Abstract
Trp53 (p53) loss of function has previously been shown to rescue tissue maintenance and developmental defects resulting from DNA damage or DNA repair gene mutations1–12. Herein, we report that p53 deficiency significantly exacerbates tissue degeneration caused by mosaic deletion of the essential genome maintenance regulator ATR. Combined loss of ATR and p53 (p53−/−ATRmKO) led to severe defects in hair follicle regeneration, localized inflammation (Mac1+Gr1+ infiltrates), accelerated deterioration of the intestinal epithelium, and synthetic lethality in adult mice. Tissue degeneration in p53−/−ATRmKO mice was characterized by the accumulation of cells maintaining high levels of DNA damage. Moreover, the elevated presence of these damaged cells in both progenitor and downstream compartments in the skin coincided with delayed compensatory tissue renewal from residual ATR-expressing cells. Together, our results indicate that combined loss of ATR and p53 in adult mice leads to the accumulation of highly damaged cells, which consequently impose a barrier to regeneration from undamaged progenitors.
Keywords: ATR, p53, DNA damage, tissue regeneration, aging, cancer
Tissue development, homeostasis, and renewal are strongly influenced by processes that control the accumulation of damaged, unproductive cells. Through its functions in cell cycle regulation and apoptosis, the p53 transcription factor plays a central role in these processes, particularly in response to DNA damage. For example, developmental arrest, tissue homeostatic failures, and premature cellular senescence in numerous mouse models harboring DNA repair gene mutations are partially rescued by p53 deficiency1–12. Furthermore, p53 pathway hyperactivation leads to tissue deterioration and promotes the appearance of several age-associated pathologies in mice13,14, arguing that unregulated p53 engagement can present a significant barrier to tissue renewal. However, mice expressing additional copies of p53 and Cdkn2a (Arf) under normal regulatory control (s-Arf/p53 mice) exhibit enhanced antioxidant gene expression, decreased accumulation of mutated cells, and a significantly extended lifespan15. This study and others have indicated that augmented p53 pathway activity can enhance tissue renewal by reducing ROS levels15 and by limiting the accumulation of damaged cells that impede repopulation from competent progenitors15–18.
Deletion of the ATR checkpoint kinase leads to an increase in double strand break formation in the course of normal DNA replication19–22 and to the activation of p53 (Supplementary Fig. 1). Previously, we have demonstrated that such deletion in the presence of wild-type p53 leads to the rapid elimination of ATR-deleted (ATRΔ/−) cells from adult tissues that maintain high rates of cellular proliferation; however, cellularity in most tissues is quickly restored by concomitant regeneration from progenitors that have escaped ATR deletion23. Despite this initial reconstitution, ATR-mosaic knockout (ATRmKO) mice ultimately fail to maintain tissue homeostasis and exhibit age-related phenotypes23. In light of the proposed functions for p53 in tissue homeostasis, p53 activation in ATRmKO mice could drive acute tissue degeneration by reducing cellularity or, alternatively, facilitate immediate regeneration by limiting the accumulation of damaged cells, thus promoting renewal from undamaged progenitors.
To investigate these models, systemic mosaic deletion of ATR in p53+/+ and p53−/− mice was performed as described previously23 by tamoxifen (TAM)-induced acute activation of a ubiquitously expressed form of Cre recombinase (UBC-Cre-ERT2). The rates of Cre-mediated lox recombination of the ATRflox allele were identical regardless of p53 status, as demonstrated by equivalent ATRflox recombination rates in the brain, bone marrow and intestines of ATRflox/+ (ATRmHet) mice, and by deletion rates in the brains of ATRflox/− (ATRmKO) mice (Fig. 1f), where ATR-null cells demonstrate no selective disadvantage over time23. Consistent with previous studies23, a rapid deterioration of the intestinal epithelium was observed in ATRmKO mice 6 days after TAM treatment (Fig. 1e), reaching a nadir a few days later (7–14 days after TAM treatment, ref. 23). This degeneration coincided with a 28% mortality of ATRmKO animals within 2 weeks of ATR deletion (Fig. 1a), after which time compensatory tissue renewal from residual ATR-expressing progenitors allowed ATRmKO mice to survive similar to controls for up to one year23.
Figure 1.
Mosaic ATR deletion in adult mice is synthetic lethal with p53 deficiency. (a) Kaplan-Meier representation of survival following acute, mosaic ATR deletion in p53+/+ and p53−/− mice. Approximately 72% of ATRmKO mice survive the immediate period following TAM treatment, while less than 6% of p53−/−ATRmKO mice (1 in 17) were viable beyond this time. (b) The single surviving p53−/−ATRmKO mouse (from part a) and a comparably treated ATRmKO mouse 1 month after TAM treatment. (c) H&E stained sections of the humeral bone from mice of the indicated genotypes 6 days after TAM treatment, 200×; magnification. (d) Absolute number of myeloid cells (Gr1+) and T cells (CD3, CD4, or CD8+) obtained from 4 hindlimb bones 6 days after TAM treatment (n = 4–5 mice per genotype). The loss of myeloid cells from the bone marrow of ATRmKO and p53−/−ATRmKO relative to each respective ATRflox/+ control was significant (P = 0.01 and 0.03, respectively). (e) H&E stained sections of intestines from mice of the indicated genotypes 6 days after TAM treatment, 200× magnification. (f) Abundance of the ATRflox-recombined allele (ATRΔ) in the brain, bone marrow, and intestines 6 days after TAM treatment (n = 7–12 mice per genotype). The frequency of ATRflox recombination was determined by quantitative PCR amplification of the ATRflox allele from genomic DNA isolated from each tissue (described in Methods). P values from t tests comparing the mean frequencies of ATRΔ in p53−/−ATRmKO and ATRmKO mice were 0.047 and 0.149 in the bone marrow and intestines, respectively. Error bars in both d and f represent the S.E.M. of each data set.
Surprisingly, p53 absence failed to prevent short-term mortality following ATR mosaic deletion and, in contrast, led to an accelerated and highly penetrant lethal phenotype, with greater than 94% of p53−/−ATRmKO mice dying within 2 weeks of TAM treatment at a median time of 8 days (Fig. 1a). The single surviving p53−/−ATRmKO mouse in these studies had an abnormally low deletion rate (60% brain deletion). However, this mouse exhibited hair-graying within one month of TAM treatment (Fig. 1b), a phenotype observed in p53+/+ATRmKO mice only at significantly later time points (3–6 months)23. Furthermore, histological analysis of mice sacrificed just prior to median mortality (6 days after TAM) revealed that p53 absence failed to rescue acute bone marrow degeneration and exacerbated the acute deterioration of the intestinal epithelium observed in ATRmKO mice (Fig 1c–e). Consistent with ATR’s essential role during DNA replication19–22, decreased cellularity in the bone marrow was primarily attributable to loss of highly proliferative populations (myeloid) instead of largely quiescent subsets (T cells, Fig. 1d). Synthetic lethality and regenerative deficiencies were also observed in p53−/−ATRmKO mice under conditions of decreased TAM treatment and lower-than-average deletion rates (Fig. 1b and data not shown). These data indicate that p53 deficiency fails to rescue acute bone marrow and intestinal degeneration seen in ATRmKO mice and that p53 is required for organismal survival following ATR deletion.
The multiple checkpoint defects conferred by ATR and p53 loss (ATR, S/G2; p53, G1) might permit redundant S phase entry and additive genome destabilization. Indeed, a synthetic-lethal interaction between ATR-Chk1 pathway inhibition and p53 deficiency is observed in cultured cells (ref. 24–26 and data not shown). However, accelerated depletion of ATR-deleted cells did not appear to be the primary cause of tissue failure in p53−/−ATRmKO mice, as ATRΔ/− cells were represented at similar or higher frequencies in the intestines and bone marrow in comparison to ATRmKO mice (Fig. 1f). In addition, mitotic spreads from freshly isolated p53−/−ATRmKO bone marrow exhibited mild and extensive chromatid breaks at an elevated frequency and significantly increased severity (Supplementary Fig. 2). Such levels of genomic instability in the absence of exogenous DNA-damaging treatments are hallmarks of ATR deletion19–22. Together, these data argue that tissue homeostasis in p53−/−ATRmKO mice is compromised despite the continued maintenance of ATRΔ/− cells.
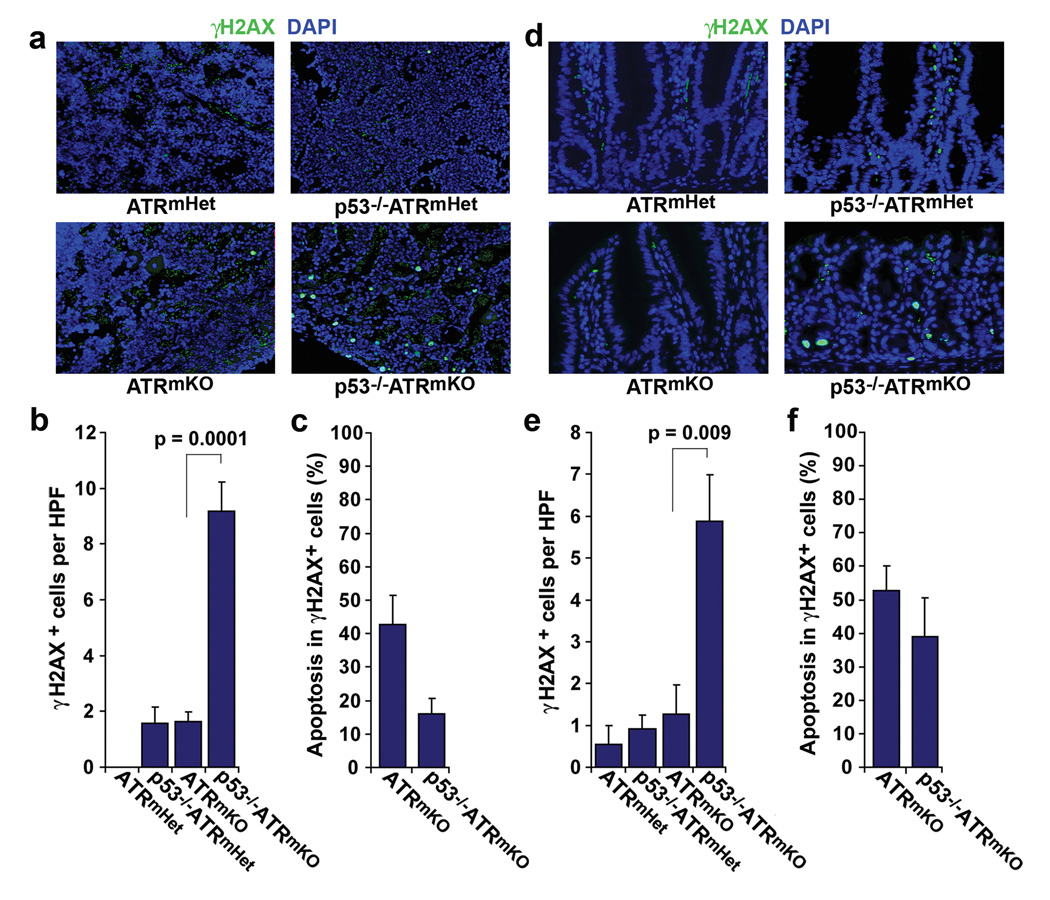
ATR deletion leads to dramatic increases in genomic instability during DNA synthesis, resulting in H2AX phosphorylation (γH2AX)20,21. Notably, the frequency of cells exhibiting H2AX phosphorylation in the bone marrow and intestines of p53−/−ATRmKO mice was dramatically increased compared to ATRmKO controls (Fig. 2). γH2AX staining in the bone marrow was most prominent in cells with aberrantly large nuclei, which were morphologically distinguishable from multinucleated megakaryocytes or condensed apoptotic bodies (Fig. 2a). Indeed, p53-independent apoptosis (cleaved caspase-3 staining) was observed in minority fractions of γH2AX-positive cells in both the bone marrow and intestines of p53−/−ATRmKO mice (Fig. 2c,f), leading to insignificant increases in total numbers of apoptotic cells (Supplementary Fig. 3c). Given that ATRΔ/− cells were maintained at high levels in each of these tissues (Fig. 1f), these data are consistent with p53 deficiency permitting the accumulation of ATRΔ/− cells that have acquired DNA damage in the course of expansion.
Figure 2.
Accumulation of damaged cells in the bone marrow and intestines of p53−/−ATRmKO mice. (a) Representative γH2AX (green) and DAPI (blue) stained sections of the humeral bones from mice of the indicated genotypes 6 days after TAM treatment. Autofluorescent red blood cells (green, DAPI negative) in the sinusoidal spaces of the marrow were excluded from further analysis. (b) Quantification of γH2AX-positive cells in the bone marrow. The abundance of γH2AX-positive cells was determined from 6–10 high power field images (HPF, 200×) of indicated mice (n = 3–6 mice per genotype). Only nucleated (DAPI-positive) cells were scored in this analysis. (c) Frequency of γH2AX-positive cells staining positive for cleaved caspase-3, as determined by co-immunostaining (also see Supplementary Fig. 3a). (d) Representative γH2AX (green) and DAPI (blue) stained intestinal sections from mice of the indicated genotypes 6 days after TAM treatment. Autofluorescent red blood cells (green, DAPI negative) in the vasculature and stroma were excluded from quantifications. (e) Quantification of γH2AX-positive cells in intestinal epithelium (n = 2–6 mice per genotype). The abundance of γH2AX-positive cells was determined as in b. (f) Frequency of γH2AX-positive cells staining positive for cleaved caspase-3, as determined by co-immunostaining (also see Supplementary Fig. 3b). Error bars in b, c, e and f represent the S.E.M. of each data set.
