Abstract
To contribute to the question of the putative role of cystatins in Alzheimer disease and in neuroprotection in general, we studied the interaction between human stefin B (cystatin B) and amyloid-β-(1–40) peptide (Aβ). Using surface plasmon resonance and electrospray mass spectrometry we were able to show a direct interaction between the two proteins. As an interesting new fact, we show that stefin B binding to Aβ is oligomer specific. The dimers and tetramers of stefin B, which bind Aβ, are domain-swapped as judged from structural studies. Consistent with the binding results, the same oligomers of stefin B inhibit Aβ fibril formation. When expressed in cultured cells, stefin B co-localizes with Aβ intracellular inclusions. It also co-immunoprecipitates with the APP fragment containing the Aβ epitope. Thus, stefin B is another APP/Aβ-binding protein in vitro and likely in cells.
Keywords: Diseases/Neurodegeneration, Methods/Mass Spectrometry, Methods/Surface Plasmon Resonance, Protein/Folding, Protein/Protein-Protein Interactions, Alzheimer Disease, Amyloid
Introduction
Neurodegenerative diseases present a huge burden in the developed world's aging population. They are all in one way or another connected to aberrant protein folding and aggregation of the proteins involved (1). Various protein conformational disorders of the central and peripheral nervous system are known, which often appear sporadically but also run in families. These are among others: Parkinson and Alzheimer diseases, dementia with Lewy bodies, vascular and fronto-temporal dementia, and amyotrophic lateral sclerosis.
The Aβ peptide implicated in Alzheimer disease pathology is a cleavage product of the membrane Aβ precursor protein (APP).3 It is the main constituent of extracellular amyloid plaques, however, together with its oligomers, it also resides intracellularly (2). It has been shown that Aβ oligomers prepared in vitro and those extracted from living cells exert cytotoxicity and cause symptoms of reversible memory loss in animal models (3).
Amyloid protein oligomers have special structural properties, which are reflected in a common antioligomer antibody (4). This antibody not only binds the oligomers against which it was raised but also binds chaperones and some other proteins involved in disaggregating protein aggregates in cells (5). Aβ-binding proteins, the so called “amateur chaperones,” were suggested to have a potential in Alzheimer disease therapy (6, 7).
It has been shown before that human cystatin C is an Aβ-binding protein (8). Cystatins are single chain proteins that inhibit cysteine cathepsins (9). Human stefin B (also known as cystatin B) is a member of subfamily A of cystatins, classified as family I25 in the MEROPS scheme (10). Stefin B, a protein of 98 amino acid residues and 1 Cys, is predominantly intracellular, whereas cystatin C, a protein of 120 residues and 2 disulfide bonds, is a secretory protein. Three-dimensional structures of stefins and cystatin C have been determined, among others, the solution structure of stefin A (11) and cystatin C (12, 13).
Human cystatin C has been found as a constituent of senile plaques of Alzheimer disease patients (14) and stefins A and B have also been reported to localize to amyloid plaques of various origin (15, 16). It has been suggested that cystatins play a role in Alzheimer disease (17, 18).
Stefin B has been used as a model protein to study amyloid fibril formation, often in comparison to the more stable stefin A (19–21). Stefin B has a much higher tendency than stefin A to form dimers, higher oligomers, and amyloid fibrils, nevertheless, the stefin A domain-swapped dimer could be prepared under more extreme solution conditions and its structure was determined by NMR (22, 23). The model for the mechanism of stefin B fibrillization involves off-pathway lower oligomer formation (probably involving domain-swapping, due to a high energetic barrier) and a larger nucleus in the order of 30 dimers, explaining both an unusual behavior at higher protein concentrations and a relatively long lag phase (21).
Similarly to some other amyloid proteins, it has been demonstrated that stefin B interacts predominantly with acidic phospholipids and that higher oligomers, distinct from monomers, dimers, and tetramers, are toxic to cells (24, 25). Moreover, studies on the pore forming characteristics of this protein have confirmed the suggestion that toxicity of oligomers is related to membrane perforation (26).
As already said, Aβ peptide interacts with quite a number of amyloid proteins (6, 7). A few of them are gelsolin, α2-macroglobulin, and crystallin-αB. In this work we probed the interaction of Aβ with human stefin B. We chose conditions under which Aβ fibrillizes and stefin B does not. The interaction of Aβ with stefin B has been confirmed directly by using surface plasmon resonance (SPR) where Aβ was bound to the sensor chip and samples of separated monomers, dimers, tetramers, and higher oligomers of the wild-type Glu31 stefin B or the dimeric Tyr31 stefin B were injected across. Interaction of the Tyr31 stefin B dimers with Aβ was additionally confirmed by electrospray ionization-mass spectrometry (ESI-MS). As further proof of the interaction, fibrillization of Aβ was completely inhibited by exactly those oligomers, which showed binding by SPR. Co-localization and co-immunoprecipitation cellular experiments affirm the possibility that the two proteins interact in the cell.
EXPERIMENTAL PROCEDURES
Preparation and Isolation of Recombinant Human Stefin B
Recombinant human stefin B was expressed in Escherichia coli and purified by affinity and gel chromatography (27) and on size exclusion chromatography using a Superdex 75 column, the wild-type Glu31 stefin B eluted as a set of well defined oligomers (28), allowing isolation of monomers, dimers, tetramers, and higher oligomers. Tyr31 stefin B eluted from the Superdex 75 column predominantly as a dimer. The recombinant proteins have Ser at position 3 instead of Cys to prevent covalent disulfide bond formation.
