Abstract
DNA and RNA oligonucleotides having formacetal internucleoside linkages between uridine and adenosine nucleosides have been prepared and studied using UV thermal melting, osmotic stress and X-ray crystallography. Formacetal modifications have remarkably different effect on double helical RNA and DNA – the formacetal stabilizes RNA helix by +0.7 °C, but destabilizes DNA helix by -1.6 °C per modification. The apparently hydrophobic formacetal has little effect on hydration of RNA but decreases the hydration of DNA, which suggests that at least part of the difference in thermal stability may be related to differences in hydration. A crystal structure of modified DNA shows that two isolated formacetal linkages fit almost perfectly in an A-type helix (decamer). Taken together, the data suggest that RNA may tolerate nonionic backbone modifications better than DNA. Overall, formacetal appears to be an excellent mimic of phosphate linkage in RNA and an interesting modification for potential applications in fundamental studies and RNA based gene control strategies, such as RNA interference.
The potential of RNA interference (RNAi) to become a new therapeutic approach stimulates interest in chemical modifications to optimize the efficacy of RNA-based drugs.1 Several modifications, developed earlier to improve the properties of antisense oligonucleotides, have also shown promising results in RNAi.1,2 We are interested in developing RNA analogues having non-ionic internucleoside linkages.3,4 Among such, we have found that replacement of selected phosphates with amide linkages (CH2-CO-NH-5′) does not change the overall conformation and thermal stability of RNA.3 Recently, Iwase et al.5 reported that introduction of two such amide linkages at the overhanging uridines of an siRNA did not significantly change the RNAi activity. The formacetal linkage 3′-O-CH2-O-5′ is another interesting modification that has served as a phosphate mimic in structural studies on a photolyase bound to a DNA lesion6 and may have broader use in fundamental studies and practical applications involving modified RNA.
Previous studies suggested that formacetal modification might have different effect on DNA and RNA oligonucleotides. Van Boom and co-workers found that formacetal modifications strongly destabilized DNA duplexes by -1.4 to -2.4 °C per modification.7 Gao and co-workers reported similar findings (-3 °C per modification), however, their preliminary NMR studies suggested that formacetal linkage caused little structural perturbation of DNA helix.8 Matteucci found that formacetals in the DNA strand only slightly decreased the stability of DNA-RNA heteroduplexes.9 In contrast, our preliminary studies showed that formacetals were somewhat stabilizing in all RNA duplexes (+0.2 to +0.8 °C per modification).4 However, each of these studies used different model sequences, which makes it difficult to compare and rationalize the results. In all earlier studies formacetal linkages were inserted between uridine or thymidine nucleosides only, which reduced the synthetic effort but also limited the sequences that could be studied.
To rationalize the earlier results and to gain more insight in structural and thermodynamic properties of formacetal-modified RNA and DNA we prepared formacetal (f) linked dinucleotides r(UfA) and d(TfA). The self-complementary UA and TA motifs allowed us to prepare a series of RNA and DNA oligonucleotides for thermodynamic (UV melting and osmotic stress) as well as X-ray crystallographic studies. Herein, we show that the formacetal modification has a surprisingly different effect on double helical RNA compared to DNA. Although formacetal stabilizes RNA helix by +0.7 °C per modification, it strongly destabilizes DNA helix (-1.6 °C per modification). Interestingly, the apparently hydrophobic formacetal has little effect on hydration of RNA but decreases the hydration of DNA, as judged by osmotic stress experiments. X-Ray crystal structure shows that two formacetal linkages do not distort the conformation of an A-type decamer. Our results suggest that the formacetal linkage fits remarkably well in a double helical RNA and may be an interesting modification to test in short interfering RNAs (siRNAs). Taken together with our earlier studies on amide3 and formacetal4 modified RNA, the current results also suggest that RNA may tolerate nonionic backbone modifications better than DNA.
Results
Synthesis of formacetal-modified oligonucleotides
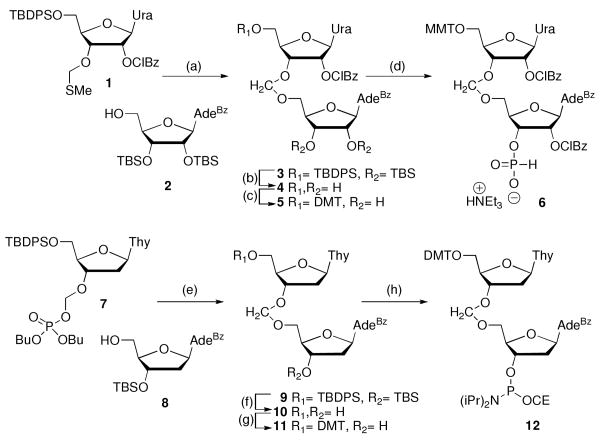
Synthesis of formacetal-modified RNA and DNA was done by adopting and modifying literature procedures by us4 and van Boom and co-workers,10 respectively. N-Iodosuccinimide and triflic acid (TfOH) mediated coupling11 of thioacetal 14 with adenosine acceptor 212 gave r(UfA) dimer 3 (Scheme 1). Deprotection of silyl groups, selective 5′-tritylation and one-pot 2′-benzoylation and 3′-phosporylation following our previously reported procedures4 gave the phosphonate dimer 6. Analogous synthesis of d(TfA) was not successful, apparently because of low stability of deoxyadenosine 8 under the acidic (TfOH) coupling conditions.
Scheme 1.
Synthesis of formacetal linked dimers. a
a Steps: (a) 1, 2, NIS, TfOH, THF, -40 to -25 °C, 1.5 h, 72%; (b) TBAF, THF, rt 18 h; (c) p-methoxytrityl chloride, pyridine, 35 °C, 1.5 h 39% (two steps); (d) 2-chlorobenzoyl chloride, THF/pyridine, -78 °C, 1.5 h, then PCl3, imidazole, NEt3, THF/pyridine/CH2Cl2, -78 °C, 0.5 h, 73%; (e) TMSOTf, ClCH2CH2Cl, rt, 40 min, 43%; (f) TBAF, THF, rt, 1 h, 50%; (g) p-dimethoxytrityl chloride, pyridine, rt, 20 h 75%; (h) NC(CH2)2OP(Cl)N(iPr)2, DIEA, CH2Cl2, rt, 1 h 20 min, 50%.
However, trimethylsilyl triflate mediated coupling of dibutyl phosphate 710 with deoxyadenosine acceptor 812 gave r(UfA) dimer 9 in acceptable yield (Scheme 1). Deprotection of silyl groups, selective 5′-tritylation and standard phosphoramidite synthesis gave dimer 12. Using the dimers 6 and 12 in standard H-phosphonate and phosphoramidite solid-phase synthesis protocols we initially prepared two self-complementary model sequences r(UfA)6 OL2 and d(TfA)6 OL4 (Table 1). The choice of the sequences was based on our earlier UV thermal melting and osmotic stress study,13 that had characterized the non-modified RNA (OL1) and DNA (OL3) controls.
Table 1.
Thermal melting and osmotic stress results of formacetal-modified oligonucleotides. a
| No | Sequence | tm °C | −ΔH kcal/mol b | −ΔH kcal/mol c | ΔS eu c | −ΔG310 kcal/mol c | Ethylene glycol ΔnWd | Glycer ol ΔnWd | Acetam ide ΔnWd |
|---|---|---|---|---|---|---|---|---|---|
| OL1 e | r(UpApUpApUpApUpApUpApUpA) | 30.0 ± 0.3 | 82.4 ± 4.2 | 70.3 ± 4.0 | 206 ± 13 | 6.4 ± 0.1 | 36 ± 6 | 40 ± 6 | 67 ± 6 |
| OL2 | r(UfApUfApUfApUfApUfApUfA) | 38.3 ± 0.1 | 77.1 ± 0.7 | 82.2 ± 1.6 | 238 ± 5 | 8.4 ± 0.1 | 37 ± 3 | 34 ± 3 | 100 ± 2 |
| OL3 e | d(TpApTpApTpApTpApTpApTpA) | 22.9 ± 0.7 | 39.5 ± 3.3 | 41.4 ± 4.1 | 113 ± 14 | 6.2 ± 0.2 | 22 ± 7 | 26 ± 8 | 38 ± 6 |
| OL4 | d(TfApTfApTfApTfApTfApTfA) | <10 | - | - | - | - | - | - | - |
| OL5 | d(CpGpTpApTpApTpApTpApTpApCpG) | 46.2 ± 0.4 | 30.6 ± 1.1 | 37.9 ± 2.0 | 93 ± 6 | 9.2 ± 0.1 | 15 ± 2 | 19 ± 2 | 34 ± 3 |
| OL6 | d(CpGpTfApTfApTfApTfApTfApCpG) | 30.4 ± 0.3 | 34.1 ± 1.0 | 32.8 ± 1.5 | 82 ± 5 | 7.4 ± 0.1 | 13 ± 2 | 9 ± 3 | 19 ± 2 |
Oligonucleotide (2 μM) in 10 mM sodium cacodylate (pH 7.4), 0.1 mM EDTA, and 300 mM NaCl. Results ± standard deviations.
From δα/δ(Tm -1) vs. Tm, ref. 13.
From van't Hoff analysis of melting curves.
ΔnW = number of water molecules released upon melting. Results ± error estimates.
Data from ref. 13.