Previous studies have indicated that tissue maintenance in ATRmKO mice following acute deletion is dependent on compensatory renewal from cells that have escaped lox recombination and continue to express ATR23. We sought to directly investigate whether the increased abundance of DNA-damaged cells in p53−/−ATRmKO mice detrimentally affects this regenerative response. To both accurately monitor the process of regeneration and circumvent the lethality associated with systemic ATR and p53 combined deletion, we studied the effect of localized ATR deletion on depilation-induced hair follicle regeneration in the skin. Localized ATR deletion was accomplished through topical treatment with 4-hydroxytamoxifen (4-OHT). Four days after cessation of 4-OHT treatment, hair follicle regeneration was synchronously induced by telogen-phase depilation (plucking) of the hair shafts in the central portion of a larger 4-OHT treatment (ATR-deleted) region.
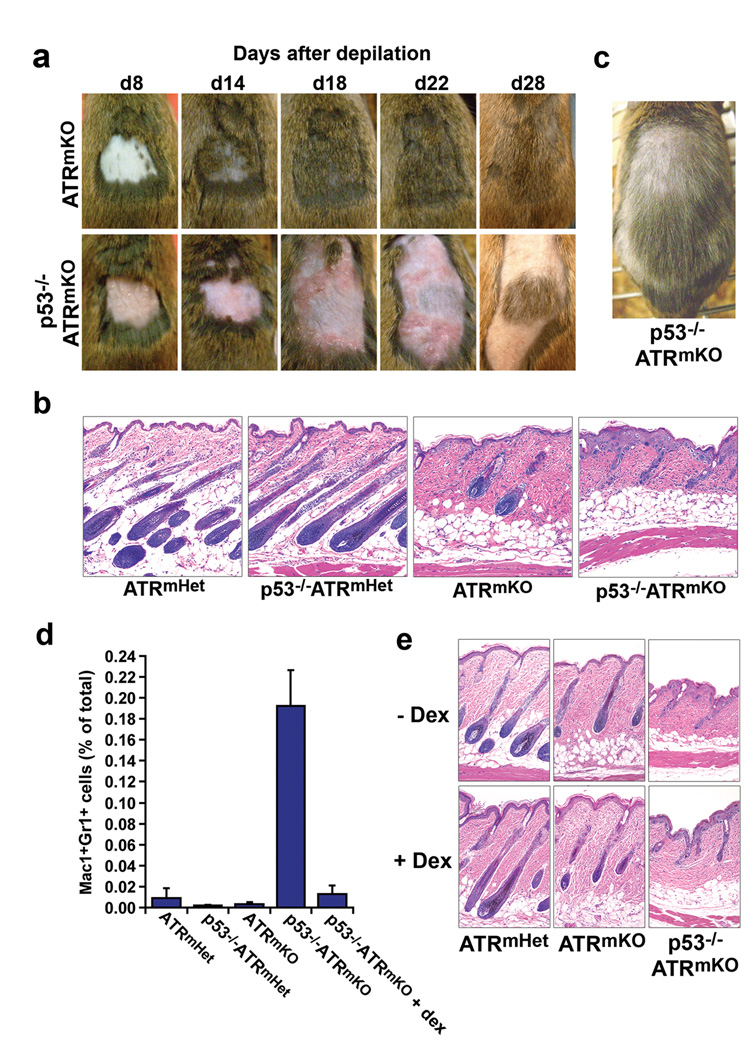
As previously described, only a short delay in hair regeneration (3–4 days) was observed in ATRmKO skin in comparison to ATRmHet controls following a single round of depilation (ref. 23 and data not shown). In sharp contrast, hair regeneration in p53−/−ATRmKO skin was severely delayed compared to ATRmKO skin, as indicated both by shaft regeneration (Fig. 3a) and formation of the hair follicle bulb (Fig. 3b). Closer examination of p53−/−ATRmKO skin revealed distinct malformations of the epithelium, root sheath and hair bulb, each of which was characterized by the presence of aberrant cells with enlarged nuclei (Fig. 3b). Notably, following hair shaft regeneration, an increased abundance of gray hair was observed in p53−/−ATRmKO skin both in the depilated region and the surrounding ATR-deleted region (Fig. 3c); this increase far exceeded that observed after a single round of depilation in ATRmKO skin (Fig. 3a and ref. 23). These results indicate that combined loss of ATR and p53 dramatically impedes hair follicle renewal and leads to long-term deficiencies in regenerated hair shafts (hair graying).
Figure 3.
p53 deficiency dramatically delays hair follicle regeneration following localized ATR deletion in the skin and leads to acute inflammation. (a) Time course following depilation of 4-OHT-treated skins of ATRflox/− and p53−/−ATRflox/− mice. An appreciable delay in hair regeneration after depilation was observed in p53−/−ATRmKO skin and was accompanied by the outward manifestations of inflammation (redness, swelling, exfoliation). These phenotypes were consistently observed in p53−/−ATRmKO mice (n = 6–12 mice per genotype). (b) Representative H&E stained sections of the epidermis 8 days after depilation. The number and quality of regenerating follicles were compromised in the p53−/−ATRmKO skin relative to ATRmKO or ATRflox/+ control skins (n = 5–7 mice per genotype). (c) Regenerated hair in p53−/−ATRmKO skin 48 days after depilation. Appreciable hair graying was observed in p53−/−ATRmKO skin in the depilated and surrounding 4-OHT treatment regions (n = 6 mice). (d) Quantification of Gr1+Mac1+ inflammatory cells in the epidermis 8 days after depilation (n = 4–5 mice per genotype). Total epidermal cells were isolated (Methods), stained with antibodies to Gr1 and Mac1 (CD11b), and the frequency of Gr1+Mac1+ cells was quantified by flow cytometry. Gr1+Mac1+ cells accumulated significantly in p53−/−ATRmKO skin (P = 3 × 10−7), and dexamethasone pretreatment of p53−/−ATRmKO skin (+ dex) prevented Gr1+Mac1+ cell accumulation, P = 0.003. Error bars, s.e.m. (e) H&E stained sections of skin 8 days after depilation in the presence or absence of dex. Mice treated with dex or left untreated were sacrificed 8 days after depilation. Delayed follicle regeneration in p53−/−ATRmKO skin was unaffected by dexamethasone treatment (n = 4 mice per genotype).
The short-term regenerative defect observed in p53−/−ATRmKO skin was accompanied by a severe inflammatory response, as characterized by reddening, exfoliation, and the recruitment of Gr1+Mac1+ innate immune cells (Fig. 3a,d). Inflammation initially occurred at the site of depilation and spread outwardly to eventually cover the entire ATR-deleted region (Fig. 3a). This phenotype was not observed in ATRmKO or ATRmHet controls (Fig. 3a,d and ref. 23). To determine whether delayed follicle regeneration in p53−/−ATRmKO skin was a consequence of acute inflammation, the topical immunosuppressant dexamethasone (dex) was applied 4 days after depilation and subsequently. This treatment was sufficient to prevent skin reddening and inhibit Gr1+Mac1+ innate immune cell infiltration into p53−/−ATRmKO epithelium and did not affect hair follicle generation in controls (Fig. 3d and Supplementary Fig. 4). However, dex treatment did not rescue hair follicle regeneration in p53−/−ATRmKO skin (Fig. 3e and Supplementary Fig. 4). Interestingly, cessation of dex treatment 22 days after depilation in p53−/−ATRmKO skin was immediately followed by a severe inflammatory response (Supplementary Fig. 4), indicating that the cause of inflammation in p53−/−ATRmKO skin continued to be present in the tissue for up to 3 weeks after depilation. In summary, while it is possible that innate immune infiltration and the gross pathological effects of inflammation in p53−/−ATRmKO skins may ultimately impact tissue maintenance in some manner (e.g. hair graying), they do not appear to be the primary cause of short-term delays in hair follicle regeneration.
To assess original ATR deletion rates prior to depilation (telogen phase) and quantify the representation of ATRΔ cells during follicle regeneration, the recombination status of the ATRflox allele was determined both before (Day 0) and 4 and 8 days after depilation. The frequencies of ATRΔ cells (either ATRΔ/+ or ATRΔ/−) were similar in all genotypes both prior to depilation (Day 0) and in early anagen phase (4 days after depilation, Fig. 4a). These results indicate that deletion rates (approximating 90% in the epithelium) were unaffected by p53 status prior to depilation. While the frequencies of ATRΔ/− cells were similar in early anagen, representation of these cells significantly decreased later in anagen (8 days after depilation) in both ATRmKO and p53−/−ATRmKO skins (P = 0.0003, Fig. 4a). Similar to other tissues (Fig. 1f), this decline was not accentuated by p53 deficiency (Fig. 4a), indicating that the regenerative delay in p53−/−ATRmKO skin occurred despite the continued maintenance of ATRΔ/− cells.
Figure 4.
p53−/−ATRmKO skin is characterized by the persistent accumulation of γH2AX-positive cells. (a) Frequency of the ATRflox-recombined allele (ATRΔ) in epidermal isolates at different time points after depilation (n = 3–13 mice per genotype per time point). Recombination of the ATRflox allele was quantified by qPCR amplification of genomic DNA isolated from total epidermal cells (described in Methods). (b) Frequency of γH2AX-positive epidermal stem and progenitor cells (CD34+α-6-integrin+) isolated from skin 4 days and 8 days following depilation. The difference in mean abundance of γH2AX-positive cells between p53−/−ATRmKO and ATRmKO skins 8 days post-depilation was significant (P = 0.023). (c) Representative γH2AX (brown) and hematoxylin (blue) stained sections of the skin 4 days and 8 days after depilation. (d) Quantification of γH2AX-positive cells in the hair follicles and epidermis of depilated skin. Frequency of γH2AX-positive cells was determined in 6–10 sections per mouse (n=3–5 mice per genotype for each time point). Note the elevated frequency of γH2AX-positive cells in both ATRmKO and p53−/−ATRmKO skins 4 days post-depilation; however, this significant increase was maintained only in p53−/−ATRmKO skin 4 days later (day 8 post-depilation). The difference in mean abundance of γH2AX-positive cells between p53−/−ATRmKO and ATRmKO skins 8 days post-depilation was significant (P = 0.013). Increased frequency of γH2AX-positive cells in p53−/−ATRmKO skin was observed both in the presence and absence of dexamethasone (Supplementary Fig. 4).
We hypothesized that the regenerative defect in p53−/−ATRmKO skin may be caused by the persistent accumulation of highly damaged ATRΔ/− cells, which would continue to occupy functional space and impede regeneration from residual ATR-expressing progenitors. To investigate this possibility, γH2AX-positive cells were quantified in the general epithelium and in CD34+α-6-integrin+ stem and progenitor cells, which are among the earliest populations of cells to proliferate and contribute to hair follicle renewal27–29. In early anagen (day 4 post-depilation), the abundance of γH2AX-positive cells was increased to similar extents in ATRmKO and p53−/−ATRmKO skins, both in progenitor populations and in the general epithelium (Fig. 4b–d). Therefore, the generation of damaged ATRΔ/− cells soon after exiting quiescence is not strongly influenced by p53 function. However, at later stages of hair follicle regeneration (day 8 post-depilation), the frequency of γH2AX-positive cells significantly declined in ATRmKO skins and was maintained only in p53−/−ATRmKO skins (Fig. 4c,d). Interestingly, continued proliferation during differentiation from progenitors may amplify differences in the frequency of γH2AX-positive cells between ATRmKO and p53−/−ATRmKO skins (Fig. 4c,d). γH2AX-positive cells exhibited enlarged nuclei that were morphologically distinct from apoptotic cells, consistent with the low frequency of cells staining positive for cleaved caspase-3 (Supplementary Fig. 5). These data indicate that ATR deletion in the absence of p53 leads to the persistent accumulation of genetically unstable ATRΔ/− cells.