Thioflavin T (ThT) Fluorescence Measurements
A stock solution of Aβ peptide in distilled water was prepared at 476 μm concentration and kept on ice. An aliquot was added to the reaction mixture of stefin B in PBS, pH 7.3. The order of adding the proteins is very important, as Aβ starts to fibrillize immediately at pH 7.3 (PBS) and at 40 °C, with only a short lag phase. The fibrillization reaction took place at 40 °C and at different molar ratios, stefin B to Aβ (1:1 to 1:8). The starting concentration of stefin B (of any oligomeric form) was 17 μm, to which Aβ was added to final concentrations of 17, 34, 68, and 136 μm. During the reaction, at several time points, an aliquot was taken from the fibrillization mixture and added to a 0.025 m phosphate buffer, pH 7.5, with ThT dye dissolved to A416 = 0.66. ThT fluorescence spectra were measured from 455 to 550 nm (λex = 440 nm) with a luminescence spectrometer LS 50B (PerkinElmer Life Sciences) and the intensity at 482 nm was read. To probe the status of Aβ before the CM5 chip preparation, a continuous time course of ThT fluorescence was measured at 482 nm (λex = 440 nm) for 7 min, after Aβ was incubated for 1 min in pH 4.0 buffer (as for coupling) with the ThT dye dissolved.
Transmission Electron Microscopy (TEM)
Protein samples were taken at certain time points of Aβ fibrillization and at the plateau of the reaction. 15 μl of a 17–34 μm solution, diluted 10 to 50 times as appropriate, were applied to a formvar and carbon-coated grid. If not indicated otherwise, after 5 min the sample was soaked away and the grid was stained with 1% uranyl acetate. To check the initial status of Aβ on the sensor chip, a sample of Aβ (after 1 min of incubation at pH 4.0) was put on the grid (with 5 more min left on the grid surface) and TEM was recorded.
Philips CM 100 transmission electron microscope was used and at the applied voltage of 80 kV, magnifications were from ×10,000 to 130,000. Images were recorded by a Gatan Bioscan CCD camera, using Digital Micrograph software.
SPR
SPR experiments were performed using a Biacore X biosensor instrument. Aβ peptide was covalently bound to the sensor chip CM5. Aβ peptide was coupled to the surface of the sensor flow cell activated with 0.4 m 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and 0.1 m N-hydroxysuccinimide. The 10 mm acetate buffer, pH 4.0, was used for coupling. The excess of the reactive groups were blocked with ethanolamine. Monomers, dimers, tetramers, and higher oligomers of wild-type Glu31 stefin B and dimeric iso-form Tyr31 stefin B were used as analytes. Kinetic measurements were performed at 25 °C at a flow rate of 5 μl/min in PBS buffer, pH 7.3. To obtain the concentration range where stefin B protein interacts with Aβ peptide bound on the chip, concentrations of the Tyr31 variant (0.5–200 μm) and Glu31 variant (20–200 μm) were applied. The SPR response was compared with a control sensor flow cell on the same chip, which was activated and blocked in the same way as the flow cell with ligand, omitting the injection of Aβ peptide. Regeneration of the sensor surface was performed with 3 μl of 10 mm glycine, pH 3.0, in the case of the Tyr31 variant or 5 μl of 2 m NaCl in the case of the Glu31 variant.
Preparation of Samples for ESI- MS
Stefin B (Tyr31 variant, which is predominantly dimeric) was purified by SEC on a SuperdexTM 75 GL column (10 × 300 mm) (GE Healthcare) connected to an ÄKTA Purifier system (GE Healthcare). Protein was eluted with 20 mm ammonium acetate, pH 7.5, at a flow rate of 1 ml/min. 0.2-ml fractions were collected. UV detection at 230, 280, and 360 nm was used to monitor elution.
ESI-MS Studies of Stefin B-Aβ Interaction
The SEC-purified sample of stefin B (the dimer of the Tyr31 variant) was diluted with 20 mm ammonium acetate, pH 7.5, to 2 μm and injected at the flow rate of 6 μl/min into the ESI-Q-TOF mass spectrometer (QSTAR Elite, AB Sciex instruments). MS spectra were recorded in the m/z region from 500 to 5000 Da using the following instrument parameters: ion spray voltage, 4500 V; source gas, 25 liters/min; curtain gas, 20 liters/min; declustering potential, 60 V; focusing potential, 320 V; detector voltage, 2450 V. Similar conditions were used for ESI-TOF MS analysis of 2 μm Aβ-(1–40) and the mixture of 2 μm stefin B, 2 μm Aβ-(1–40) in 20 mm ammonium acetate.
Cell Lines and Transfection Experiments
Chinese hamster ovary (CHO-K1) cells expressing wild-type APP (APP-WT) or APP with the codon 670/671 “Swedish” mutation (APP-Sw), as described (29), were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and selected in 200 mg/ml of G418. All reagents were from Invitrogen. To induce expression of stefin B, cells stably expressing APP were seeded in 6-well plates and incubated overnight to reach 70–80% confluence and 10 μg of plasmid (the multiple cloning site pcDNA3 vector (Invitrogen)) was transfected with LipofectamineTM 2000 according to the manufacturer's instructions.
Assessment of Levels of Full-length APP and APP Metabolites in CHO Cells
Cells were lysed in lysis buffer (1% Triton X-100, 150 mm NaCl, Tris, pH 7.4) containing protease and phosphatase inhibitor mixtures (Roche Diagnostics). For Western blotting, equal amounts of protein were loaded onto 4–12% Tris glycine gradient gels, transferred onto nitrocellulose membranes, probed with the antibodies described below, and analyzed using the Odyssey Infrared Imaging System (LI-COR Biotechnology, Lincoln, NB). For APP and detection of its metabolites, polyclonal antiserum CT15, which recognizes the C terminus of APP, was used (30).
Aβ Quantitation by ELISA
Aβ sandwich ELISAs were carried out using a human Aβ-(1–40) kit (Invitrogen) following the manufacturer's instructions. Experiments were performed in triplicate and repeated three times. Statistical analysis was carried out in SPSS version 16.0. Data are expressed as the mean ± S.E. and p < 0.05 was considered statistically significant.
Immunostaining
The CHO-K1 cell line expressing APP-Sw, grown on 12-mm coverslips, was transiently transfected with wild-type stefin B (pcDNA3- stBwt) using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Cells were then washed with PBS and fixed in 4% paraformaldehyde. Paraformaldehyde was quenched with 150 mm glycine. Cells were permeabilized with 0.2% (v/v) Triton X-100 in PBS. After quenching and permeabilization, coverslips were washed with PBS. Coverslips were then blocked in Ab buffer (PBS, with 1% bovine serum albumin (w/v) and 1% fish skin gelatin (w/v)). Rabbit polyclonal anti-stefin B (31) and mouse monoclonal anti-Aβ (DE2B4) from Abcam were used for co-localization studies. The 1 mg/ml concentrations of the primary antibodies were used. They were diluted in Ab buffer as follows: DE2B4 mouse monoclonal anti-Aβ (1:500) and rabbit polyclonal anti-stefin B (1:800). All secondary antibodies were diluted 1:500 in Ab buffer. Incubation was either overnight at 4 °C for primary antibodies or at room temperature for 2 h for secondary antibodies. After incubation coverslips were once again washed as described above and mounted on slides in Prolong antifade reagent. All Alexa Fluor-conjugated secondary antibodies and Prolong Antifade reagent were from Molecular Probes (Invitrogen).
Immunoprecipitation and Western Blotting
Cells co-expressing APP-WT or APP-Sw and stefin B were lysed in RIPA buffer. The lysates were applied to the immunoaffinity Sepharose-protein A resin prepared with polyclonal anti-rabbit stefin B antibodies, after which 15% SDS gels and Western blotting with anti-Aβ antibodies (DE2B4) were performed. To detect bound proteins the same antibodies were used as described above, only, dilution was 1:400 for the primary anti-Aβ antibodies.
Confocal Microscopy
Confocal images were taken with a Carl Zeiss LSM510 Axio observer microscope equipped with a Polychrome V monochromator using an Imago Type QE CCD camera at the Reference Center for Confocal Microscopy (LN-MCP, Institute of Pathophysiology, School of Medicine, Ljubljana, Slovenia). Sequential acquisition was used to minimize cross-talk between red and green channels. The hardware was configured with two control samples, one with single labeled/expressing green cells and the other with single labeled/expressing red cells. The background of the collected images was corrected by the ImageJ rolling ball algorithm plug in and quantitative co-localization analysis was performed by JACoP (32), an ImageJ co-localization plug in.
RESULTS
Initial Conformation and Oligomeric State of Stefin B Variants
Two variants (iso-forms) of wild-type human stefin B were reported thus far. Tyr31 stefin B, derived from protein sequencing (33) was later cloned (27) and its three-dimensional structure in complex with papain was determined (34). The Tyr31 variant is predominantly dimeric in solution, whereas the more common wild-type, as reported in GenBank, Glu31 stefin B, exists as a mixture of oligomers (25, 28).
The two variants differ in stability; the Tyr31 variant proved a more labile protein (by 11 kJ/mol) as shown by urea denaturation (35) and, it transformed into a molten globule conformation, either by reducing pH (36) or by mutation of P74S (37).
The P79S mutant of the Tyr31 variant forms a tetramer. The crystal structure and NMR structure in solution shows that two domain-swapped dimers “intercalate” by exchanging loops to form the tetramer (37). The peptide bond preceding Pro74 is cis in the tetramer, although apparently not in the monomeric stefin B in complex with papain (34).
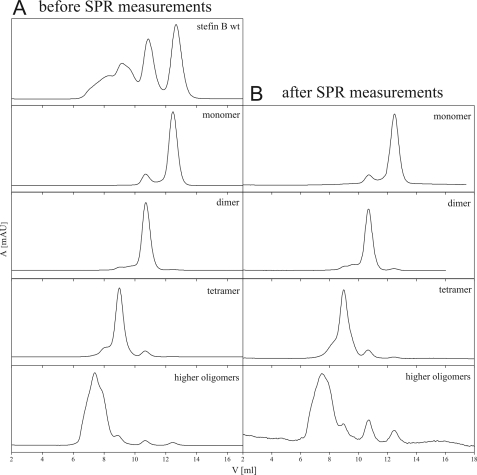
Separate oligomers of wild-type Glu31 stefin B were isolated (Fig. 1) by SEC. The elution diagrams of monomers, dimers, and higher oligomers are shown in Fig. 1A. After the SPR experiment, standing 14 days at 4 °C, the samples were checked for equilibration of the oligomeric species (Fig. 1B). As elutions did not change much, one can judge that oligomers are rather stable. From the initial samples as applied to SEC and SPR, one can say that in the monomer sample there is <10% dimers; in the dimer sample there are <5% monomers and <10% tetramers; in the tetramer sample there are <5% dimers and <20% higher oligomers; in the higher oligomers there are <10% of other species (>5% tetramers). Oligomers, which are surprisingly stable, were additionally checked by ESI-MS (not shown), which confirmed their molecular weight.
FIGURE 1.
Oligomeric state of the wild-type Glu31 stefin B samples before (A) and after (B) SPR measurements. A Superdex 75 column (GE Healthcare) was used to perform size exclusion chromatography. The column was equilibrated in phosphate buffer, pH 7.0, containing 0.15 m NaCl. The flow rate was 0.5 ml/min at room temperature. 50 μl of the Glu31 stefin B monomers, dimers, tetramers, and higher oligomers were injected.
Stefin B Inhibits Aβ Fibril Growth in an Oligomer-specific Manner
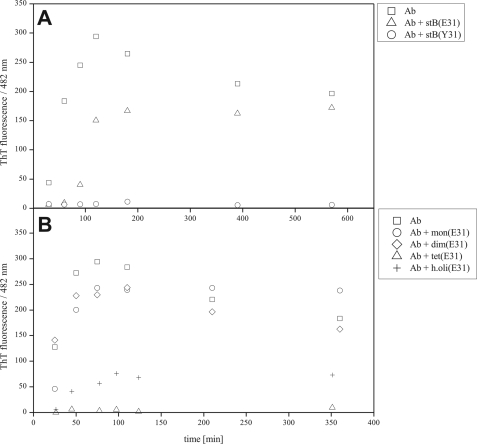
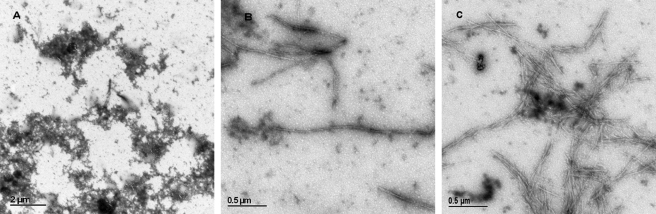
Conditions were established under which Aβ fibrillizes regularly and stefin B does not. The mixture undergoing fibrillization was left unperturbed between measurements of ThT fluorescence. Amyloid-fibril formation by Aβ, alone and with stefin B in a 1:1 molar ratio, are shown in Fig. 2. Dimeric Tyr31 stefin B completely inhibits Aβ fibril growth (Fig. 2A), which was also confirmed by TEM (Fig. 3A). Wild-type Glu31 stefin B, which is composed of monomers and different oligomers, shows no comparable inhibitory effect (Figs. 2A and 3B).
FIGURE 2.
Inhibition of Aβ fibril formation by stefin B measured by ThT fluorescence. The Aβ peptide concentration was 17 μm throughout, pH 7.3, 40 °C. A, Aβ alone, 1:1 molar ratio of Aβ to Tyr31 stefin B (complete inhibition) and 1:1 molar ratio of Aβ to Glu31 stefin B. B, Aβ alone, and 1:1 molar ratios to Glu31 stefin B monomers, dimers, tetramers, and higher oligomers. The protein concentrations of stefin B were 17 μm.
FIGURE 3.
Morphology of the aggregates and fibrils as observed by TEM. After Aβ fibril formation reached steady state (about 100 h, pH 7.3, 40 °C), a 15-μl sample was spread on the carbon-coated grid, 1% uranyl acetate was added for a contrast, and TEM images were recorded. At the same time point samples were taken of mixtures of Aβ with the following proteins: A, Tyr31 stefin B; B, Glu31 stefin B; and C, Aβ alone.
To see the effect of individual oligomers of wild-type Glu31 stefin B on fibril growth of Aβ, monomers, dimers, tetramers, and higher oligomers of stefin B were isolated by SEC (Fig. 1, A and B). Each was then separately mixed with Aβ. It was observed that only tetramers inhibited Aβ fibril growth completely (Fig. 2B).
Stefin B Interacts with Aβ Depending on the Oligomeric State of the Former
SPR Measurements
Oligomer-specific binding between stefin B and Aβ was demonstrated using SPR. Aβ peptide was immobilized on the surface of sensor chip CM5 and samples of the two stefin B variants and isolated oligomers were injected across the surface. By several control experiments we confirmed that Aβ bound to the chip was not considerably aggregated or fibrillar as judged from low ThT fluorescence of the Aβ sample in the buffer used for coupling before application to the chip and TEM taken at the same time (supplemental Fig. S1). Although some fibrils formed in the first 5-min incubation at pH 4 for coupling, the majority of the peptide (70%) does not bind ThT and is thus not in a fibrillar form. Furthermore, after immobilization of Aβ, the first injection of 25 mm NaOH desorbs a significant amount of the peptide, which possibly represents noncovalently associated Aβ into oligomers or fibrils. We cannot exclude that a minor amount of Aβ is in the aggregated or fibrillar forms and thus the exact determination of rate or equilibrium affinity constants of the interaction was not feasible. The sensorgrams, however, showed clear interaction as depicted below and were reproducible.
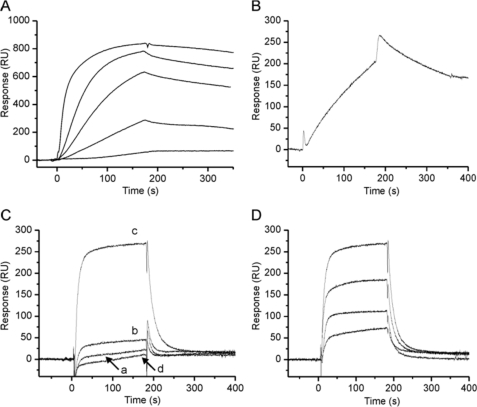
Dimeric Tyr31 stefin B reacted with Aβ in a concentration-dependent manner (Fig. 4A). To verify the interaction, we also immobilized Tyr31 stefin B and were able to observe the binding of Aβ (Fig. 4B). Tetramers of wild-type Glu31 stefin B bound strongly to Aβ, but not so to monomers, dimers, and higher oligomers (Fig. 4C), thus explaining the specific inhibitory effect of tetramers on Aβ fibril growth (Fig. 2B). This interaction of wild-type tetramers with Aβ was also concentration dependent, as shown in Fig. 4D.
FIGURE 4.
Binding experiments by SPR. The interaction of Aβ with the two stefin B iso-forms (Tyr31 and Glu31 stefin B) and isolated oligomers of the wild-type Glu31 stefin B were assessed at 5 μl/ml in PBS buffer, pH 7.3. A, binding of Tyr31 stefin B (analyte) to chip-immobilized Aβ. The concentration of Tyr31 stefin B was 0.5, 1, 2, 4, and 20 μm (from bottom to top). B, binding of 10 μm Aβ to chip-immobilized Tyr31 stefin B. C, binding of monomers (curve a), dimers (b), tetramers (c), and higher oligomers of wild-type Glu31 stefin B (d) to chip-immobilized Aβ. All forms were assessed at 100 μm. D, binding of the isolated tetramers of wild-type Glu31 stefin B at 10, 25, 50, and 100 μm (from bottom to top).
ESI-MS Measurements
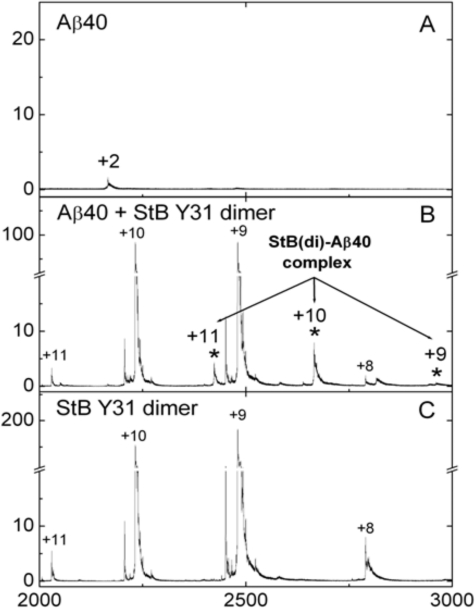
Formation of the complex between stefin B and Aβ was monitored directly in an ESI-MS experiment. The ESI-MS spectrum of Aβ-(1–40) displayed 3 major peaks (with charges +5, +4, and +3), which correspond to a molecular mass species of 4329.91 Da (theoretical mass 4329.89 Da). The isolated Tyr31 stefin B dimer displayed 2 major peaks corresponding to a molecular mass of 22,314.2 Da (theoretical Mr 22,301.4). In the spectrum of stefin B and the Aβ-(1–40) mixture, 3 new peaks with molecular mass of 26,644.4 (charges +11, +10, and +9, respectively) were observed, which corresponds to the theoretical mass of the complex formed between one Tyr31 stefin B dimer and one molecule of Aβ-(1–40).
The ESI-MS results (Fig. 5) clearly show formation of a complex between the Tyr31 stefin B dimer and Aβ-(1–40). It coexists together with uncomplexed components and therefore can be considered as weak. A similar complex between the wild-type Glu31 stefin B tetramer and Aβ could not be observed, due to the lower intensity of MS peaks corresponding to higher oligomers.
FIGURE 5.
Complex detected by ESI-MS. ESI-MS spectra of Aβ-(1–40), stefin B dimer (Tyr31 variant) and their mixtures were recorded: A, 2 μm Aβ-(1–40); B, a mixture of 2 μm Aβ and 2 μm stefin B; and C, 2 μm stefin B. Peaks corresponding to the Aβ-stefin B complex are denoted with an asterisk and numbers above the peaks denote charge state of the ions.
Effect of Stefin B on APP Metabolism and Aβ Generation in Cells
To explore a possible functional link between stefin B and Aβ in vivo, we studied the effect of co-expressing stefin B in the cell culture expressing APP.
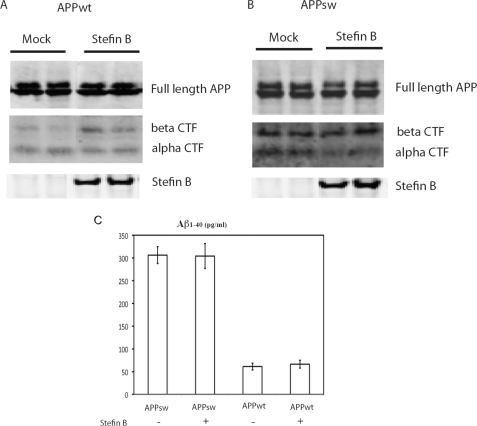
Transfection of stefin B into CHO cells expressing either wild-type or Swedish mutant APP had no impact on cell toxicity, as determined by lactate dehydrogenase release levels (not shown). Furthermore, it was shown (Fig. 6) that the presence of stefin B did not modify levels of wild-type or Swedish full-length APP. Similarly, stefin B did not affect Aβ-(1–40) secretion levels. In the absence of stefin B we observed a ∼6-fold increase in Aβ levels in cells bearing the Swedish mutation (29), a difference that did not change in the presence of stefin B (Fig. 6).
FIGURE 6.
Western blots and ELISA of APP and its metabolites upon stefin B co-expression. Western blots showing APP, C-terminal fragments aCTF, bCTF, and stefin B in CHO-K1 cells expressing APP wild-type (A) and APP Swedish mutant (B) and stefin B, respectively, in comparison to mock vector expression. Antibodies to stefin B, aCTF, bCTF, and APP were used (see ”Experimental Procedures“). C shows levels of Aβ-(1–40) measured by ELISA in cells expressing APP wild-type, and the same in cells co-expressing stefin B. Also shown are levels of Aβ-(1–40) in cells expressing APP-Sw or APP-Sw together with stefin B, which in both cases is 6-fold higher, as expected (26).
Nevertheless, the presence of stefin B did have an impact on APP metabolism. Specifically, in cells expressing wild-type APP, it led to an increase in the levels of C-terminal fragments of APP cleaved by β-secretase (bCTF) but not those derived from α-secretase (aCTF). By contrast, in cells expressing APP-Sw, stefin B reduced the level of aCTF without affecting bCTF (Fig. 6).
Co-immunoprecipitation Experiment
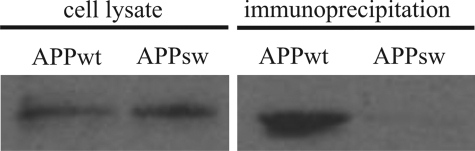
To examine if APP/Aβ and stefin B associate in cells, we transfected stefin B cDNA into CHO cells expressing either wild-type or Swedish mutant APP. 24 h after transfection the cells were lysed and immunoprecipitated with polyclonal antibodies against stefin B. The fraction bound to the anti-stefin B antibodies and the lysate were then run on 15% SDS gels followed by Western blotting with antibodies against Aβ (DE2B4) (Fig. 7) and stefin B (not shown). As shown in Fig. 7, a complex between stefin B and the ∼15-kDa C-terminal fragment of APP, comprising the Aβ sequence, was detected in APP-WT cells but not in cells expressing APP-Sw.
FIGURE 7.
Stefin B-Aβ/APP co-immunoprecipitation. CHO-K1 cells, expressing either wild-type or Swedish mutant APP, were transfected with the vector encoding stefin B and lysed after 24 h. The lysates were immunoprecipitated with polyclonal anti-rabbit stefin B antibodies, after which 15% SDS gels and Western blotting with anti-Aβ antibodies (DE2B4) were performed (see ”Experimental Procedures“ for more details). First and second lanes show results obtained on whole cell lysates of APP-WT and APP-Sw, respectively, and the third and fourth lanes show the corresponding samples after co-immunoprecipitation on anti-stefin B antibodies. The anti-Aβ antibodies (DE2B4) clearly detect a fragment of 15 kDa (according to molecular mass standards) in the APP-WT cells.
Co-localization of Stefin B and Aβ Aggregates in Cells
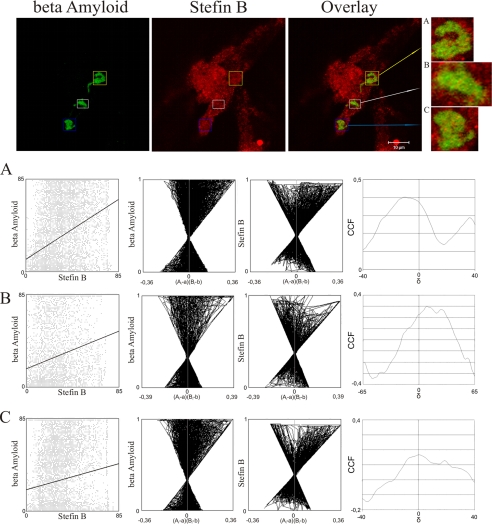
The possibility that Aβ aggregates and stefin B co-localize intracellularly was explored next. The CHO-K1 cell line expressing APP-Sw, with the level of Aβ increased ∼6-fold (29), was transiently transfected with stefin B. Staining with mouse anti-Aβ antibodies and rabbit polyclonal anti-stefin B antibodies showed that stefin B and Aβ peptide associate with Aβ inclusions (Fig. 8). Visual inspection of the staining pairs suggested co-localization of the two proteins in Aβ inclusions, possibly Aβ aggresomes (38). The regions of interest were defined by a quality co-localization analysis. We used the public domain tool JACoP for quantitative co-localization (Table 1). Dispersed scatter plots, the ICA plots (Fig. 8, graphs A–C), where most of the pixels are found on the right side of the plot, indicate partial co-localization of Aβ and stefin B.
FIGURE 8.
Aβ peptide co-localization with overexpressed stefin B in Aβ inclusions. Upper line represents confocal images of CHO-K1 cells immunostained with an antibody against Aβ (green) and stefin B (red). Overlay represents the overlay of two corresponding images. The scale bar is indicated. Inclusion bodies were marked with different colored squares (blue, white, and yellow). A zoomed view of the selected regions of interest on the side of the color panels are marked as A–C. Only the selected regions of interest were used for quantitative co-localization analysis, which was performed by JACoP, an Image J plug in. The three lines in boxes below the color images marked A–C represent the results of the JACoPs analysis (see Table 1).
TABLE 1.
Quantitative co-localization analysis performed with different intensity correlation coefficient based tools, which are grouped in public domain tool JACoP
A, B, and C represent selected regions of interest used in quantitative co-localization analysis. Rr, Pearson correlation coefficient; Rr(random), Pearson correlation coefficient determined with Costes randomization based co-localization between random images of the green channel and original image of the red channel with 200 randomization rounds; p value expressed as a percentage; R, overlap coefficient; kaβ, overlap coefficient determined for Aβ (green channel); kstB, overlap coefficient determined for stefin B (red channel); M, Manders coefficient; Maβ, Manders coefficient determined for Aβ (green channel); MstB, Manders coefficient determined for stefin B (red channel); CCF, Van Steensel cross-correlation function; ICQ, Li intensity correction quotient.
| Retion of Interest | Rr | Rr(random) | p value | r = kstB × kaβ | M(threshold) | CCF | ICQ | |||
|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.37 | 0.0 ± 0.166 | 100% | kaβ | 0.923 | 0.739 | Maβ | 0.46 | 0.402 | 0.171 |
| kstB | 0.591 | MstB | 0.563 | |||||||
| B | 0.222 | 0.001 ± 0.185 | 100% | kaβ | 0.857 | 0.651 | Maβ | 0.318 | 0.301 | 0.096 |
| kstB | 0.495 | MstB | 0.463 | |||||||
| C | 0.164 | 0.001 ± 0.155 | 100% | kaβ | 0.762 | 0.656 | Maβ | 0.416 | 0.261 | 0.072 |
| kstB | 0.565 | MstB | 0.43 | |||||||
DISCUSSION
Conclusions from in Vitro Studies
The first indication for in vitro interaction between Aβ and stefin B has been obtained by ThT fluorescence (Fig. 2, A and B) and TEM data (Fig. 3A), which show complete inhibition of Aβ fibril growth by wild-type Glu31 stefin B tetramers and dimeric Tyr31 variant. In accordance, SPR measurements show binding of exactly these two oligomeric forms of stefin B to Aβ (Fig. 4, A–D). This contrasts with monomers, dimers, and higher oligomers of wild-type Glu31 stefin B, which do not show considerable binding by SPR (Fig. 4C) and no inhibition of Aβ fibrillization. Small inhibition observed by the samples of higher oligomers (Fig. 2B) can be ascribed to the presence of small amounts of the tetramer in the sample (Fig. 1A). A complete separation of the higher oligomers from monomers, dimers, and tetramers is not possible (there are always about 5% tetramers present) yet transitions are slow and have always been checked before and after the measurements.
The fact that dimers of Tyr31 stefin B behave differently from those of Glu31 stefin B could be due to lower stability of the Tyr31 variant (35) and its tendency to form a molten globule (36). Thermal denaturation experiments4 indicate that Tyr31 stefin B dimers could be domain-swapped, similarly to stefin A dimers (22, 23) and cystatin C dimers (39).
Conclusions from Cellular Studies
Our studies using cultured cells (Fig. 6) show that the presence of stefin B does not change the steady-state levels of full-length wild-type or Swedish mutant APP, in line with a previous report showing that cystatin C had no effect on APP levels or processing (40).
There are some interesting observations on the influence of stefin B co-expression on the ratio of the APP C-terminal fragments, which seems worthwhile to explore in future studies. Namely, the aCTF gets reduced in the APP-Sw case and the bCTF was increased in APP-WT expressing cells. Furthermore, co-immunoprecipitation experiments on lysates of CHO-K1 cells expressing APP-WT or APP-Sw (Fig. 7) demonstrates a complex between stefin B and the APP fragment around 15 kDa in wild-type APP expressing cells. This kind of fragment was reported some time ago by Greengard (41). The 15-kDa C-terminal fragment is the major membrane-bound APP fragment and is most likely non-amyloidogenic (41). Obviously, monoclonal mouse anti-Aβ antibody DE2B4 recognizes only soluble peptides comprising the Aβ segment. This also would explain why there is no soluble fraction observed in the Aβ-rich APP-Sw cells (Fig. 7, lanes 3 and 4). The co-localization experiments in the CHO-K1 cell line expressing APP-Sw, supported by quantitative correlation analysis (see Fig. 8 and Table 1), shows that stefin B partially co-localizes with Aβ intracellular inclusions, which represent aggregated Aβ. Together, these results support the possibility that stefin B interacts with Aβ and APP C-terminal fragments in vivo.
Possible Chaperone Function of Stefin B Domain-swapped Oligomers
There is some evidence, currently under debate, that stefin B (cystatin B) might have additional function(s) other than cysteine protease inhibition. The protein was found as a component of a multiprotein complex specific to the cerebellum, together with a number of cytoskeleton proteins (42). Oligomerization of stefin B in cells was recently reported by Melli and co-workers (43) and the same phenomenon has also been observed in vitro (28, 25). Stefin B oligomers and aggregates in cells (43), which were confirmed and characterized further by our group,5 have no known function in the cell. On the basis of the in vitro and cell culture results presented in this work we propose that stefin B tetramers (and likely domain-swapped dimers) may bind to Aβ in vivo in such a way as to prevent its fibril formation. This so called ”armature“ chaperone action was recently ascribed to some other Aβ-binding proteins (7). This hypothesis seems supported by TEM data (Fig. 3, A–C), which show longer and more regular fibrils of Aβ in the presence of wild-type Glu31 stefin B (Fig. 3B); in comparison to those of Aβ fibrils alone (Fig. 3C). A complete inhibition of fibril growth at the granular aggregate stage happens in the presence of the dimeric Tyr31 variant (Fig. 3A). The tetramers of Glu31 stefin B, which also completely inhibit ThT fluorescence (Fig. 2B) and bind strongly to Aβ (Fig. 4, C and D) are expected to block fibrillization at the same stage.
From the three-dimensional structures of stefin B monomer (34), tetramer (37), and protection patterns of the fibrils (44) it should be possible to derive possible Aβ binding sites. The binding surface is not present in higher oligomers, which showed no binding by SPR. Their three-dimensional structure has not been solved as yet. In the future, as crystallization of the complex between stefin B and Aβ is a difficult task, their interaction could possibly be observed by NMR.
Binding of Aβ to the β sheet edge was proposed for other Aβ-binding proteins (5). Such a surface in stefin B could represent unpaired strand 5 or, alternatively, an exposed surface formed by strands 2 and 3 by domain-swapping.
The feature common to chaperones is their transient interaction with non-native conformations of other proteins, which is followed by preventing aggregation, correct folding or unfolding and, subsequent targeting to proteasomal or lysosomal degradation (45). Small heat shock proteins are made typically of 12–24 repeats of α-crystallin domains, which form dimers. The small heat shock proteins, αB-crystallins, bind to and inhibit amyloid fibril formation of Aβ, β2-microglobulin (46), and α-synuclein (47). In all these cases the interaction between interacting proteins was weak.
In Conclusion
Mutations in the stefin B (cystatin B) gene cause EPM1 (48). For the most part they lead to reduced protein expression, however, some are functional and lead to changed proteins, with different stability and aggregation properties (28). Lack of stefin B causes knock-out mice to undergo extensive loss of neurons in the granule layer of the cerebellum (49). They also show signs of ataxia and myoclonus. Stefin B gets overexpressed in many conditions of neurodegeneration, such as amyotrophic lateral sclerosis and Alzheimer disease but as shown recently also in conditions of oxidative stress (50). The loss of inhibitory function might be related to misregulation of the stefin B-cathepsin B axis of proteolysis and cell death. However, as the results presented here seem to suggest, aberrant protein folding of stefin B or, aggregation of other proteins, stefin B acting as a chaperone, might also be an explanation for increased oxidative stress, when stefin B is missing.
Supplementary Material
Acknowledgment
We are grateful to Louise Kroon Žitko (Jožef Stefan Institute, Ljubljana) for help with isolation of the recombinant stefin B protein.
This work was supported by Program P1-0140 (proteolysis and its regulation, led by B. Turk) via the Slovenian Research Agency (ARRS) and Estonian Science Foundation Grant 9171 (to P. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
E. Žerovnik, unpublished data.
S. Čeru, R. Layfield, V. Bergant-Zavašnik, U. Repnik, N. Kopitar-Jerala, V. Turk, and E. Žerovnik, submitted for publication.
- APP
- Aβ precursor protein
- SPR
- surface plasmon resonance
- ESI-MS
- electrospray ionization-mass spectrometry
- ThT
- thioflavin T
- PBS
- phosphate-buffered saline
- TEM
- transmission electron microscopy
- CHO
- Chinese hamster ovary
- ELISA
- enzyme-linked immunosorbent assay
- WT
- wild-type
- bCTF
- C-terminal fragments of APP cleaved by β-secretase
- aCTF
- C-terminal fragments of APP cleaved by α-secretase.
REFERENCES
- 1.Ross C. A., Poirier M. A. (2004) Nat. Med. 10, S10–S17 [DOI] [PubMed] [Google Scholar]
- 2.Walsh D. M., Selkoe D. J. (2004) Protein Pept. Lett. 11, 213–228 [DOI] [PubMed] [Google Scholar]
- 3.Cleary J. P., Walsh D. M., Hofmeister J. J., Shankar G. M., Kuskowski M. A., Selkoe D. J., Ashe K. H. (2005) Nat. Neurosci. 8, 79–84 [DOI] [PubMed] [Google Scholar]
- 4.Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 5.Yoshiike Y., Minai R., Matsuo Y., Chen Y. R., Kimura T., Takashima A. (2008) PLos ONE 3, e3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilhelmus M. M., Boelens W. C., Otte-Höller I., Kamps B., de Waal R. M., Verbeek M. M. (2006) Brain Res. 1089, 67–78 [DOI] [PubMed] [Google Scholar]
- 7.Wilhelmus M. M., de Waal R. M., Verbeek M. M. (2007) Mol. Neurobiol. 35, 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sastre M., Calero M., Pawlik M., Mathews P. M., Kumar A., Danilov V., Schmidt S. D., Nixon R. A., Frangione B., