Abstract
Interleukin (IL)-31 is a recently described cytokine, preferentially produced by T helper 2 lymphocytes and associated with skin diseases, such as atopic dermatitis. IL-31 is a member of the four α-helix bundle cytokine family and is related to the IL-6 subgroup. Its heterodimeric membrane receptor is composed of the gp130-like receptor (GPL) subunit associated to the oncostatin M receptor subunit. We identified critical amino acids implicated in the ligand receptor interaction by computational analysis combined with site-directed mutagenesis. Six IL-31 residues selected for their putative involvement in cytokine receptor contact sites were alanine-substituted, and the corresponding proteins were expressed in mammalian and bacterial systems. Biochemical, membrane binding, cell signaling, and cell proliferation analyses showed that mutation E44A, E106A, or H110A abolished IL-31 binding to GPL and the subsequent signaling events. A second ligand receptor-binding site involved Lys134, with alanine substitution leading to a protein that still binds GPL, but is unable to recruit the second receptor subunit and the subsequent signaling pathways. The results indicate that IL-31 recognizes its receptor complex through two different binding sites, and we propose a three-dimensional model for IL-31.
Keywords: Cytokines/Interleukins, Protein/Conformation, Protein/Protein-Protein Interactions, Protein/Structure, Receptors/Cytokine, Interleukin-31
Introduction
Interleukin-31 (IL-31)4 is a newly described T cell-derived cytokine mostly produced by T helper type 2 cells and involved in promoting skin disorders and regulating other allergic diseases such as asthma (1–5). Mice treated with intradermal injection of IL-31 or transgenic mice overexpressing IL-31 present increased scratching behavior and develop severe dermatitis (1). An association of IL-31 with atopic dermatitis was found in the IL-31 transgenic mouse phenotype that mimics the human pathology (6).
In humans, atopic individuals have an increased tendency to produce high levels of IL-31 in response to external trigger factors, which may contribute to the development of pruritus. Moreover, IL-31 mRNA is increased in circulating and skin-infiltrating cutaneous lymphocyte antigen-positive T cells of these patients (7). This cytokine is related to the IL-6 cytokine family by its structure and receptor complex (8). IL-31 is a four-helix bundle cytokine comprising two long cross-over loops and one short loop (1, 9). The mRNA encodes for a 15.5-kDa protein displaying an apparent molecular mass of 24 kDa in its glycosylated form. Based on its overall length, IL-31 is quite close to the IL-2 or granulocyte colony-stimulating factor short chain groups of cytokines but does not display apparent homology or functional properties with them.
The IL-31 heterodimeric receptor is composed of gp130-like receptor (GPL, also known as GLM-R or IL-31RA) (1, 10, 11) and oncostatin M receptor (OSMR) (12, 13). GPL was cloned as a member of the type I cytokine receptor family (10, 11). Its name reflects its close homology to gp130, the shared receptor subunit of the IL-6-type cytokines (14, 15). GPL displays common structural motifs with the family of type I cytokine receptors, i.e. the cytokine-binding domain (CBD) with two pairs of conserved cysteine residues and a WSXWS box in its extracellular region (16, 17). IL-31 directly binds GPL and recruits OSMR to form the high affinity receptor (18, 19). OSMR is a 150-kDa protein composed in its external portion of a half CBD followed by an immunoglobulin-like domain, a second complete CBD, and a region consisting of three fibronectin type III domain repeats. The OSMR subunit also contributes together with gp130 to the formation of the type II OSM receptor, specifically recognizing OSM (12, 13).
The binding of IL-31 to its receptor leads to intracellular activation via recruitment of the Jak1, Jak2, STAT-1, STAT-3, and STAT-5 signaling pathways, as well as the phosphatidylinositol 3-kinase/Akt cascade. The SHP-2 and Shc adapter molecules are also recruited and contribute to an increased activation of the MAPK pathway in response to IL-31 (18–20).
Cytokines of the IL-6 family are four-helix bundle proteins, with an up-up down-down topology. In addition to IL-6, this family encompasses IL-11, IL-27, OSM, leukemia inhibitory factor, ciliary neurotrophic factor, cardiotrophin-like cytokine, and neuropoietin (21). The α-helices and loops composing theses cytokines are named A–D from the N terminus to the C terminus. Crystallographic structures and site-directed mutagenesis studies have shown that these cytokines interact with their receptor through three different binding sites, numbered 1–3 by analogy with IL-6 (22, 23). Cytokines requiring a small α-chain, like IL-6, IL-11, ciliary neurotrophic factor, cardiotrophin-like cytokine, and neuropoietin, bind first to the receptor through a site composed of residues from the C-terminal parts of the AB loop and the αD helix.
Site 2 is located on solvent-exposed faces of the αA and αC helices and is important for binding to gp130. Site 3 is specific to the IL-6 family and corresponds to an additional signaling receptor-binding site for the recruitment of gp130, leukemia inhibitory factor receptor, or OSMR (15). Binding site 3 is characterized by a conserved aromatic residue located at the N terminus of the αD helix (24).
The aim of the present study was to determine the critical binding determinants of IL-31 for interaction with and activation of its receptor components. Our results imply that Glu44 in the αA helix and Glu106 and His110 in αC helix contribute to site 2, whereas Lys134 in αD helix contributes to site 3. These residues are key residues for efficient recruitment of GPL and OSMR and for functional activity of IL-31.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Cos-7 and GO-G-UVM cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. The culture medium of Ba/F3 cells modified to express human gp130, OSMR, and GPL was supplemented with 1 ng/ml murine IL-3. Human recombinant OSM was purchased from R & D Systems (Oxon, UK). The OSMR-Fc and GPL-Fc proteins, and IgG1 isotype control antibody were produced in the laboratory. The polyclonal anti-human IL-31 antibodies used for ELISA were from R & D Systems. The anti-V5 mAb was from Invitrogen (Carlsbad, CA). The anti-STAT-3 polyclonal antibody and the mAb specific for Tyr(P)705-STAT-3 were from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling (Beverly, MA), respectively. Anti-phospho-MAPK (Thr202/Tyr204) and phospho-Akt (Ser473) mAbs and anti-MAPK and anti-Akt polyclonal antibodies were from Cell Signaling.
IL-31 Site-directed Mutagenesis and Protein Expression
The cDNA encoding wt human IL-31 was cloned in the pcDNA3.1 vector (Invitrogen) in frame with a C-terminal V5 epitope followed by a His tag and was subjected to site-directed mutagenesis using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. The mutations were verified by automatic DNA sequencing. Cos-7 cells were transfected with wt and mutant forms of human IL-31 cDNAs using the Exgen transfection reagent (Euromedex, Souffelweyersheim, France). After a 72-h transfection period, the cell supernatants were collected, cleared by centrifugation, and stored at −20 °C.
For bacterial production, the cDNA encoding wt human IL-31 was subcloned in the pET41a plasmid (Novagen) coding for the glutathione S-transferase-histidine tag thrombin-sensitive site at the 5′ side of the inserted cDNA. The 3′ side contained a V5 tag followed by a second histidine tag. To obtain the mutant forms of IL-31, site-directed mutagenesis was performed using the QuikChangeTM site-directed mutagenesis kit (Stratagene). After fermentation, the bacteria were disrupted, and the lysate was loaded on a nickel-chelating column (Qiagen). After imidazole elution, the fusion protein was cleaved with thrombin and loaded on a glutathione column. The unbound fraction was loaded onto a second nickel-chelating column to obtain the purified recombinant protein. Proteins were then submitted to SDS-PAGE and silver staining analysis.
Quantification of IL-31 V5 His wt and Mutant Supernatants by ELISA
A 96-well microtiter plate was coated with a polyclonal anti-human IL-31 antibody at 2 μg/ml overnight at 4 °C. After saturation with 20% sucrose-Tris, the wells were incubated with supernatant dilutions of wt and mutant IL-31 for 6 h at 37 °C. After three washing steps using 0.05% Tween 20 with phosphate-buffered saline, the wells were incubated with a biotinylated polyclonal anti-human IL-31 antibody (0.25 μg/ml) in 0.1% bovine serum albumin, 0.01% Tween 20 with phosphate-buffered saline overnight at 4 °C. After three additional washing steps, streptavidin-poly-horseradish peroxidase (Sanquin, Amsterdam, The Netherlands) (diluted 1:10,000 in 0.1% bovine serum albumin, 0.01% Tween 20 with phosphate-buffered saline) was added to the wells for 1 h at 37 °C. After the washing steps, binding of the streptavidin-horseradish peroxidase was visualized by the addition of ABTS substrate solution. The absorbance at 405 nm was determined using an ELISA reader.
Co-immunoprecipitation of Ligand-Receptor Complexes
For co-immunoprecipitation experiments, OSM, wild type, and mutant forms of IL-31 proteins were diluted to a final concentration of 300 ng/ml. GPL-Fc and OSMR-Fc are two fusion proteins made of the extracellular part of the membrane receptor coupled to the Fc portion of human IgG1. OSMR-Fc, kindly provided by B. Mosley (Immunex/Amgen, Seattle, WA), and GPL-Fc were subcloned in the pME18S expression vector. The soluble receptors were incubated with the indicated cytokines overnight at 4 °C. The complexes were then isolated using Sepharose beads coupled to protein A (Amersham Biosciences) and were submitted to Western blotting analyses as described above. IL-31 or OSM association with the soluble receptors was detected using an anti-V5 mAb or a biotinylated polyclonal anti-OSM antibody (R & D Systems), respectively. Soluble receptors fused to the IgG1 Fc fragment were detected with an anti-Fc antibody. The reaction was visualized by ECL using an image Master camera from Amersham Biosciences (Uppsala, Sweden).
Binding Analysis by Flow Cytometry
For the binding analysis of wt and mutant IL-31, GO-G-UVM cells were incubated for 1 h at 4 °C with wt or mutant IL-31-V5 His (700 ng/ml), followed by an additional 30-min incubation step with the anti-V5 mAb (10 μg/ml) before incubation with a phycoerythrin-conjugated anti-mouse antibody (Dako). Fluorescence was subsequently analyzed on a FACScalibur flow cytometer (Becton Dickinson, Mountain View, CA).
Phosphorylation Assays
GO-G-UVM cells were activated for 10 min with the indicated cytokines before being lysed in 62.5 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mm dithiothreitol, 0.1% bromphenol blue. The lysates were subjected to SDS-PAGE and immunoblot analysis with an antibody specific for the phosphorylated form of signaling proteins. The membranes were stripped in 0.1 m glycine, pH 2.5, for 2 h and neutralized in 1 m Tris-HCl, pH 7.6, before being reblotted with the antibody recognizing all isoform proteins.
Proliferation Assays
Ba/F3 gp130-OSMR-GPL cells were seeded in 96-well plates at a concentration of 5.103 cells/well in RPMI 1640 medium containing 5% fetal calf serum. The cells were incubated with wild type and mutants IL-31 diluted to 100 ng/ml, and then serial dilutions of the cytokines were performed in triplicate. After a 72-h incubation period, 0.5 μCi of [3H]Tdr was added to each well for the last 4 h of the culture, and the incorporated radioactivity was determined by scintillation counting.
Bioinformatic Analyses
Bioinformatic analyses were carried out using the following software: T-Coffee, PsiPred, pMUT, Weblogo, and I-TASSER. IL-31 ortholog sequences were retrieved from the Uniprot and GenBankTM data bases.