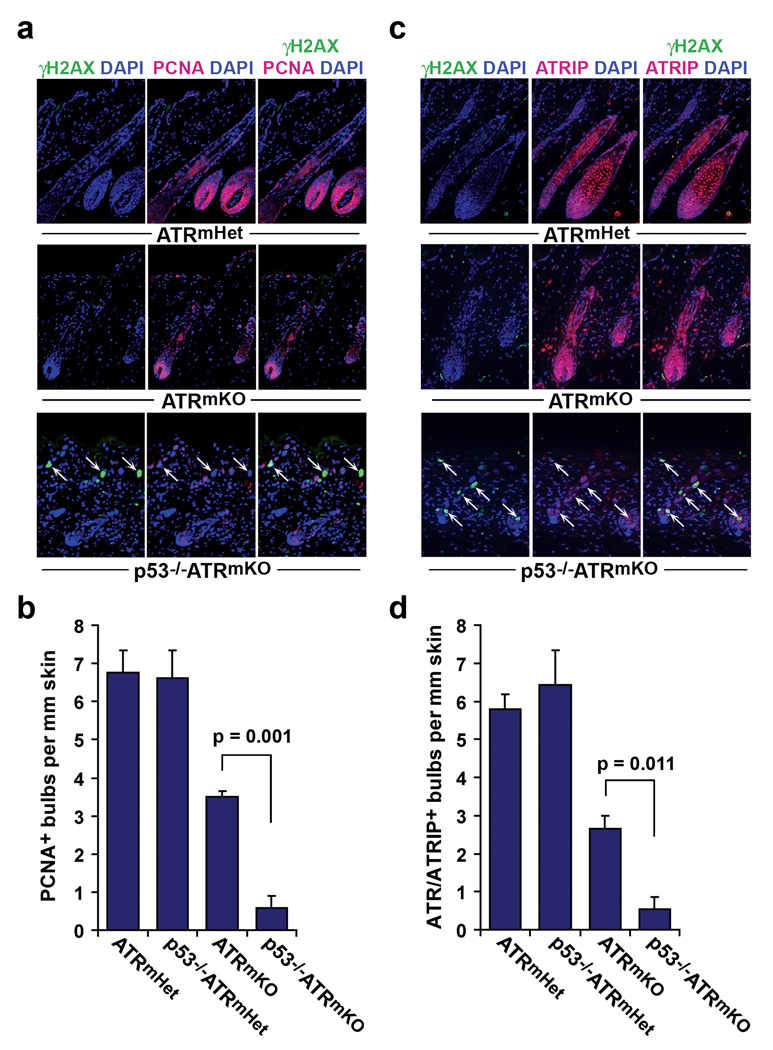
We then examined whether the accrual of γH2AX-positive cells in p53−/−ATRmKO skin was associated with defective compensatory renewal from residual cells that escaped ATR deletion. Consistent with previous studies23, a large fraction of ATRmKO hair follicles began to develop hair bulbs within 8 days of depilation (Fig. 5). These bulbs exhibited cellular proliferation (nuclear PCNA staining) and were largely comprised of cells that expressed the ATR/ATRIP complex (Fig. 5). In contrast, p53−/−ATRmKO skin largely failed to undergo compensatory regeneration, as indicated by the absence of cellular proliferation in the hair bulb and the lack of expansion of ATR/ATRIP-positive cells (Fig. 5). These data indicate that the compensatory renewal driven by residual ATR-expressing cells is strongly impeded in p53−/−ATRmKO skin.
Figure 5.
p53 is required for efficient compensatory renewal following ATR deletion. (a) Representative PCNA (red), γH2AX (green), and DAPI (blue) stained sections of skin 8 days after depilation. γH2AX-positive cells are indicated (white arrows). (b) Quantification of regenerating PCNA-positive bulbs per mm skin. (c) Follicle regeneration in ATRmKO skin by ATR/ATRIP-expressing cells. Representative ATRIP (red), γH2AX (green), and DAPI (blue) stained sections of skin 8 days after depilation. ATR and ATRIP form an interdependent complex in which ATR is required for ATRIP stability32. γH2AX-positive cells are indicated (white arrows). (d) Quantification of ATR/ATRIP-expressing bulbs per mm skin. Analyses and quantifications for b and d were performed on 6–12 sections per mouse (n=3 mice per genotype). Error bars represent S.E.M. for each data set.
Previously, we have shown that ATR-deleted cells are rapidly eliminated from proliferative tissues and replaced through compensatory renewal by undeleted, ATR-expressing progenitors23. The studies described herein demonstrate that p53 limits the accumulation of highly damaged cells following mosaic ATR-deletion and argue that loss of this constraint delays tissue regeneration. These results strongly support the model that p53 plays a key role in maintaining adult tissue homeostasis by preventing functionally compromised cells from acting as a barrier to progenitor-driven renewal15–18. Occupancy of tissues with genetically unstable cells might delay tissue renewal either by obstructing competent progenitors from interacting with key environmental cues or by causing the generation of secreted factors30,31 that inhibit renewal. Such mechanisms may operate as a tissue homeostatic checkpoint, delaying regeneration until damaged cells have been effectively cleared.
