Abstract
Indian Plant Anticancer Compounds Database (InPACdb) is a web-based open access database of phytochemicals. The objective of this initiative is to project the potential of anticancer phytochemicals from Indian pharmacopoeia in an integrated environment. This database is unique in providing comprehensive information covering cancer type, molecular target, 3D Stereochemical structures (tautomers, stereoisomers, conformers and resonance structures) and Chemical descriptors etc. for each entry, enabling effective cheminformatics analysis. The complete dataset of InPACdb encompasses 32 descriptive fields for each entry, and is freely available for download at http://www.inpacdb.org.
Keywords: Biological database, anticancer compounds, Indian plants
Background
Ancient Indian medicinal science was based on the natural products of plant origin. Recently, numerous bioactive compounds have been isolated from plant sources and several of them are currently in clinical trials. Plant-derived compounds have been a vital source of varied clinically useful anti-cancer agents: Camptothecin derivatives [1], Topotecan and Irinotecan, Etoposide, derived from Epipodophyllotoxin [2], and Paclitaxel (taxol) [3]. Furthermore, other potent molecules include Vinca alkolloids (Vinblastine, Vincristine) [4], Flavopiridol, a semi-synthetic analogue of the chromone alkaloid and Rohitkine, a pyridoindole alkaloid derived from leaves of Ochrosia species [5]. Presently, research on anticancer drug development is largely dependent on exploring potential phytochemicals. Indian sub-continent is rich in its diversity of flora, being a tropical country with a large spread of rain forests and river basins. It is floristically rich with about 33 percent of its botanical wealth (over 15,000 species of higher plants) being endemic [6]. Hence, analyzing this potential, InPACdb initiative is taken to expose the potent anticancer phytochemicals of Indian Origin in international arena.
Methodology
InPACdb's search engine is based on Google's proprietary search technology which utilizes java scripting and custom indexing [7]; making search comparatively faster and user friendly. The database presently comprises a dataset of 144 compounds (Table 1 in supplementary material) which has been manually sorted from 990 compounds; 200 from National Cancer institute drug list and 690 from Asian anticancer material databank and other 100 compounds from other miscellaneous sources. The Cancer type, Molecular target and Mechanism of action for each segregated compound were documented from elsewhere [8]. The chemical descriptors and 3D structure for each compound were calculated using Marvinsketch [9] and Chemsketch [10], respectively. Finally, each compound's entry is indexed via Google Custom Search for database information retrieval. A detailed flowchart summarizing the methodology implemented is shown in Figure 1.
Figure 1.
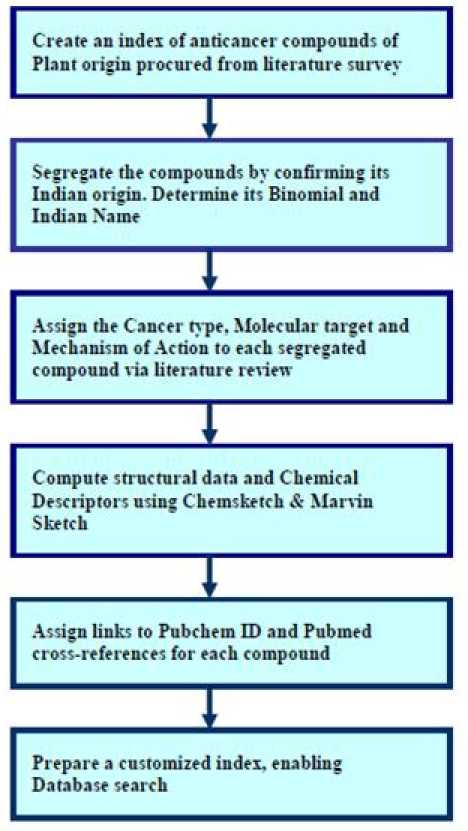
Flowchart showing the methodology implemented in database development
InPACdb features:
Ayurveda is an ancient Indian herbal medicine system and is followed till date for anticancer treatment. This medicinal system is also proven to be at par with traditional Chinese Medicinal System. Currently, there are numerous herbal databases that provide information on herbal anticancer compounds: Traditional Chinese Medicine Database System [11], Asian Anticancer material database [12] etc. These databases largely concentrate on Chinese Traditional Medicine system and are limited to data like taxonomical classification, pictures of plants and SMILES notation of bioactive compounds. InPACdb is unique in providing comprehensive information on phytochemicals of Indian origin with anticancer activity via 32 descriptive fields; encompassing the basic data like plant image, nomenclature etc. to advanced data like stereochemical properties, chemical descriptors etc. The customized search engine of this database enables wildcard querying, which shall include SMILES string, Cancer type and names of organs, Compounds and plants etc. Each entry in the database is subdivided into 6 parts: General information, origin, target, structure information, Computed Chemical Descriptors and Literature references (Figure 2).