RESULTS
Computational Analysis of the Human IL-31 Sequence
Mature IL-31 encompasses 141 residues and is related to the short chain cytokine subgroup, whereas at the same time, IL-31 signals through OSM receptor β and gp130-like receptor, both belonging to the long chain cytokine group of receptors. In addition, IL-31 has no apparent homology to other known four helical bundle cytokines, and identification of its key residues using related cytokine alignments or homology molecular modeling was therefore limited. To identify amino acids potentially involved in the cytokine receptor-binding sites, we first searched for evolutionarily conserved IL-31 residues. For this purpose, 13 IL-31 orthologs (hedgehog, horse, mouse lemur, mouse, rat, microbat, pika, rabbit, megabat, squirrel, tarsier or Microcebus murinus, tree shrew, or Tupaia belangeri, dolphin) were retrieved from the Ensembl data base (25) and then aligned using a combination of local and global alignment algorithms computed by T-Coffee (for simplicity the presented alignment only shows four relevant and distantly related species sequences) (Fig. 1A). We also determined the structural environments of conserved residues by predicting IL-31 secondary structures with PsiPred. This software predicts secondary structure by scoring matrices that are mathematical representations of sequence alignment generated by PSI-BLAST. Important sequence plasticity was observed between the orthologs studied in our alignment, and most of the conserved residues were apolar amino acids located in the α-helices forming the hydrophobic core of proteins.
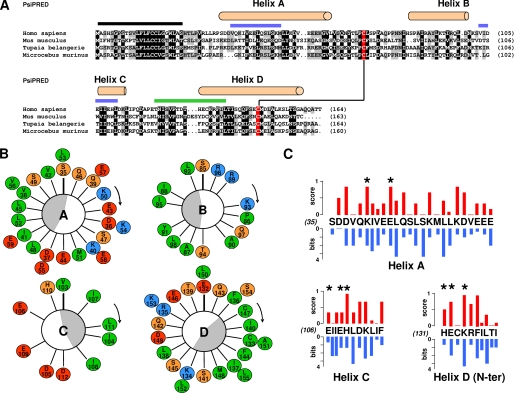
FIGURE 1.
Sequence analysis of IL-31. A, sequence alignment of different IL-31 orthologs selected to be the most divergent. The α-helices predicted by PsiPred are indicated on top of the sequences, and the predicted leader sequence is highlighted by a black line. A disulfide bridge is predicted between two conserved cysteine underlined in red. 100 and 80% conserved or type-conserved residues are highlighted in black and gray, respectively. Sites 2 and 3 are indicated with blue and green lines, respectively, above the alignment. B, Edmundson helical wheel representation of α A, B, C, and D helices. Hydrophobic residues are in green, neutral and hydrophilic amino acids are in orange, positively charged residues are in blue, and negatively charged residues are in red. The amphiphilicities of the α-helices are indicated by gray crescents. C, plot representation of residue conservation scores in blue versus pMUT residue scores in red for amino acid forming α A, C helices, and the N-terminal part of the D helix.
In addition, two cysteine residues appeared to be strictly conserved over all the known IL-31 orthologs. These amino acids are positioned in the predicted AB loop and αD helix, suggesting a bridging pattern analogous to the ones observed for leukemia inhibitory factor and OSM. Crystallographic structures and site-directed mutagenesis studies have shown that IL-6-related cytokines interact through two or three different binding sites numbered 1–3 by analogy with IL-6 (23). In the first helical turn of the predicted αD helix, we looked for conserved aromatic amino acids, which is the known hallmark of cytokine-binding site 3 in the IL-6/IL-12 family of proteins (15, 26, 27). We failed to identify the presence of such a motif in the IL-31 sequences. Then we determined α-helices amphipathicity by representing the predicted α-helix segments of human IL-31 in a helical wheel format (Fig. 1B). This representation allowed us to identify periodically buried hydrophobic amino acids. Interestingly, the residues that contribute to the solvent-exposed faces of the αA and αC helices and that form binding site 2 were mostly negatively charged amino acids. No key residue related to the IL-6 family binding site could be determined.
To predict potential IL-31 hot spots, pMUT scoring was used. This software simulates exhaustive mutations throughout the protein sequence. It combines multiple alignments, secondary structure predictions, and extracted information from human mutational data bases to determine protein integrity indexes. Values greater than 0.5 indicate potentially pathological mutation (Fig. 1C, red bars). Then the WebLogo software, which calculates the difference between maximum entropy and the entropy of the observed amino acid distribution, was also used to score residue variability among IL-31 orthologs (Fig. 1C, blue bars).
The work was focused on αA, αC, and the N-terminal portion of αD helices, which are regions known to contain the receptor-binding sites for the IL-6 cytokine family. To determine an alternative consensus score, we compared the pMUT and the conservation scores for each amino acid and identified residues with the highest scores determined by the two methods. These data, combined with the solvent exposure prediction (buried hydrophobic amino acids were not retained), led us to select the following hot spot residues as candidates: Asp37, Lys40, and Glu44 in the αA helix; Asp105, Glu106, Glu109, and His110 in the αC helix; and His131, Glu132, and Lys134 in the αD helix.