The degeneration of tissue integrity following combined loss of ATR and p53 is somewhat unexpected given that p53 deficiency generally attenuates the effects of mutations in other genome maintenance regulators1–12. While p53 loss may be sufficient to accommodate lesser degrees of genomic instability and permit mildly damaged cells to functionally contribute to tissue homeostasis, it is likely that the severe cell cycle and genome destabilizing consequences of ATR deficiency are not alleviated by p53’s absence and, rather, are exacerbated by it. According to this model, replication fork instability in concert with near-complete loss of cell cycle checkpoint functions without ATR and p53 might allow redundant or aberrant S phase entry, leading to further instability and the inappropriate propagation of terminally damaged cells. Indeed, the persistent accumulation of genetically unstable cells in p53−/−ATRmKO tissues compared to p53 wild-type ATRmKO controls supports this model (Fig. 2, Fig. 4c,d and Supplementary Fig. 2). Therefore, in addition to supporting a general function for p53 in facilitating regeneration of adult tissues, this study provides the first evidence of a specific synthetic interaction between ATR and p53 inhibition in adult mammals. These results suggest that ATR-Chk1 inhibitors may be uniquely useful for the treatment of p53-deficient cancers by causing synergistic increases in genomic instability, which may consequentially inhibit regeneration from undamaged bystanders.
METHODS
Methods and associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Gregory Beatty, Karlene Cimprich, George Cotsarelis and Avinash Bhandoola for reagents, protocols and helpful advice and the AFCRI Histology Core for tissue processing. These studies were supported by the National Institute on Aging (R01AG027376 and F30AG034027) and the Abramson Family Cancer Research Institute.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
Y.R., D.W.S. and E.J.B. designed and interpreted the experiments and wrote the manuscript. Y.R. and D.W.S. performed all of the experiments, with assistance from A.A. (mouse maintenance, qPCR and histological analysis) and C.C. and R.H.V. (quantification of Gr1+Mac1+ cell infiltration).
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Wlodarski P, et al. Role of p53 in hematopoietic recovery after cytotoxic treatment. Blood. 1998;91:2998–3006. [PubMed] [Google Scholar]
- 2.Bender CF, et al. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16:2237–2251. doi: 10.1101/gad.1007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin L, et al. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- 4.Frank KM, et al. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell. 2000;5:993–1002. doi: 10.1016/s1097-2765(00)80264-6. [DOI] [PubMed] [Google Scholar]
- 5.Gao Y, et al. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature. 2000;404:897–900. doi: 10.1038/35009138. [DOI] [PubMed] [Google Scholar]
- 6.Hakem R, de la Pompa JL, Elia A, Potterx J, Mak TW. Partial rescue of Brca1 (5–6) early embryonic lethality by p53 or p21 null mutation. Nat Genet. 1997;16:298–302. doi: 10.1038/ng0797-298. [DOI] [PubMed] [Google Scholar]
- 7.Lim DS, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim DS, et al. Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol. 2000;20:3772–3780. doi: 10.1128/mcb.20.11.3772-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Yang EM, Brugarolas J, Jacks T, Baltimore D. Involvement of p53 and p21 in cellular defects and tumorigenesis in Atm−/− mice. Mol Cell Biol. 1998;18:4385–4390. doi: 10.1128/mcb.18.7.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, et al. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat Genet. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- 11.Botchkarev VA, et al. p53 is essential for chemotherapy-induced hair loss. Cancer Res. 2000;60:5002–5006. [PubMed] [Google Scholar]
- 12.Orii KE, Lee Y, Kondo N, McKinnon PJ. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc Natl Acad Sci U S A. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 14.Maier B, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18:306–319. doi: 10.1101/gad.1162404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 16.Serrano M, Blasco MA. Cancer and ageing: convergent and divergent mechanisms. Nat Rev Mol Cell Biol. 2007;8:715–722. doi: 10.1038/nrm2242. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Cao I, et al. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 2006;7:546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 20.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chanoux RA, et al. ATR and H2AX Cooperate in Maintaining Genome Stability under Replication Stress. J Biol Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4:216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 25.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A. 2001;98:9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nghiem P, Park PK, Kim Ys YS, Desai BN, Schreiber SL. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J Biol Chem. 2002;277:4428–4434. doi: 10.1074/jbc.M106113200. [DOI] [PubMed] [Google Scholar]
- 27.Morris RJ, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 28.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Coppe JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- 32.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.