UV thermal melting and osmotic stress
To gain insight into the effect of formacetal modification on thermal stability and hydration of DNA and RNA, we studied the model sequences OL2 and OL4 using the UV thermal melting and osmotic stress method as previously reported.13,14 Substitution of 12 out of 22 phosphates with formacetals increased the thermal stability of the formacetal-modified RNA duplex OL2 (Table 1, +0.7 °C per modification). Most remarkably, the relatively hydrophobic formacetal linkages did not decrease the hydration of RNA, as judged by little change in ΔnW, the number of water molecules released upon melting. In fact, in acetamide the formacetal duplex OL2 was more hydrated than the non-modified control OL1. In contrast, the DNA OL4 was strongly destabilized by the formacetal modification (at least -1 °C per modification). Because OL4 was not suitable for osmotic stress due to the low thermal stability, we prepared OL6 having five formacetal linkages between the central T and A nucleosides and two closing non-modified CG base pairs. Compared to the non-modified control OL5, replacement of 10 out of 26 phosphates with formacetals resulted in strong destabilization of DNA (Table 1, -1.6 °C per modification). Most remarkably, the glycerol and acetamide series showed substantial loss of hydration upon formacetal modification of DNA.
Crystal structure of formacetal-modified DNA
To gain more insight into structural features of formacetal-modified oligonucleotides we synthesized and attempted crystallization of a series of self-complementary DNA sequences having the TfA dimer inserted at a central position. The DNA decamer with sequence GCGTATACGC does not form crystals suitable for high-resolution diffraction studies. Accordingly, our attempts to solve crystal structures of GCGTATACGC having a formacetal modification so far have not been successful. However, replacement of a single 2′-deoxynucleotide at various positions of GCGTATACGC with a ribonucleotide or a 2′-O-modified nucleotide analogue typically yields well diffracting crystals.15 Following such a design, we succeeded in solving the structure of d(GCGTfAUMeACGC) with a formacetal linkage between residues T4 and A5 and 2′-O-methyl-uridine (UMe) at position 6 (OL7; residues in the strand are numbered 1 to 10). The crystals diffracted to 1.75 Å resolution and were of space group P41212, (a=b=33.20Å; c=68.51Å), with one strand per asymmetric unit. Selected crystal data and refinement parameters are summarized in Table 2 and an example of the quality of the final electron density is depicted in Figure 1.
Table 2.
Selected crystal and refinement data.
| Space group | Tetragonal P41212 |
|---|---|
| Unit cell constants [Å] | a = b = 33.20, c = 68.51 |
| Resolution [Å] | 1.75 |
| No. of unique reflections | 4,006 |
| Completeness (1.78 – 1.75 Å) [%] | 95.2 (72.4) |
| R-merge (1.78 – 1.75 Å) [%] | 5.8 (18.7) |
| R-work [%] | 22.8 |
| R-free [%] | 28.2 |
| No. of DNA atoms | 201 |
| No. of waters | 25 |
| R.m.s. deviations bonds [Å] | 0.022 |
| R.m.s. deviations angles [°] | 3.0 |
| Average B-factor [Å2] | 35.2 |
Figure 1.
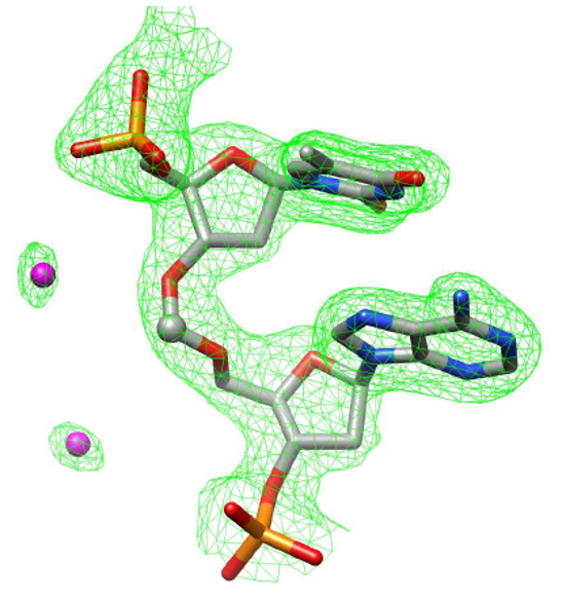
Quality of the electron density. Final Fourier sum 2Fo-Fc electron density (1σ threshold) around the T4fA5 dimer is shown as a green meshwork and water molecules are magenta spheres.
In the crystal the duplex adopts an A-form conformation (Figure 2), with the paired strands exhibiting an identical geometry due to the dyad symmetry.
Figure 2.