Expression and Quantification of IL-31 Mutants
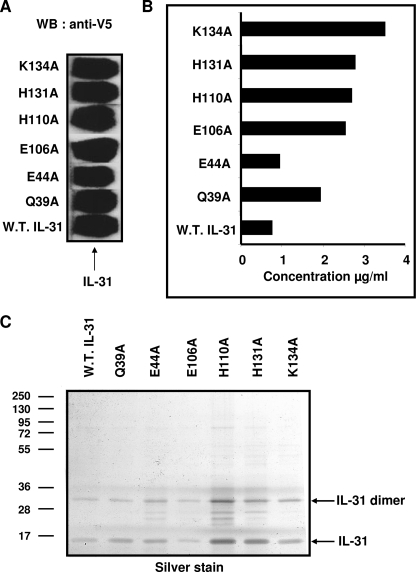
Based on our predictions, we introduced the following mutations in human IL-31 to alter its binding properties: D37A, K40A, E44A, D105A, E106A, E109A, and H110A are predicted to disrupt IL-31-binding site 2 and are located at the αA and αC helices; and H131A, E132A and K134A are predicted to disrupt IL-31 binding site 3 and are located at the N-terminal part of the αD helix. In addition, the Gln39 amino acid, located in the vicinity of IL-31 binding site 2 but not predicted as a potential hot spot residue, was mutated to an alanine and used as an irrelevant control for the experiments. To allow an easier protein identification, wild type and mutant IL-31 variants were tagged with a V5 epitope followed by a His tag, and the corresponding cDNAs were expressed in the Cos-7 cell line. After a 72-h culture period, the supernatants were collected and analyzed for their IL-31 content by Western blotting using an anti-V5 mAb (Fig. 2A). Among the 11 mutants analyzed, only the Q39A, E44A, E106A, H110A, H131A, and K134A mutants were secreted with an expected molecular mass of 24 kDa proteins, and their expression level was similar to that of the wt protein. The remaining mutants were not expressed, likely reflecting some folding problems linked to the introduced mutations, and were not further analyzed in the study (data not shown). The average concentration of mutated forms of IL-31, as determined by sandwich ELISA, varied between 1 and 4 μg/ml and was similar to the wt protein concentration (Fig. 2B). Similar results were also obtained using antibodies recognizing different IL-31 epitopes (data not shown). Taken together, these results suggest that Q39A, E44A, E106A, H110A, H131A, and K134A mutations did not affect IL-31 folding and structure and could be used for further functional characterization. The wt and mutant forms of IL-31 produced in bacterial system were submitted to SDS-PAGE and silver staining analysis to control the purity (Fig. 2C).
FIGURE 2.
Expression and quantification of IL-31 mutants. Cos-7 cells were transfected with the empty pcDNA3.1 plasmid (control plasmid) or with the pcDNA3.1 plasmid encoding the V5 His-tagged wt (W.T.) or mutant IL-31. A, Western blot (WB) was performed on Cos-7 cells supernatants. An anti-V5 antibody was used for the revelation. B, concentrations of wt and mutant IL-31 were determined by ELISA as described under “Experimental Procedures.” C, purified proteins were analyzed by SDS-PAGE and silver staining revelation.
Induction of Ternary Complex Formation between IL-31 Mutants and Soluble GPL/OSMR
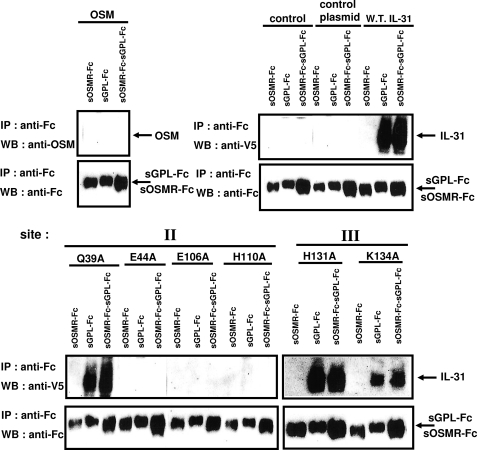
We tested the ability of IL-31 mutants to interact in vitro with soluble forms of GPL (sGPL) and OSMR (sOSMR). Soluble forms of GPL and OSMR were expressed as Fc fusion proteins (sGPL-Fc and sOSMR-Fc, respectively) and were immunoprecipitated using protein A beads. The soluble Fc receptor-loaded beads were then incubated overnight with wild type and mutated forms of IL-31 to allow the formation of binary or ternary complexes. Interaction of wt and mutant forms of IL-31 with their receptors were further detected by immunoblotting using an anti-V5 antibody (Fig. 3). OSM alone failed to recognized OSMR or GPL in agreement with the literature (13). wt IL-31 or the Q39A control mutant were able to interact with single chain GPL, whereas E44A, E106A, and H110A mutants failed to recognize GPL alone or in combination with OSMR. These results suggest that Glu44, Glu106, and His110 contribute to the definition of a binding site 2 in IL-31. The absence of any direct recognition in vitro between OSMR and the different IL-31 forms was likely to be due to weak interactions between both proteins as established in previous studies (19). We did the same experiments with H131A and K134A IL-31 mutants containing mutations directed against predicted binding site 3 of IL-31. As mentioned above, the weakness of interaction between soluble OSMR and IL-31 did not allow the detection of a signal even in the presence of wild type IL-31. Nevertheless, in contrast with the identified site 2 mutants, interaction of GPL with the H131A and K134A mutants remained unchanged. Immunoprecipitation experiments did not allow us to make conclusions about the contribution of His131 and Lys134 to the definition of an IL-31 site 3, so additional functional assays were performed to clarify this point.
FIGURE 3.
Co-immunoprecipitation of wt or mutant IL-31 with sGPL-Fc or sOSMR-Fc. All proteins were produced in the Cos-7 cell system. Cell culture supernatant of Il-31 mutants was diluted to a final concentration of 300 ng/ml. Soluble receptors were incubated in the presence of 300 ng/ml of cytokines overnight at 4 °C before being immunoprecipitated with protein A beads. IL-31 association was detected using a monoclonal anti-V5 antibody or OSM with a biotinylated polyclonal anti-OSM antibody. Soluble receptors fused to the IgG1 Fc fragment were detected with an anti-Fc antibody. WB, Western blot; IP, immunoprecipitation; W.T., wild type.