Figure 2.

Showing an entry from InPACdb
The general information part of the database entry displays the compound's unique InPACdb accession number, followed by its nomenclature, 2D-structure image, compound type (Alkaloid, Flavonoid, Phenolic etc), IUPAC name and an external link to the compound’s PUBCHEM ID (if available). Origin section provides detailed information about Indian plant origin from which the compound is isolated. The Indian nomenclature of the plant is given either in Sanskrit (if available) or general name in other Indian languages, subsequently followed by its Binomial name and respective plant's image. Target section gives information on the probable types of cancers on which the compound can act upon, which were curated based on peer-reviewed literature sources. Furthermore, the molecular targets of the respective compounds are also provided along with its bioactivity. Atomic coordinates of each compound entry is provided in the form of PDB and MOL format in structure information section. Moreover, additional information of the compounds like isomers (Sterioisomers, Tautomers, Resonance) and conformers are also given in SDF format, in addition to SMILES notation.
Properties Computed from the structure: this section of the database provides various computed descriptors like Molecular Weight, Molecular Formula, XLogP, H-Bond Donors, H-Bond Acceptors, Refractivity, Rotatable Bond Count, Tautomer Count, atomic Mass, Topological Polar Surface Area, Vander Waals surface area (3D) and Complexity. Finally, reference section lists the citations relevant to the anticancer activity of the respective compounds in the database. Moreover, the multiple links to the citations are also provided (if available).
Utility
The data presented in InPACdb can be effectively used for Cheminformatics studies like QSAR analysis, pharmacophore search, molecular docking etc. pertaining to anticancer drug discovery. InPACdb also clearly portrays the potential role of google custom search as a Biological information retrieval system.
Caveat and Future Developments
The database will be extended with more data on molecular interactions, embedded interactive visualization tools and additional chemical descriptors. The users are also welcome to contribute relevant data to the database via email to authors. The dataset and web interface InPACdb shall be upgraded periodically.
Supplementary material
Footnotes
Citation:Vetrivel et al, Bioinformation 4(2): 71-74 (2009)
References
- 1.Kepler JA, et al. J Org Chem. 1969;34:3853. doi: 10.1021/jo01264a026. [DOI] [PubMed] [Google Scholar]
- 2.Utsugi, et al. Cancer Research. 1996;56:2809. [PubMed] [Google Scholar]
- 3.Cragg GM, Newman DJ. J Ethnopharmacol. 2005;100:72. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Johnson IS, et al. Cancer Res. 2001;23:1390. [PubMed] [Google Scholar]
- 5.Arguello F, et al. Blood. 1998;91:2482. [PubMed] [Google Scholar]
- 6. http://www.bioversityinternational.org/publications/Web_version/174/ch06.htm.
- 7. http://www.google.com/coop/cse.
- 8. http://pubchem.ncbi.nlm.nih.gov.
- 9. http://www.chemaxon.com.
- 10. http://www.acdlabs.com.
- 11. http://www.cintcm.ac.cn.
- 12. http://asiancancerherb.info.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