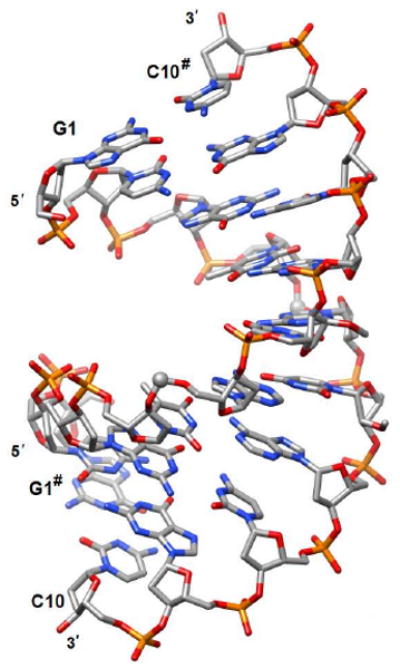
Crystal structure of the duplex [d(GCGTfAUMeACGC)]2 (OL7). The formacetal carbon is highlighted with a sphere and the hash symbol indicates a symmetry-related strand. The view is across the major (top) and minor grooves (bottom).
In the formacetal moiety all torsion angles adopt a conformation that is consistent with a standard A-form backbone (Figure 3; α sc-, β ap, γ sc+, δ sc+, ε ap, ζ, sc-). The conformation of the sugars in T4, fA5 and UMe6 is C3′-endo and their backbone torsion angles are −60°, 165°, 49°, 80°, −155°, −71° (T4), −77°, 174°, 65°, 76°, −153°, −72° (fA5) and −72°, 168°, 57°, 78°, −144°, −77° (UMe6), respectively (α, β, γ, δ, ε, ζ). Overall, the crystal structure demonstrates that formacetal fits almost perfectly within the A-type duplex. This is consistent with earlier studies by us4 and others.8
Figure 3.
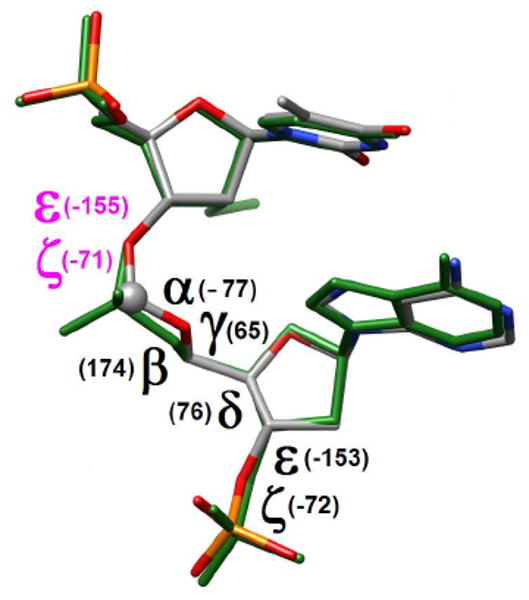
Overlay of the T4fA5 and UMe6pA7 (green bonds) dinucleotides. Torsion angles of the formacetal linkage are depicted in black. Values of the ε and ζ torsion angles in the 5′-flanking T4 residue are highlighted in purple.
The backbone around formacetal linkages is relatively dry. Several water molecules are located in the vicinity of the formacetal moiety but the distances from individual atoms exceed 3.5 Å (Figure 4A). Hydrogen bonds between water molecules and residues T4 and T5 are only established to base atoms (not shown). The phosphate groups of flanking residues are well hydrated (Figure 4B). However, at the current resolution of the crystal structure, hydration patterns around the duplex can only be visualized to a limited extent. A more complete account of the effects of the formacetal moiety on the local water structure will have to await the determination of crystal structures at higher resolution.
Figure 4.
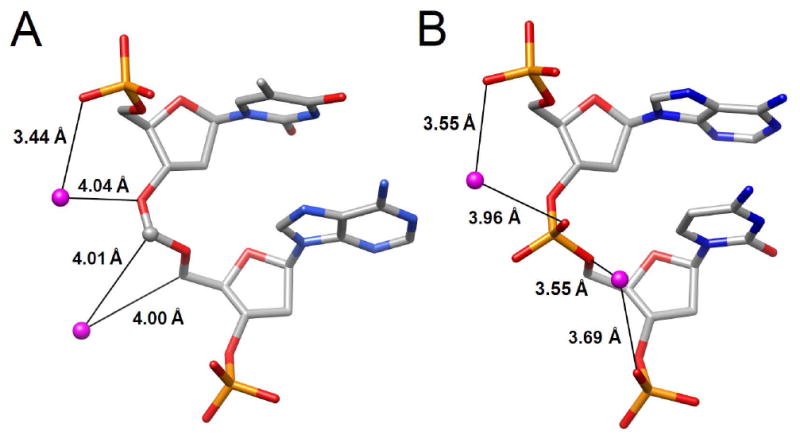
Comparison of water molecules associated with the backbones of (A) formacetal linkage in T4fA5 and (B) phosphate linkage in A7pC8. Hydrogen bonds are shown as black lines and distances are given in Å.