Binding of wt and Mutant IL-31 Proteins to GO-G-UVM Cell Line
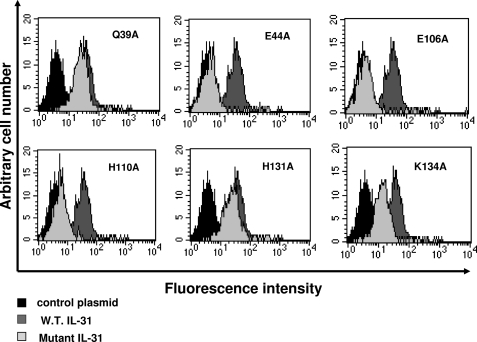
We next studied the binding properties of IL-31 mutants on the membrane receptor complex. For this purpose, we used the GO-G-UVM glioblastoma cell line, which spontaneously expresses sufficient amounts of GPL and OSMR on its surface to follow IL-31 membrane binding by flow cytometry (11, 19). The cells were incubated with 700 ng/ml wt or mutant IL-31, and the cytokine binding to the cell surface was then monitored by fluorescence-activated cell sorter analysis using a mAb directed against the V5 tag epitope added to the ligand C terminus (Fig. 4). Signals were readily detected when wt IL-31, the Q39A control mutant and, the H131A (site 3) mutant were incubated with GO-G-UVM. In contrast, we observed a nearly complete abrogation of cytokine binding when the E44A, E106A, and H110A mutants were added to GO-G-UVM cells. These results are in agreement with the above experiments. Interestingly, binding of the K134A mutant was significantly weaker compared with wt IL-31, suggesting that this mutation (which targets cytokine-binding site 3) alters to some extent the IL-31 interaction with its high affinity bipartite receptor.
FIGURE 4.
Binding detection of wt or mutant IL-31 on the GO-G-UVM cell line by flow cytometry analysis. 700 ng/ml of wild type (W.T.) or mutant proteins were incubated for 1 h with the GO-G-UVM cell line. Indirect detection of the binding was then performed using an anti-V5 mAb followed by a phycoerythrin-conjugated anti-mouse antibody.
Biological Activities of wt and Mutant IL-31
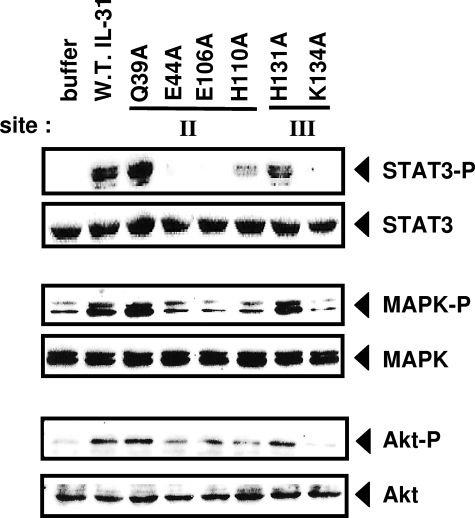
To further analyze the properties of IL-31 mutants, we studied different signaling pathways recruited in response to bipartite IL-31 receptor activation. For this purpose, we incubated the GO-G-UVM cells with wt or mutant proteins and measured STAT-3, MAPK, and Akt phosphorylation levels (Fig. 5). No or only residual STAT-3 and MAPK recruitment was detected in response to E44A, E106A, H110A, and K134A mutants, further consolidating the notion that these four residues contribute toward IL-31-binding sites (Fig. 5). The Akt pathway was mainly triggered by Q39A, H131A, and wt forms of IL-31. No or weaker signals were observed in response to the other mutants studied. Importantly, the K134A mutant located in putative site 3 of the cytokine failed to mediate STAT-3, MAPK, and Akt signaling, suggesting a loss in its ability to induce receptor chain dimerization. The K134A mutant protein, although it binds to GPL and even somewhat better to the combination of GPL and OSMR, does not behave as an antagonist (data not shown).
FIGURE 5.
Analysis of STAT-3, MAPK, and Akt phosphorylation induced by wt or mutant IL-31. The GO-G-UVM cell line was incubated for 10 min in the presence of wt (W.T.) or mutant IL-31 forms (10 ng/ml for STAT-3 analysis and 50 ng/ml for MAPK and Akt analysis), before being lysed and subjected to Western blotting using anti-phospho-STAT-3 (STAT3-P), anti-STAT-3 (STAT3), anti-phospho-MAPK (MAPK-P), anti-MAPK (MAPK), anti-phospho-Akt (Akt-P), and anti-Akt (Akt) antibodies.
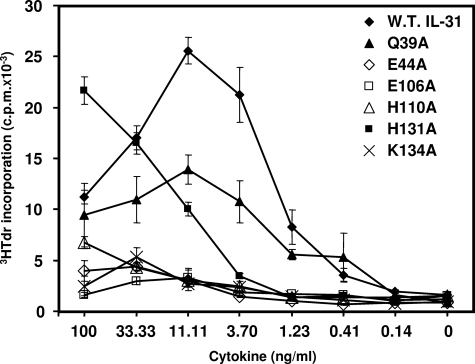
We then studied the ability of the IL-31 mutants to induce the proliferation of the murine Ba/F3 cell line stably transfected with human gp130, OSMR, and GPL (rendering this cell line responsive to both OSM and IL-31) (19). Increasing concentrations of mutated proteins were added to the cultures, and tritiated thymidine incorporation experiments were carried out (Fig. 6). This cell line proliferated in response to wt, Q39A, and H131A forms of IL-31 and to a weaker level to E44A, E106A, H110A, and K134A mutants. These results reinforce the notion that E44A, E106A, and H110A mutations located in IL-31-binding site 2 abrogated the cytokine interaction with GPL and altered IL-31 biological activities. Again, the K134A mutation, located in the predicted IL-31-binding site 3, failed to mediate signaling through the IL-31 heterocomplex receptor, suggesting that the K134A mutation disrupted IL-31 interaction with OSMR.
FIGURE 6.
Proliferation response of Ba/F3 cell lines transfected with gp130, OSMR and GPL upon stimulation with wt or mutant IL-31 proteins. The cells were cultured in triplicate with serial dilutions of proteins as indicated. W.T., wild type.
Ab Initio Modeling of IL-31 and Selection of a Potential Model
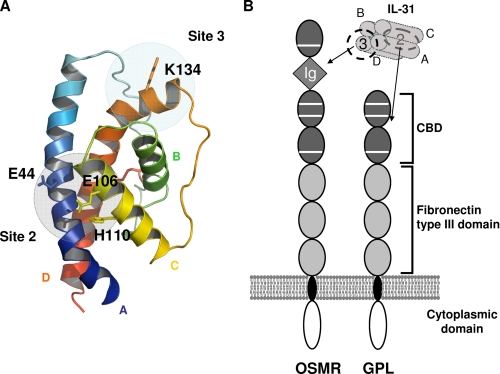
We tried to identify a three-dimensional model of human IL-31 that was compatible with our different data. For this purpose, we evaluated ab initio models of IL-31 generated by I-TASSER and selected a model that was compatible with the information provided by our results. I-TASSER combines the methods of threading, ab initio modeling, and structural refinement to predict a protein structure. The model selected and represented in Fig. 7A adopts the classical four-helix bundle fold of the IL-6 cytokine family with an up-up down-down topology. It also exhibits a predicted site 2 where all the amino acids identified by site-directed mutagenesis (Glu44, Glu106, and His110) are well clustered. Moreover, in this model, the side chain of Lys134 is exposed to solvent and can be engaged in binding site 3. In addition, the size of the α-helices was in agreement with the PsiPred prediction, and the disulfide bridge between Cys72 and Cys147 was also well predicted. Based on the accumulated knowledge, the model represented in Fig. 7A is a good low resolution model of human IL-31. Finally, the combination of our results and the literature suggests that IL-31 binds the CBD region of GPL via its site 2 and the Ig-like domain of OSMR via its site 3 (Fig. 7B) (15, 24, 28, 29).
FIGURE 7.
Model representation of IL-31. A, selected model of human IL-31 predicted by I-TASSER. α A, B, C, and D helices are colored in blue, green, yellow, and orange, respectively. The predicted disulfide bridge is highlighted in red. Binding sites 2 and 3 are indicated by gray circles. B, schematic view of the domain structure of IL-31, OSMR, and GPL and representation of the IL-31/IL-31 receptor complex interaction. Gray square, Ig-like domain; dark gray oval, CBD; light gray oval, fibronectin type III domain.
DISCUSSION
Because IL-31 has been associated with pathologies such as atopic dermatitis, the development of proteins able to neutralize IL-31 in vivo may have potential clinical interest (3, 5, 30–33). The design of mutated antagonistic forms of cytokines often requires the identification of amino acids involved in ligand receptor interactions (21).
The aim of the present study was to characterize the receptor-binding epitopes of human IL-31. Considering the large evolutionary distance between IL-31 and the members of the conventional four-helical cytokine family, we applied a new strategy to predict the IL-31 structure and select amino acid potentially implicated in the protein-binding sites. Residue selection was based on IL-31 ortholog conservation, analyses of α-helices amphipathicity and charge distribution, and simulation of mutations along the whole IL-31 sequence to analyze its stability and resulting functional implications. In a second step these residues have been mutated, and the biological activities of these proteins have been studied.
The structure of four α-helix bundle proteins, with an up-up down-down topology, is characteristic of type I cytokines. Their structural and functional analyses have allowed a classification of most of them into two families: long chain or short chain cytokines. Long chain cytokines (encompassing the IL-6/IL-12 family of mediators, granulocyte colony-stimulating factor, and leptin) have a length of ∼180–200 amino acids, and each helix is made of 20–30 amino acids (34). Short chain cytokines display a length of ∼140 residues with 20 amino acids/helix (35, 36). They also contain two small β-sheets: one in the CD loop and the other one in the AB loop located behind the D helix (21, 37).
IL-31 is a cytokine related to the IL-6 family in part because of the 28% sequence homology between its binding chain, GPL, and gp130 but also because it recruits OSMRβ to form its functional receptor complex (1, 11, 19). The different IL-31 orthologs have a length of 164 amino acids. However, this cytokine displays a specific feature; the A and D antiparallel α-helices display a length of 25–28 amino acids, whereas its two other B and C helices have a smaller length of 10–16 amino acids. Moreover, no β-sheet can be predicted on the AB and CD loops in the IL-31 structure. Despite its link to the IL-6/IL-12 cytokine family, IL-31 displays a secondary structure related to both the long and short chain families of cytokines. The data based on three-dimensional studies and site-directed mutagenesis complete the initial sequence information and two-dimensional analyses.
As shown by Dillon et al. (1), the functional IL-31 receptor associates two membrane subunits GPL (gp130-like receptor) and OSMRβ. There is no evidence for involvement of a small α-receptor chain (contact site 1) as observed in the IL-6 receptor complex, for example.
In addition, no evidence exists for the requirement of a soluble moiety coupled to IL-31 as observed in closely related composite cytokines such as IL-12 (38, 39), IL-23 (40), IL-27 (41), or cardiotrophin-like cytokine/cytokine-like factor (42, 43).
The IL-6/IL-12 family members bind to their receptor chains via well conserved hot spots. Experimental results have shown that the long chain cytokines bind to an Ig-like domain expressed by one of their co-receptors, defining binding site 3 (22, 44, 45). Because no Ig-like domain is present in the GPL sequence, we can predict that IL-31 site 3 most likely interacts with the OSMRβ Ig-like domain. The present study demonstrates that IL-31 site 3 is located in the N-terminal part of its αD helix.
IL-31 binding site 2 is composed of residues of the αA and αC helices and interacts with the GPL subunit. Co-precipitation experiments using site 2 mutants enabled us to show the importance of this site for direct binding of IL-31 to GPL. Moreover, engagement of binding to site 2 is a prerequisite step for the subsequent recruitment of the OSMRβ chain for receptor complex formation. This situation is similar to those reported for leukemia inhibitory factor, OSM, or granulocyte colony-stimulating factor receptor complexes (13, 28, 29, 46, 47).
The identification of the residues involved in the interaction between a cytokine and its receptor chains is essential to develop antagonist molecules or to select neutralizing monoclonal antibodies. Indeed a recent study showed a beneficial effect of a neutralizing monoclonal antibody directed against IL-31 in a model of atopic dermatitis. The authors note an improvement in animal scratching behavior following the injection of this molecule (48). Based on the knowledge of IL-31/IL-31 receptor interactions, it will now be of interest to perform saturating mutagenesis on key IL-31 residues to develop antagonist mutants having potential for clinical applications.
Acknowledgments
We thank Greg Elson and Catherine Guillet for careful review of the manuscript.
This work was supported by Grant 5176 from the Association pour la Recherche contre le Cancer and by the Ciblage Moléculaire et Applications Thérapeutiques Program of the Région Pays de la Loire.
- IL
- interleukin
- GPL
- gp130-like receptor
- OSMR
- oncostatin M receptor
- CBD
- cytokine-binding domain
- STAT
- signal transducers and activators of transcription
- ELISA
- enzyme-linked immunosorbent assay
- mAb
- monoclonal antibody
- MAPK
- mitogen-activated protein kinase
- wt
- wild type
- s
- soluble form.
REFERENCES
- 1.Dillon S. R., Sprecher C., Hammond A., Bilsborough J., Rosenfeld-Franklin M., Presnell S. R., Haugen H. S., Maurer M., Harder B., Johnston J., Bort S., Mudri S., Kuijper J. L., Bukowski T., Shea P., Dong D. L., Dasovich M., Grant F. J., Lockwood L., Levin S. D., LeCiel C., Waggie K., Day H., Topouzis S., Kramer J., Kuestner R., Chen Z., Foster D., Parrish-Novak J., Gross J. A. (2004) Nat. Immunol. 5, 752–760 [DOI] [PubMed] [Google Scholar]
- 2.Takaoka A., Arai I., Sugimoto M., Yamaguchi A., Tanaka M., Nakaike S. (2005) Eur. J. Pharmacol. 516, 180–181 [DOI] [PubMed] [Google Scholar]
- 3.Bilsborough J., Leung D. Y., Maurer M., Howell M., Boguniewicz M., Yao L., Storey H., LeCiel C., Harder B., Gross J. A. (2006) J. Allergy Clin. Immunol. 117, 418–425 [DOI] [PubMed] [Google Scholar]
- 4.Takaoka A., Arai I., Sugimoto M., Honma Y., Futaki N., Nakamura A., Nakaike S. (2006) Exp. Dermatol. 15, 161–167 [DOI] [PubMed] [Google Scholar]
- 5.Sonkoly E., Muller A., Lauerma A. I., Pivarcsi A., Soto H., Kemeny L., Alenius H., Dieu-Nosjean M. C., Meller S., Rieker J., Steinhoff M., Hoffmann T. K., Ruzicka T., Zlotnik A., Homey B. (2006) J. Allergy Clin. Immunol. 117, 411–417 [DOI] [PubMed] [Google Scholar]
- 6.Leung D. Y., Bieber T. (2003) Lancet 361, 151–160 [DOI] [PubMed] [Google Scholar]
- 7.Neis M. M., Peters B., Dreuw A., Wenzel J., Bieber T., Mauch C., Krieg T., Stanzel S., Heinrich P. C., Merk H. F., Bosio A., Baron J. M., Hermanns H. M. (2006) J. Allergy Clin. Immunol. 118, 930–937 [DOI] [PubMed] [Google Scholar]
- 8.Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bazan J. F. (1990) Immunol. Today 11, 350–354 [DOI] [PubMed] [Google Scholar]
- 10.Ghilardi N., Li J., Hongo J. A., Yi S., Gurney A., de Sauvage F. J. (2002) J. Biol. Chem. 277, 16831–16836 [DOI] [PubMed] [Google Scholar]
- 11.Diveu C., Lelièvre E., Perret D., Lak-Hal A. H., Froger J., Guillet C., Chevalier S., Rousseau F., Wesa A., Preisser L., Chabbert M., Gauchat J. F., Galy A., Gascan H., Morel A. (2003) J. Biol. Chem. 278, 49850–49859 [DOI] [PubMed] [Google Scholar]
- 12.Thoma B., Bird T. A., Friend D. J., Gearing D. P., Dower S. K. (1994) J. Biol. Chem. 269, 6215–6222 [PubMed] [Google Scholar]
- 13.Mosley B., De Imus C., Friend D., Boiani N., Thoma B., Park L. S., Cosman D. (1996) J. Biol. Chem. 271, 32635–32643 [DOI] [PubMed] [Google Scholar]
- 14.Hibi M., Murakami M., Saito M., Hirano T., Taga T., Kishimoto T. (1990) Cell 63, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 15.Bravo J., Heath J. K. (2000) EMBO J. 19, 2399–2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazan J. F. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6934–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazan J. F. (1989) Biochem. Biophys. Res. Commun. 164, 788–795 [DOI] [PubMed] [Google Scholar]
- 18.Dreuw A., Radtke S., Pflanz S., Lippok B. E., Heinrich P. C., Hermanns H. M. (2004) J. Biol. Chem. 279, 36112–36120 [DOI] [PubMed] [Google Scholar]
- 19.Diveu C., Lak-Hal A. H., Froger J., Ravon E., Grimaud L., Barbier F., Hermann J., Gascan H., Chevalier S. (2004) Eur. Cytokine Netw. 15, 291–302 [PubMed] [Google Scholar]
- 20.Chattopadhyay S., Tracy E., Liang P., Robledo O., Rose-John S., Baumann H. (2007) J. Biol. Chem. 282, 3014–3026 [DOI] [PubMed] [Google Scholar]
- 21.Wang X., Lupardus P., Laporte S. L., Garcia K. C. (2009) Annu. Rev. Immunol. 27, 29–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammacher A., Richardson R. T., Layton J. E., Smith D. K., Angus L. J., Hilton D. J., Nicola N. A., Wijdenes J., Simpson R. J. (1998) J. Biol. Chem. 273, 22701–22707 [DOI] [PubMed] [Google Scholar]
- 23.Boulanger M. J., Chow D. C., Brevnova E. E., Garcia K. C. (2003) Science 300, 2101–2104 [DOI] [PubMed] [Google Scholar]
