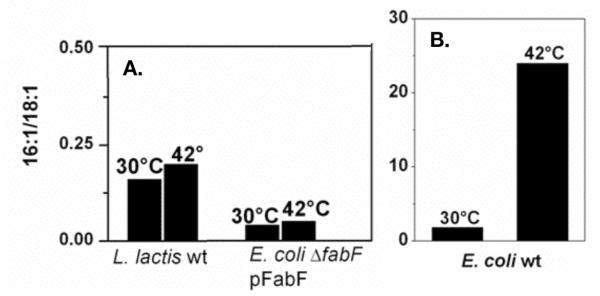
Cultures E. coli grown at higher temperatures exhibit significantly lower levels of UFA and an accumulation of palmitoleic acid due to the intrinsic temperature sensitivity of FabF (de Mendoza and Cronan 1983; de Mendoza et al. 1983; Edwards et al. 1997) (Fig. 1) Similar FabF-dependent regulation has been observed in other bacteria (Kutchma et al. 1999; Allen and Bartlett 2000). To test if L. lactis FabF was able to restore thermal regulation to an E. coli FabF mutant strain, we determined the fatty acid compositions of cultures of a ΔfabF strain expressing L. lactis FabF grown at either 30°C or 42°C. These cultures showed only minimal changes in UFA with growth temperature (Fig. 4) indicating that unlike E. coli FabF, the activity of L. lactis FabF is not regulated by temperature. This finding suggested that L. lactis lacks a mechanism to alter its fatty acid composition with growth temperature and this was confirmed. Cultures of L. lactis grown at 30°C or 42°C had essentially identical fatty acid compositions (Fig. 4).
Fig. 4.
Ratios of palmitoleate to cis-vaccenate in a E. coli fabF strain expressing L. lactis FabF grown at either 30 °C or 42 °C (panel A). The compositions of wild type strains of L. lactis (panel A) and E. coli (panel B) grown at the same temperatures are shown for comparison.
Overexpression of L. lactis FabF complements growth of the L. lactis ΔfabH bypass strain
A mutant strain of L. lactis that lacks the function of FabH, 3-ketoacyl-ACP synthase III was previously reported by our laboratory (Lai and Cronan 2003). Although the ΔfabH strain was unable to grow in the absence of supplementation with long chain fatty acids, it retained about 10% of the normal fatty acid synthetic capacity. This residual synthetic ability was suggested to result from FabF-catalyzed decarboxylation of malonyl-ACP to acetyl-ACP (Lai and Cronan 2003). The acetyl-ACP produced was postulated to act as the primer forming the methyl end of the fatty acid chain in place of the butyryl-ACP normally produced when FabH is present (the long chain KAS enzymes of E. coli are known to accept acetyl-ACP in vitro) (Alberts et al. 1972). A mutant derivative called strain CL114 had previously been selected from the ΔfabH strain (Lai and Cronan 2003). This strain grew, albeit slowly, when supplemented with octanoic acid, although it remained unable to grow in the absence of fatty acid supplementation (Lai and Cronan 2003). In contrast, the original ΔfabH strain showed only barely detectable growth with octanoate (Lai and Cronan 2003). Since strain CL114 retained the ΔfabH lesion, we refer to it as the ΔfabH bypass strain. Growth of the ΔfabH bypass strain was specific to octanoate. Supplementation with a variety of other short (C4-C7) and medium (C9, C10) chain length fatty acids failed to allow growth ((Lai and Cronan 2003), data not shown). We tested whether or not overexpression of L. lactis FabF could functionally replace FabH in L. lactis. For this purpose we constructed a Lactococcus plasmid (pRK51) that expressed L. lactis FabF from a strong constitutive Lactococcus promoter. Although this plasmid was readily transformed into both the L. lactis wild type strain IL1403 and the ΔfabH bypass strain CL114, we were repeatedly unable to introduce plasmid pRK51 into the original ΔfabH strain, CL112. Therefore, we have necessarily limited our analyses to the wild type and ΔfabH bypass strains.