Circular dichroism of formacetal-modified oligonucleotides
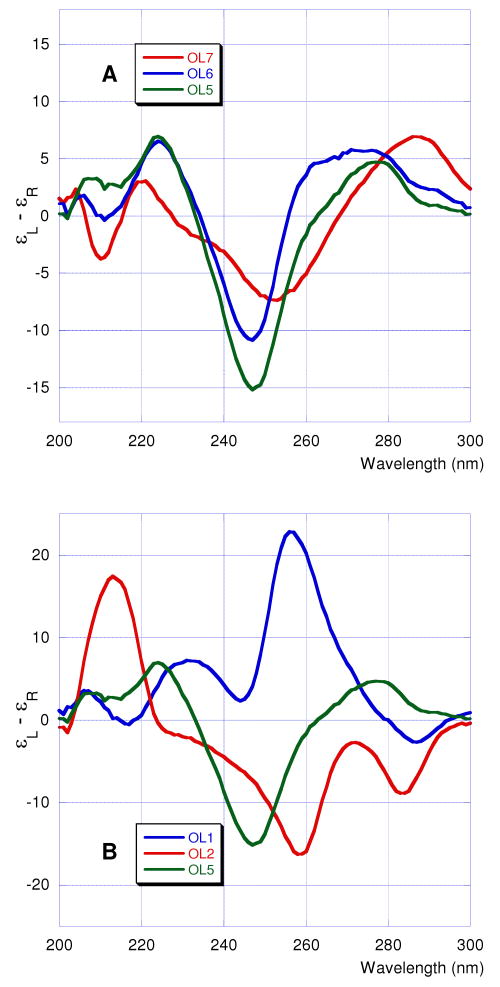
To gain insight into overall helix conformation of formacetal-modified oligonucleotides in solution we studied the circular dichroism (CD) spectra of RNA OL1 and OL2 and DNA OL5, OL6, and OL7. All DNA oligonucleotides gave CD spectra (Figure 5A) typical for B-form helical conformations – positive bands at 260-290 nm, strong negative bands around 250 nm and positive bands at 220-225 nm accompanied with weaker bands around 210 nm (negative).16 Consistent with our previous findings in the RNA series,4 the CD spectra of formacetal-modified DNA OL6 were similar to the non-modified control OL5.
Figure 5.
CD spectra of oligonucleotides.
The non-modified RNA OL1 showed CD spectrum typical for an A-form helical conformation with a strong positive band at 250-260 nm (Figure 5B).16 However, the CD spectrum of the formacetal-modified RNA OL2 was different from the spectra of either typical A or B helices and displayed a set of unique bands – negative around 285 and 260 nm and positive around 215 nm. This result was surprising because our previous studies had shown that the CD spectra of formacetal-modified RNA were virtually identical to CD spectra of non-modified RNA.4 However, the RNAs in our earlier studies contained less formacetal modifications (up to six formacetals out of 24 linkages) compared to OL2 (12 formacetals out of 22 linkages).
Discussion
The present UV thermal melting study confirms the earlier literature data that formacetal modification has different effect on double helical DNA and RNA (Table 1). In the present study, formacetal modification strongly destabilizes double helical DNA by -1.6 °C per modification, which is consistent with the results of van Boom and co-workers7 (-1.4 to -2.4 °C per modification) and Gao et al.8 (-3 °C per modification). In contrast, formacetal modification stabilizes double helical RNA by +0.7 °C per modification, which is consistent with our earlier studies4 (+0.2 to +0.8 °C per modification).
Taken together with our earlier results on amide modified RNA,3 the present data suggest that RNA may tolerate nonionic backbone modifications better than DNA. If this phenomenon holds true for other modifications, there are two implications important for design of RNA analogues, for example such as, modified siRNAs. First, it is conceivable that the phosphate backbone may be in general an excellent site to introduce nonionic modifications in RNA. Second, many nonionic backbone modifications have been introduced in DNA and found to destabilize DNA-RNA heteroduplexes.17 One may conclude that such modifications are not useful for applications involving RNA as well. However, our formacetal data suggest that modifications that are even strongly destabilizing in DNA may actually be stabilizing in RNA. Therefore, the data obtained on modified DNA17 should not be used to predict the biophysical properties of the same modification in RNA without additional experimental support. It is possible that other nonionic backbone modifications that may not look promising in DNA, may in fact be well tolerated in RNA.
Osmotic stress results show that formacetal modification causes little change in RNA's hydration. This result is somewhat unexpected because more than half (12 out of 22) of the polar phosphates are replaced by the relatively hydrophobic formacetals in OL2. As could have been expected, formacetal modification causes significant loss of hydration in DNA OL6. These results suggest an intriguing possibility that at least part of the observed differences in thermal stability of formacetal-modified OL2 and OL6 may be caused by favorable hydration of formacetal-modified RNA and unfavorable hydration of formacetal-modified DNA. However, direct comparison is complicated by the fact that the CD spectra suggest that formacetal-modified RNA OL2 may not be in the expected A-form helix. This result is unexpected because in earlier studies formacetal modifications caused virtually no change in CD spectra of RNA. Unfortunately, all attempts to crystallize OL2 were not successful. Work on structural aspects of formacetal linkages in RNA by crystallography and solution phase NMR is currently in progress in our laboratories and should clarify the unexpected CD result.
The crystal structure (Figures 2 and 3) demonstrates that formacetal (two substitutions per decamer OL7) fits almost perfectly within an A-form helix, the conformation adopted by double-stranded RNA. This is consistent with earlier studies by us4 and others8 and is further confirmed by the modeled structure of a DNA duplex with several formacetal linkages per strand that exhibits an overall A-form (see Figure S2 in Supporting Information). Although, OL7 exists as a B-form helix in solution (Figure 5A), it crystallizes in the A-form (Figure 2). This is likely due to an intrinsic tendency of this decamer to relatively easily flip into the A-form as well as to dehydration and packing interactions in the crystal. Moreover, the decamer sequence here features a 2′-O-methyl-uridine residue and incorporation of a single ribonucleotide into a DNA oligonucleotide was previously demonstrated to convert the B- to the A-form in the solid state.15a, 18
The crystal structure of OL7 confirms our current and previous4 thermal melting results that the formacetal linkages are well tolerated in A-form helix. Thus, the overall conformation of the decamer duplex is little perturbed by substitution of two phosphate linkages with formacetals. The bridging C-O bonds in formacetal are somewhat shorter (∼1.4 Å) than the P-O bonds in phosphate (∼1.6 Å). However, the O-C-O angle (∼110°) is wider than the analogous O-P-O angle (102-104°). The wider angle compensates for the shorter bonds in formacetal and, thus, the distance between the 3′-O and the 5′-O of the next nucleoside changes only from ∼2.5 Å in phosphate to ∼2.3 Å in formacetal. The backbone of nucleic acid is flexible enough to accommodate such a relatively small change. Taken together, our data suggest that isolated formacetal linkages are remarkably good mimics of the internucleoside phosphate linkages in RNA, but not necessarily in DNA.
Conclusions
The present study is to our knowledge the first attempt at direct comparison of nonionic backbone modification in RNA and DNA. Our results suggest that RNA may tolerate nonionic backbone modifications better than DNA. Therefore, conclusions based on thermal stability of modified DNA17 should be used with caution when designing chemically modified RNA analogues. Taken together the excellent thermal stability and the fact that two formacetal linkages do not disturb the A-form decamer duplex suggest that formacetals may be excellent mimics of phosphate for certain biochemical studies of RNA. For example, RNA fragments having single formacetal modification as a non-cleavable phosphate analogue may be useful as substrates for co-crystallization with RNA-processing enzymes. Similar application have been demonstrated in DNA.6 Our studies also suggest that formacetal may be an interesting modification to test in siRNAs, where the charge reduction may help cellular uptake and biodistribution. However, the structural consequences of extensive formacetal modification of RNA need to be further investigated.
Experimental Section
Synthesis of formacetal modified oligonucleotides
Formacetal containing RNAs and DNAs were synthesized by following and modifying the reported procedures.4,10 For details, see the Supporting Information.
UV thermal melting and osmotic stress
Melting of each oligonucleotide (2 μM) was done in 10 mM sodium cacodylate, 0.1 mM EDTA, and 300 mM NaCl in presence of 0, 5, 10, 15, and 20 % weight/volume of each of the four osmolytes in Table 1. Oligonucleotide concentration were calculated using extinction coefficients obtained using the nearest-neighbor approximation.19 Absorbance vs. temperature profiles were measured at 260 nm on a Varian Bio 100 spectrometer equipped with a six-position Peltier temperature controller. The temperature was increased at 0.5 °C per minute. Five samples were measured concurrently in the double-beam mode. At temperatures below 15 °C the sample compartment was flushed with dry nitrogen gas.
The melting temperatures and thermodynamic parameters were obtained using Varian Cary software (Version 02.00). The experimental absorbance vs. temperature curves were converted into a fraction of strands remaining hybridized (α) vs. temperature curves by fitting the melting profile to a two-state transition model, with linearly sloping lower and upper base lines. The melting temperatures (tm) were obtained directly from the temperature at α = 0.5. The final tm was an approximation of at least five to eight measurements. The thermodynamic parameters were determined using two different methods20 as described bellow:
from the width at the half-height of differentiated melting curve (Table 1, column 4). The fraction of strands remaining hybridized (α) vs. temperature curves were converted into differentiated melting curves (δα/δ(Tm-1) vs. Tm) using Varian Cary software (Version 02.00). The width of the of the differentiated melting curve at the half-height is inversely proportional to the van't Hoff transition enthalpy; for a bimolecular transition ΔH = 10.14/(T1-1 - T2-1) where T1 is the lower temperature at one-half of (δα/δ(Tm-1)) and T2 is the upper temperature at one-half of (δα/δ(Tm-1)).20 The final -ΔH is the average of at least 10 measurements. Full experimental data are given in Supporting Information Tables S2-S4 and in Ref 13 for OL1 and OL3.
from the van't Hoff plot of ln K vs 1/Tm (Table 1, columns 6-7). For a bimolecular transition of self-complementary strands the equilibrium constant K = α/[2C(1-α)2] where C is the total strand concentration (C = 2 × 10-6 M). The van't Hoff plot (ln K vs. 1/Tm) is linear with -ΔH/R as the slope and ΔS/R as the intercept (R is the universal gas constant 1.986 cal/mol/K). All fitting and calculation operations were done using Varian Cary software (Version 02.00) using settings for a bimolecular transition of self-complementary strands. The final -ΔH and -ΔS is the average of at least 10 measurements. Full experimental data are given in Supporting Information Tables S2-S4 and in Ref 13 for OL1 and OL3.
The changes in the number of water molecules associated with the melting process ΔnW were determined as described previously by Spink and Chaires14 and by us13: ΔnW = (-ΔH/R)[d(Tm-1)/d(lnaw)], where -ΔH is the enthalpy determined from the width at the half-height of differentiated melting curve in pure buffer and R is the universal gas constant (1.986 cal/mol/K). The experimentally determined values of water activity (lnaw) at given cosolute concentrations were provided by Drs. Spink and Chaires. The slope of the plot of reciprocal temperature (in K) of melting vs. the logarithm of water activity (lnaw) at different concentrations (0, 5, 10, 15, and 20%) of small cosolutes gave the value of d(Tm-1)/d(lnaw). The final ΔnW were obtained by linear fitting using KaleidaGraph software (Version 3.51) with a confidence level usually better than 98%. The experimental uncertainties in the final ΔnW were calculated as previously described by us.13
Crystallization and data collection
Crystals of 5′-GCGTfAUMeACGC-3′ (UMe = 2′-OMe-U) were grown by the hanging-drop vapor diffusion technique using the Nucleic Acid Miniscreen (Hampton Research, Aliso Viejo, CA).21 Droplets (2 μL) containing oligonucleotide (0.6 mM), sodium cacodylate (20 mM, pH 6.0), potassium chloride (40 mM), magnesium chloride (10 mM), spermine tetrahydrochloride (6 mM), and 2-methyl-2,4-pentanediol (MPD; 5% (v/v)) were equilibrated against a reservoir of MPD (1 mL, 35%). Crystals were mounted in nylon loops without further cryo-protection and frozen in liquid nitrogen. Diffraction data were collected on the undulator beamline X25 at the National Synchrotron Light Source (NSLS). Diffraction data were processed with the program HKL2000.22 Selected crystal data and refinement parameters are listed in Table 2.
Structure refinement
The structure was determined by the Molecular Replacement (MR) technique with the program MOLREP,23 using a single strand of an A-form DNA (PDB ID 411D) as the search model. Initially the single strand was refined as an all-DNA model and the refinement was carried out with the program Refmac,24 randomly setting aside 8% of the reflections for calculating the R-free.25 Water molecules were added into regions of superimposed (2Fo-Fc) sum and (Fo-Fc) difference Fourier electron density. The refinement was carried out in both space groups P41 and P41212. However, refinement in the former space group resulted in higher values for R-free and R-work (typically around 3-4% difference), thereby indicating the higher symmetry is correct. At a later stage refinement was continued and the phosphorus and the two non-bridging oxygen atoms between the T4 and A5 residues replaced by a carbon. The refinement of the structure clearly reveals all the atoms of the formacetal linkage.
Data deposition
Final coordinates and structure factors have been deposited in the Protein Data Bank (http://www.rcsb.org): PDB ID code 3HR3.
Supplementary Material
Acknowledgments
We thank Binghamton University, NIH (R01 GM071461) and NSF-NATO (fellowship DGE-0410935 to AK) for support of this research. We also thank Dr. Annie Héroux for the X-ray data collection at beamline X25 of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, New York. Financial support for the beamline comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
Footnotes
Supporting Information Available: Experimental procedures, details of thermal melting and osmotic stress experiments, and copies of 1H and 13C NMR data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Watts JK, Deleavey GF, Damha MJ. Drug Discovery Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]; Corey DR. J Clin Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rozners E. Curr Org Chem. 2006;10:675–692. [Google Scholar]; Manoharan M. Curr Opin Chem Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Rozners E, Katkevica D, Bizdena E, Strömberg R. J Am Chem Soc. 2003;125:12125–12136. doi: 10.1021/ja0360900. [DOI] [PubMed] [Google Scholar]
- 4.Rozners E, Strömberg R. J Org Chem. 1997;62:1846–1850. [Google Scholar]; Rozners E, Katkevica D, Strömberg R. ChemBioChem. 2007;8:537–545. doi: 10.1002/cbic.200600515. [DOI] [PubMed] [Google Scholar]
- 5.Iwase R, Toyama T, Nishimori K. Nucleosides, Nucleotides & Nucleic Acids. 2007;26:1451–1454. doi: 10.1080/15257770701542389. [DOI] [PubMed] [Google Scholar]; Iwase R, Miyao H, Toyama T, Nishimori K. Nucleic Acids Symposium Series. 2006:175–176. doi: 10.1093/nass/nrl087. [DOI] [PubMed] [Google Scholar]
- 6.Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, Essen LO. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 7.Quaedflieg PJLM, Pikkemaat JA, Van der Marel GA, Kuyl-Yeheskiely E, Altona C, Van Boom JH. Recl Trav Chim Pays-Bas. 1993;112:15–21. [Google Scholar]
- 8.Gao X, Brown FK, Jeffs P, Bischofberger N, Lin KY, Pipe AJ, Noble SA. Biochemistry. 1992;31:6228–6236. doi: 10.1021/bi00142a009. [DOI] [PubMed] [Google Scholar]
- 9.Matteucci M. Tetrahedron Lett. 1990;31:2385–2388. [Google Scholar]
- 10.Quaedflieg PJLM, Timmers CM, van der Marel GA, Kuyl-Yeheskiely E, van Boom JH. Synthesis. 1993:627–633. [Google Scholar]
- 11.He GX, Bischofberger N. Tetrahedron Lett. 1997;38:945–948. [Google Scholar]
- 12.Zhu XF, Williams HJ, Scott AI. Perkin. 2000;1:2305–2306. [Google Scholar]
- 13.Rozners E, Moulder J. Nucleic Acids Res. 2004;32:248–254. doi: 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spink CH, Chaires JB. Biochemistry. 1999;38:496–508. doi: 10.1021/bi9820154. [DOI] [PubMed] [Google Scholar]
- 15.(a) Egli M, Usman N, Rich A. Biochemistry. 1993;32:3221–3237. [PubMed] [Google Scholar]; (b) Egli M. In: Advances in Enzyme Regulation. Weber G, editor. Vol. 38. Elsevier Science Ltd.; Oxford, UK: 1998. pp. 181–203. [DOI] [PubMed] [Google Scholar]; (c) Egli M, Minasov G, Tereshko V, Pallan PS, Teplova M, Inamati GB, Lesnik EA, Owens SR, Ross BS, Prakash TP, Manoharan M. Biochemistry. 2005;44:9045–9057. doi: 10.1021/bi050574m. [DOI] [PubMed] [Google Scholar]; (d) Egli M, Pallan PS. Annu Rev Biophys Biomol Struct. 2007;36:281–305. doi: 10.1146/annurev.biophys.36.040306.132556. [DOI] [PubMed] [Google Scholar]
- 16.Sosnick TR, Fang X, Shelton VM. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]; Gray DM, Ratliff RL, Vaughan MR. Methods Enzymol. 1992;211:389–406. doi: 10.1016/0076-6879(92)11021-a. [DOI] [PubMed] [Google Scholar]
- 17.Freier SM, Altmann KH. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ban C, Ramakrishnan B, Sundaralingam M. J Mol Biol. 1994;236:275–285. doi: 10.1006/jmbi.1994.1134. [DOI] [PubMed] [Google Scholar]
- 19.Puglisi JD, Tinoco I., Jr Methods in Enzymology. 1989;180:304–324. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 20.Breslauer KJ. Methods in Enzymology. 1995;259:221–242. doi: 10.1016/0076-6879(95)59046-3. [DOI] [PubMed] [Google Scholar]
- 21.Berger I, Kang CH, Sinha N, Wolters M, Rich A. Acta Cryst D. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Vagin AA, Teplyakov A. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]; Collaborative Computational Project, Number 4. Acta Cryst D. 1994;50:760–763. [Google Scholar]
- 24.Murshudov GN, Vagin AA, Dodson EJ. Acta Cryst D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Brünger AT. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.