- 24.Plun-Favreau H., Perret D., Diveu C., Froger J., Chevalier S., Lelièvre E., Gascan H., Chabbert M. (2003) J. Biol. Chem. 278, 27169–27179 [DOI] [PubMed] [Google Scholar]
- 25.Hubbard T. J., Aken B. L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. (2009) Nucleic Acids Res. 37, D690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson K. R., Vernallis A. B., Heath J. K. (1996) J. Biol. Chem. 271, 11971–11978 [DOI] [PubMed] [Google Scholar]
- 27.Di Marco A., Gloaguen I., Graziani R., Paonessa G., Saggio I., Hudson K. R., Laufer R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 9247–9252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deller M. C., Hudson K. R., Ikemizu S., Bravo J., Jones E. Y., Heath J. K. (2000) Structure 8, 863–874 [DOI] [PubMed] [Google Scholar]
- 29.Boulanger M. J., Bankovich A. J., Kortemme T., Baker D., Garcia K. C. (2003) Mol. Cell 12, 577–589 [DOI] [PubMed] [Google Scholar]
- 30.Schulz F., Marenholz I., Fölster-Holst R., Chen C., Sternjak A., Baumgrass R., Esparza-Gordillo J., Grüber C., Nickel R., Schreiber S., Stoll M., Kurek M., Rüschendorf F., Hubner N., Wahn U., Lee Y. A. (2007) J. Allergy Clin. Immunol. 120, 1097–1102 [DOI] [PubMed] [Google Scholar]
- 31.Dambacher J., Beigel F., Seiderer J., Haller D., Göke B., Auernhammer C. J., Brand S. (2007) Gut 56, 1257–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jawa R. S., Chattopadhyay S., Tracy E., Wang Y., Huntoon K., Dayton M. T., Baumann H. (2008) J. Interferon Cytokine Res. 28, 207–219 [DOI] [PubMed] [Google Scholar]
- 33.Lei Z., Liu G., Huang Q., Lv M., Zu R., Zhang G. M., Feng Z. H., Huang B. (2008) Allergy 63, 327–332 [DOI] [PubMed] [Google Scholar]
- 34.Boulanger M. J., Garcia K. C. (2004) Adv. Protein Chem. 68, 107–146 [DOI] [PubMed] [Google Scholar]
- 35.Rozwarski D. A., Gronenborn A. M., Clore G. M., Bazan J. F., Bohm A., Wlodawer A., Hatada M., Karplus P. A. (1994) Structure 2, 159–173 [DOI] [PubMed] [Google Scholar]
- 36.Hill E. E., Morea V., Chothia C. (2002) J. Mol. Biol. 322, 205–233 [DOI] [PubMed] [Google Scholar]
- 37.Aasland D., Oppmann B., Grötzinger J., Rose-John S., Kallen K. J. (2002) J. Mol. Biol. 315, 637–646 [DOI] [PubMed] [Google Scholar]
- 38.Wolf S. F., Temple P. A., Kobayashi M., Young D., Dicig M., Lowe L., Dzialo R., Fitz L., Ferenz C., Hewick R. M., Kelleher K., Herrmann S. H., Clark S. C., Azzoni L., Chan S. H., Trinchieri G., Perussia B. (1991) J. Immunol. 146, 3074–3081 [PubMed] [Google Scholar]
- 39.Gearing D. P., Cosman D. (1991) Cell 66, 9–10 [DOI] [PubMed] [Google Scholar]
- 40.Oppmann B., Lesley R., Blom B., Timans J. C., Xu Y., Hunte B., Vega F., Yu N., Wang J., Singh K., Zonin F., Vaisberg E., Churakova T., Liu M., Gorman D., Wagner J., Zurawski S., Liu Y., Abrams J. S., Moore K. W., Rennick D., de Waal-Malefyt R., Hannum C., Bazan J. F., Kastelein R. A. (2000) Immunity 13, 715–725 [DOI] [PubMed] [Google Scholar]
- 41.Pflanz S., Timans J. C., Cheung J., Rosales R., Kanzler H., Gilbert J., Hibbert L., Churakova T., Travis M., Vaisberg E., Blumenschein W. M., Mattson J. D., Wagner J. L., To W., Zurawski S., McClanahan T. K., Gorman D. M., Bazan J. F., de Waal Malefyt R., Rennick D., Kastelein R. A. (2002) Immunity 16, 779–790 [DOI] [PubMed] [Google Scholar]
- 42.Elson G. C., Lelièvre E., Guillet C., Chevalier S., Plun-Favreau H., Froger J., Suard I., de Coignac A. B., Delneste Y., Bonnefoy J. Y., Gauchat J. F., Gascan H. (2000) Nat. Neurosci. 3, 867–872 [DOI] [PubMed] [Google Scholar]
- 43.Plun-Favreau H., Elson G., Chabbert M., Froger J., deLapeyrière O., Lelièvre E., Guillet C., Hermann J., Gauchat J. F., Gascan H., Chevalier S. (2001) EMBO J. 20, 1692–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Layton J. E., Shimamoto G., Osslund T., Hammacher A., Smith D. K., Treutlein H. R., Boone T. (1999) J. Biol. Chem. 274, 17445–17451 [DOI] [PubMed] [Google Scholar]
- 45.Layton J. E., Hall N. E., Connell F., Venhorst J., Treutlein H. R. (2001) J. Biol. Chem. 276, 36779–36787 [DOI] [PubMed] [Google Scholar]
- 46.Skiniotis G., Lupardus P. J., Martick M., Walz T., Garcia K. C. (2008) Mol. Cell 31, 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huyton T., Zhang J. G., Luo C. S., Lou M. Z., Hilton D. J., Nicola N. A., Garrett T. P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12737–12742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimstad O., Sawanobori Y., Vestergaard C., Bilsborough J., Olsen U. B., Grønhøj-Larsen C., Matsushima K. (2009) Exp. Dermatol. 18, 35–43 [DOI] [PubMed] [Google Scholar]