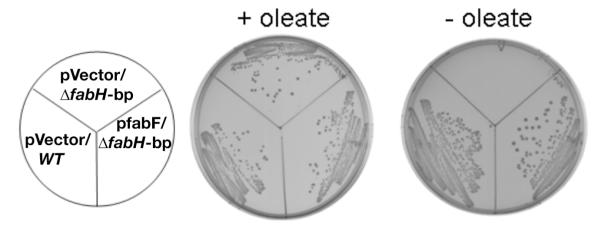
Overexpression of FabF in the ΔfabH bypass strain allowed growth in the absence of fatty acid thereby showing that FabF overproduction functionally replaced FabH (Fig. 5). Indeed, upon overexpression of FabF, the bypass mutant strain grew as well as the wild type strain in the absence of fatty acid. As expected, FabF overexpression restored fatty acid synthesis to the ΔfabH bypass strain as shown by [1-14C]acetate incorporation into phospholipid acyl chains (Fig. 6). A dramatic increase in the rate of fatty acid synthesis was seen compared to the strain carrying the empty vector. Indeed, upon FabF overproduction the ΔfabH bypass strain attained a fatty acid composition that was almost identical to that of the wild type strain as determined by mass spectroscopy (Table 3).
Fig. 5.
Growth of the L. lactis ΔfabH bypass (bp) strain in the presence (pRK51) or absence (vector pOR123) of FabF overexpression. Strains were grown for 36 h at 30°C on GM17 plates supplemented with 5 μg/ml erythromycin and in the presence or absence of 100 μM oleic acid as given.
Fig. 6.
Fatty acid synthesis in the L. lactis ΔfabH bypass strain carrying plasmid pRK51. Cultures were grown to mid log phase at 30 °C in the presence of 5 μCi [1-14C]-acetate and in the presence or absence of 100 μM oleic acid (FA). Fatty acid methyl esters were generated via base-catalyzed hydrolysis of phospholipids and separated by argentation TLC as described in Materials and Methods. The entirety of each sample was loaded on the TLC plate. The abbreviations are: fabF denotes the presence (pRK51) or absence (vector pORI23) of the L. lactis fabF gene and FA denotes oleic acid.
Table 3.
Complementation of the L. lactis ΔfabH bypass strain by FabF overproduction.
| Strain | Phenotype | Plasmid | FA Supple ment* |
Fatty acid distribution (percent total) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 | C16:1 | C16:0 | C18:1 | C18:0 |
16:1 18:1 |
SFA UFA |
||||
| IL1403 | Wild type | Vector | − | 7.9 | 5.0 | 40.1 | 45.4 | 3.9 | 0.22 | 1.1 |
| CL114 | ΔfabH bypass | Vector | − | 5.8 | 20.6 | 26.3 | 15.8 | 31.3 | 1.30 | 1.7 |
| CL114 | ΔfabH bypass | pRK51 | − | 16.0 | 4.2 | 33.2 | 38.5 | 7.6 | 0.11 | 1.3 |
| CL114 | ΔfabH bypass | Vector | + | 6.1 | 14.2 | 16.9 | 17.6 | 13.7 | 0.81 | 1.1 |
| CL114 | ΔfabH bypass | pRK51 | + | 6.0 | 3.4 | 13.8 | 31.4 | 1.0 | 0.11 | 0.60 |
The strains were supplemented with 0.02% CFA. CFA was not included in yield calculations.
Given that overproduction of FabF greatly improved fatty acid synthetic ability of the ΔfabH bypass strain (Fig 6), it seemed possible that the bypass mutation could be a mutationally altered FabF able to use exogenous octanoyl moieties as the primer for fatty acid synthesis. To test this possibility the fabF genes of several independently isolated fabH bypass mutants were amplified using a high fidelity polymerase and the PCR products were directly sequenced. All bypass mutants gave sequences that were identical to the fabF sequence of the wild type L. lactis strain IL1403 (data not shown), indicating that the bypass phenotype was not due to an altered KAS II protein. Moreover, in the original bypass strain CL114 we sequenced the intergenic region between FabF and the upstream gene fabG1 (plus about 200 bp of the 3′ end of fabG1) and found that the sequence obtained was identical to that of wild type strain. These findings are consistent with our further characterization of the ΔfabH bypass strain which indicates that the ability to grow in the presence of octanoate is due to acquisition of a means to assimilate this fatty acid (see below).
Further characterization of the ΔfabH bypass strain
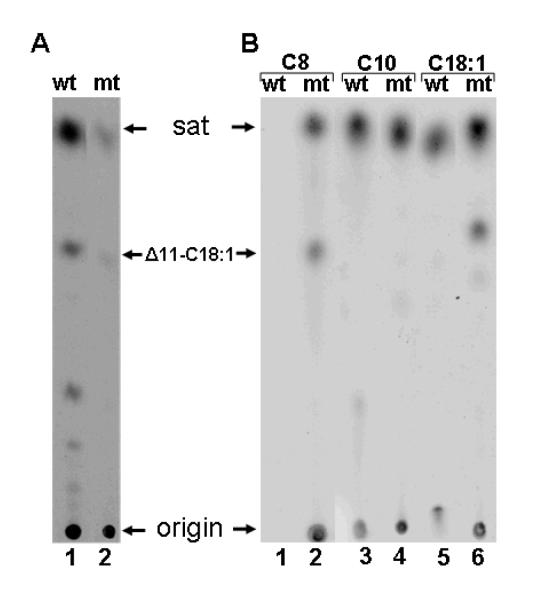
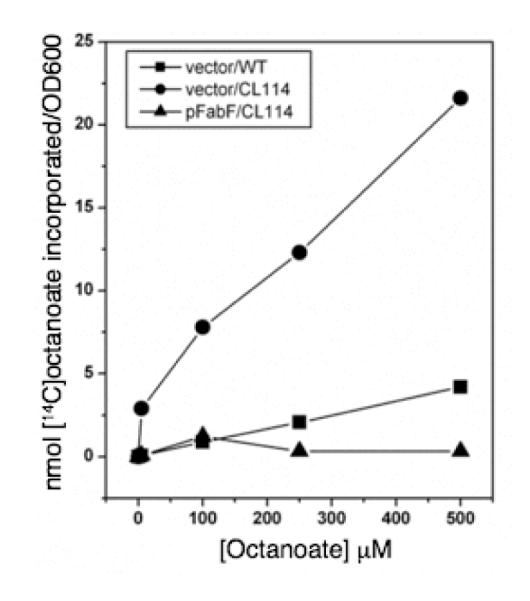
The dependence of growth rate of the ΔfabH bypass strain on octanoate concentration was investigated in liquid cultures of the wild type strain L. lactis 1403 and the fabH bypass strain, CL114 (Fig. 7A). As seen previously (Lai and Cronan 2003) in the absence of octanoate supplementation, the bypass strain grew much more slowly than wild type cultures, and the growth rates of the bypass strain depended directly on the supply of exogenous octanoate up to a concentration of 250 μM (Fig. 7A). A similar pattern was seen in the incorporation of [1-14C]octanoate into the long chain phospholipid fatty acids of the bypass strain (Fig 7B). In contrast, the wild type strain was inhibited by octanoate, exhibiting a reduction in growth rate at even the lowest octanoate concentrations tested (Fig. 7A), and had much lower levels of [1-14C]octanoate incorporation into phospholipid acyl chains (Fig. 7B). Similar growth dependence experiments were also conducted with hexanoate. Addition of this six-carbon fatty acid had no effects on growth of the wild type nor the bypass mutant strains and neither strain incorporated [1-14C]hexanoate into cellular phospholipids (data not shown). These data are consistent with the indications discussed above and previously (Lai and Cronan 2003) that the bypass of the fabH-dependent fatty acid synthesis deficiency is specific to octanoate. The finding that a strain competent for fatty acid synthesis was unable to efficiently incorporate [1-14C]octanoate into cellular lipids suggested that the ΔfabH bypass strain might show a similar phenotype upon FabF overexpression. Indeed, octanoate incorporation by cultures of CL114 carrying the FabF construct pRK51 was as poor as that seen in cultures of the wild type stain IL1403 carrying the vector control (Fig. 8). Therefore, it seems that the presence of a functional fatty acid synthetic pathway prevents exogenous octanoate from efficiently entering the synthetic pathway.
Fig. 7.
Dependence of growth and octanoate incorporation into phospholipid acyl chains on octanoate concentration in the wild type (open circles) and ΔfabH bypass mutant (closed circles) L. lactis strains. Panel A. Cultures were grown in GM17 medium in the presence various concentrations of the short chain fatty acid, octanoate. Growth was monitored as optical density at 600 nm. Panel B. Incorporation of radiolabeled octanoate into the acyl chains of phospholipids of the wild type and ΔfabH bypass mutant. strains Cultures were grown in the presence of variable concentrations of [1-14C]octanoate until mid-log phase. Phospholipids were extracted as described in Materials and Methods and radiolabel incorporation into the lipids was determined by scintillation counting.
Fig. 8.
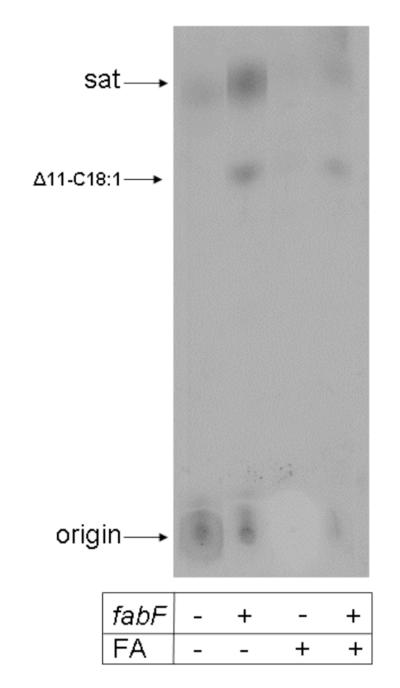
Effects of L. lactis FabF overexpression on the rates of [14C]octanoate incorporation into the phospholipids of the L. lactis wild type and fabH bypass strains. The strains carried either the fabF plasmid pRK51 or the empty vector pOR123.
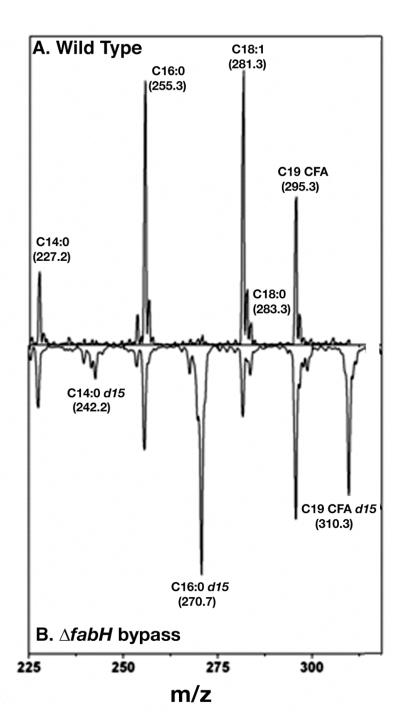
The ΔfabH bypass strain was hypothesized to convert exogenously added octanoic acid to octanoyl-ACP that would become the primer for synthesis of long chain fatty acids (Lai and Cronan 2003). If so, the octanoic acid moiety should be incorporated intact as the last eight carbon atoms of the long chain fatty acids of this strain (the methyl group plus the seven preceding methylene groups). This prediction was tested by supplementation with octanoic-d15 acid followed by mass spectral analysis of the long chain fatty acids of the cellular phospholipids. The bypass mutant strain efficiently incorporated octanoic-d15 acid into each of the cellular fatty acid species as shown by the appearance of species of myristic (C14:0), palmitic (C14:0) and cis-11,12-methyleneoctadecanoic acids (C19 CFA) that had masses 15 mass units greater than the species synthesized de novo (Fig. 9). Since degradation to acetyl-CoA and its reuse to synthesize fatty acids would result in almost complete loss of the deuterium as 2H2O, these data indicated that the octanoic-d15 acid was incorporated intact into the acyl chains. As expected from the radioactive labeling data (Fig. 8), the wild type strain failed to incorporate more than traces of octanoic-d15 acid (Fig. 9). Further evidence that the methyl ends of the fatty acids of the ΔfabH bypass strain are derived from the exogenous fatty acids was obtained by supplementation with decanoic-d19 acid or nonanoic acid (data not shown). Although neither acid could support growth of the ΔfabH bypass strain, decanoic-d19 acid was efficiently incorporated into both the wild type and ΔfabH bypass strains to give saturated fatty acid species of 19 mass units greater than those of the de novo synthesized fatty acids. Acids having masses of 246.4, 274.3 and 302.3 (the masses of d19–substituted myristic, palmitic, and stearic acids) were observed. No peaks for d19–substituted unsaturated fatty acids or their cyclopropane derivatives were observed (data not shown). This was the result expected since decanoic acid is formed after the step in the anaerobic pathway of unsaturated fatty acid synthesis where the cis double bond is introduced (Fig. 1). We also examined incorporation of nonanoic acid which, if incorporated, would give long chain fatty acids having odd numbers of carbon atoms. Although incorporation of this acid was very inefficient, the ΔfabH bypass strain converted nonanoic acid primarily to an acid having a mass of 269.7, that of a C17 saturated fatty acid. Acids having masses of 267.6 and 295.3 were also formed indicating that nonanoic acid was converted to both saturated and unsaturated fatty acids (data not shown). The deuterium labeling experiments were confirmed by labeling the ΔfabH bypass strain with either [1-14C]octanoic acid or [1-14C]decanoic acid (Fig. 10). As expected the C8 fatty acid was incorporated into both saturated and unsaturated species whereas the C10 fatty acid was incorporated only into saturated acids (Fig. 10). Labeling of the ΔfabH bypass strain with [1-14C]acetic acid (Fig. 10) showed the low residual level of fatty acid synthesis previously reported for the ΔfabH strain (Lai and Cronan 2003).
Fig. 9.
Incorporation of octanoic-d15 acid into membrane fatty acids of wild type (panel A) versus ΔfabH bypass mutant (panel B) L. lactis strains. Cultures were grown overnight in GM17 medium supplemented with 100 μM octanoic-d15 acid. Phospholipids were extracted and analyzed via mass spectrometry as described in Materials and Methods. Note that the masses of the C19 CFA lacking isotopic enrichment and cis-vaccenic acid-d15 acid are very similar (295.3 and 296.3, respectively) and thus identification of the peak in this region in the bypass mutant phospholipids is problematical. However, labeling with radioactive octanoate showed incorporation of octanoic acid into cis-vaccenic acid (data not shown).
Fig. 10.
Incorporation of acetate (panel A) or fatty acids (panel B) labeled with 14C in the carboxyl carbon into membrane phospholipids of the wild type L lactis strain IL1403 (wt) or the fabH bypass mutant strain CL114 (mt). The cultures were grown in GM17 medium overnight in the presence of either 5 μCi of [1-14C]-acetate or 100 μM [1-14C]-labeled octanoate, decanoate, or oleate. Methyl esters were transesterified from total phospholipids and separated via argentation TLC as described in Materials and Methods. Approximately 10,000 counts were loaded for each sample, with the exception of two samples (panel A, lane 2; panel B, lane 1) that failed to reach that level of incorporated radioactivity. The greater spot intensity in panel A relative to panel B is due to an increased exposure time of the film (which also increased the background).
Discussion
Expression of L. lactis FabF in E. coli fabB and fabB(Ts) fabF mutants allowed growth of these strains under otherwise non-permissive conditions indicating that the enzyme can catalyze all of the reactions required to elongate the butyryl-ACP primer resulting from FabH action to the long chain saturated and unsaturated fatty acids required for synthesis of functional membrane phospholipids and lipid A. These results readily explain how L. lactis can synthesize both saturated and unsaturated fatty acids using a single long chain KAS. Since the L. lactis FabF differs in its specificity from that of the canonical E. coli FabF it seems that the gene should be renamed as fabP. It seems surprising that Enterococcus faecalis, a close relative L. lactis (both were once classified as streptococci) encodes two long chain KAS homologues, one of which is able to functionally replace E. coli FabB whereas the other cannot. A plausible explanation for this dichotomy lies in the genome sizes of the two organisms. The genome of E. faecalis is 30% larger than that of L. lactis suggesting that the latter organism has undergone a process of genome minimization perhaps due to selection for fast growth under controlled conditions through its use in the dairy industry. It is known that the total number of paralogous genes increases with bacterial genome size (Hooper and Berg 2003) and thus the smaller genome size of L. lactis suggests that this organism may have lost the second long chain KAS gene present in E. faecalis. Indeed, loss of ancestral genes is a central trend of the evolution of lactic acid bacteria (Makarova et al. 2006). A comparison of the L. lactis and E. faecalis genomes supports this notion. The single L. lactis FabF is located within a cluster of fatty acid synthesis genes in essentially the same genome context as the E. faecalis fabF2 gene (now called fabF), the long chain KAS that fails to complement E. coli fabB mutants (Wang and Cronan 2004). The second E. faecalis gene (now called fabO) that encodes a protein with high sequence relatedness to E. coli FabF is found in a separate cluster of three genes bracketed by a fabI (enoyl-ACP reductase) homologue and the gene, fabN, that encodes the protein responsible for insertion of the cis double bond (Wang and Cronan 2004; Lu et al. 2005). In L. lactis the fabI and fabN genes are present but the long chain KAS gene is missing. Given the theme of gene loss in lactococci (Makarova et al. 2006), it seems likely that this reflects a deletion event in L. lactis rather than gene acquisition by E. faecalis. Consistent with this trend E. faecalis has two genes that encode enoyl-ACP reductases, fabI and fabK, whereas L. lactis has only fabI. As seen in the smaller gene cluster, the missing L. lactis gene appears to have been cleanly deleted from between the other two fatty acid synthetic genes.
The other major trend in the evolution of lactic acid bacteria is metabolic simplification (Makarova et al. 2006). The fact that from L. lactis FabF has the functions carried out by two enzymes in other bacteria fits this trend. However, reducing the number of enzymes necessarily increases demands on the remaining enzyme since the substrate specificity of L. lactis FabF must be set such that appropriate numbers of acyl chains flow from the branch point to the saturated and unsaturated arms of the synthetic pathway. Hence, FabP merits further study for comparison with the well-studied FabB and FabF enzymes of E. coli.
Complementation of the L. lactis fabH bypass mutant strain by overproduction of L. lactis FabF unexpectedly gave wild type growth rates. There seem two plausible mechanisms for replacement of FabH by FabF. Both scenarios postulate that FabF must be in excess of the level that is required for chain elongation. In the first scenario the scarcity of acyl-ACPs relative to FabF molecules would result in FabF active sites occupied only by malonyl-ACP. Due to the lack of bound acyl chains, the only fate of the malonyl-ACP would be decarboxylation to acetyl-ACP. FabF would then condense this acetyl-ACP with malonyl-ACP to give the 3-ketobutryl-ACP normally produced by FabH. E. coli FabF and FabB are known to perform both malonyl-ACP decarboxylation and condensation of acetyl-ACP with malonyl-ACP in vitro (Alberts et al. 1972; Arnvig Mcguire et al. 2000; McGuire et al. 2001). The second scenario is related except that the direct precursor of acetyl-ACP would be acetyl-CoA and acetyl-ACP would be formed by the acyl-CoA:ACP transacylase activity of FabF. We think this possibility is less likely than the first because acetyl-CoA was a very poor transacylation substrate when tested with a mixture of the E. coli FabF and FabB proteins (Alberts et al. 1972). It should be noted that there are prior reports that a bacterium can survive with only a single functional KAS. In the presence of thiolactomycin, a noncovalent inhibitor of all three E. coli KAS activities, overproduction of FabB (or isolation of a FabB resistant to the antibiotic) allows growth suggesting that FabB alone is sufficient for fatty acid synthesis (Tsay et al. 1992; Jackowski et al. 2002). However, the non-covalent nature of the inhibition precluded use of in vitro assays to ensure that thiolactomycin had totally disabled FabH and FabF. Moreover, FabB seems the most sensitive of the E. coli three KAS enzymes (Tsay et al. 1992).
The finding that simply increasing the level of FabF functionally replaces FabH raises the question of why L. lactis has retained the fabH gene, particularly given the theme of genome minimization. One possibility is that FabH is a more efficient means of synthesizing the primer needed for the methyl end of the acyl chain. This is because the ATP used in making a malonyl-ACP molecule (that consumed in the carboxylation of acetyl-CoA to malonyl-CoA) is wasted by decarboxylation of malonyl-ACP. If the rate of malonyl-ACP decarboxylation is in marked excess of the levels of primer needed, this seems a reasonable rationale. However if malonyl-ACP is decarboxylated in only slight excess then the energetic cost would be modest, only an eighth or ninth of the ATP required to make a 16 or 18 carbon fatty acid from acetyl-CoA
Acknowledgements
R.M-K. was supported in part by a National Science and Engineering Research Council of Canada Postdoctoral Fellowship. This work was supported by NIH grant AI15650.
References
- Alberts AW, Bell RM, Vagelos PR. Acyl carrier protein. XV. Studies of ß-ketoacyl-acyl carrier protein synthetase. J Biol Chem. 1972;247:3190–3198. [PubMed] [Google Scholar]
- Allen EE, Bartlett DH. FabF is required for piezoregulation of cis-vaccenic acid levels and piezophilic growth of the deep-sea bacterium Photobacterium profundum strain SS9. J Bacteriol. 2000;182:1264–1271. doi: 10.1128/jb.182.5.1264-1271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnvig Mcguire K, McGuire JN, von Wettstein-Knowles P. Acyl carrier protein (ACP) inhibition and other differences between ß-ketoacyl synthase (KAS) I and II. Biochem Soc Trans. 2000;28:607–610. doi: 10.1042/0300-5127:0280607. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bolotin A, et al. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Cronan JE., Jr. Bacterial fatty acid biosynthesis: targets for antibacterial drug discovery. Annu Rev Microbiol. 2001;55:305–332. doi: 10.1146/annurev.micro.55.1.305. [DOI] [PubMed] [Google Scholar]
- Christie WW. Lipid Analysis: Isolation, Separation, Identification and Structural Analysis of Lipids. 3 edn The Oily Press; Bridgwater, UK: 2003. [Google Scholar]
- de Mendoza D, Cronan JE. Temperature regulation of membrane fluidity in bacteria. Tren. Biochem. Sci. 1983;8:49–52. [Google Scholar]
- de Mendoza D, Ulrich A Klages, Cronan JE., Jr. Thermal regulation of membrane fluidity in Escherichia coli. Effects of overproduction of ß-ketoacyl-acyl carrier protein synthase I. J Biol Chem. 1983;258:2098–2101. [PubMed] [Google Scholar]
- Edwards P, Nelsen JS, Metz JG, Dehesh K. Cloning of the fabF gene in an expression vector and in vitro characterization of recombinant fabF and fabB encoded enzymes from Escherichia coli. FEBS Lett. 1997;402:62–66. doi: 10.1016/s0014-5793(96)01437-8. [DOI] [PubMed] [Google Scholar]
- Garwin JL, Klages AL, Cronan JE., Jr. ß-Ketoacyl-acyl carrier protein synthase II of Escherichia coli. Evidence for function in the thermal regulation of fatty acid synthesis. J Biol Chem. 1980;255:3263–3265. [PubMed] [Google Scholar]
- Guzman L-M, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper SD, Berg OG. Duplication is more common among laterally transferred genes than among indigenous genes. Genome Biol. 2003;4:R48. doi: 10.1186/gb-2003-4-8-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski S, Murphy CM, Cronan JE, Jr., Rock CO. Acetoacetyl-acyl carrier protein synthase. A target for the antibiotic thiolactomycin. J Biol Chem. 1989;264:7624–7629. [PubMed] [Google Scholar]
- Jackowski S, Zhang YM, Price AC, White SW, Rock CO. A missense mutation in the fabB ß-ketoacyl-acyl carrier protein synthase I) gene confers thiolactomycin resistance to Escherichia coli. Antimicrob Agents Chemother. 2002;46:1246–1252. doi: 10.1128/AAC.46.5.1246-1252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutchma AJ, Hoang TT, Schweizer HP. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD) J Bacteriol. 1999;181:5498–5504. doi: 10.1128/jb.181.17.5498-5504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Cronan JE. ß-Ketoacyl-acyl carrier protein synthase III (FabH) is essential for bacterial fatty acid synthesis. J Biol Chem. 2003;278:51494–51503. doi: 10.1074/jbc.M308638200. [DOI] [PubMed] [Google Scholar]
- Lu YJ, White SW, Rock CO. Domain swapping between Enterococcus faecalis FabN and FabZ proteins localizes the structural determinants for isomerase activity. J Biol Chem. 2005;280:30342–30348. doi: 10.1074/jbc.M504637200. [DOI] [PubMed] [Google Scholar]
- Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire KA, Siggaard-Andersen M, Bangera MG, Olsen JG, von Wettstein-Knowles P. beta-Ketoacyl-[acyl carrier protein] synthase I of Escherichia coli: aspects of the condensation mechanism revealed by analyses of mutations in the active site pocket. Biochemistry. 2001;40:9836–9845. doi: 10.1021/bi0105577. [DOI] [PubMed] [Google Scholar]
- McIntyre DA, Harlander SK. Improved electroporation efficincy of intact Lactococcus lactis subsp. lactis cells grown in defined media. App. Env. Microbiol. 1989;55:2621–2626. doi: 10.1128/aem.55.10.2621-2626.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y-A, Haefliger J-A, Francioli P, Moreillon P. Expression of Staphylococcus aureus clumping factor A in Lactococcus lactis subsp. cremoris using a new shuttle vector. Infec. Immun. 2000;68:3516–3522. doi: 10.1128/iai.68.6.3516-3522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetman G, et al. Electrospray ionization mass spectrometric analysis of phospholipids of Escherichia coli. Molecular Microbiology. 1996;20:233–238. doi: 10.1111/j.1365-2958.1996.tb02504.x. [DOI] [PubMed] [Google Scholar]
- Terzaghi BE, Sandine WE. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay JT, Rock CO, Jackowski S. Overproduction of ß-ketoacyl-acyl carrier protein synthase I imparts thiolactomycin resistance to Escherichia coli K-12. J Bacteriol. 1992;174:508–513. doi: 10.1128/jb.174.2.508-513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich AK, de Mendoza D, Garwin JL, Cronan JE., Jr. Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature-dependent regulation of membrane lipid composition. J Bacteriol. 1983;154:221–230. doi: 10.1128/jb.154.1.221-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cronan JE. Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J Biol Chem. 2004;279:34489–34495. doi: 10.1074/jbc.M403874200. [DOI] [PubMed] [Google Scholar]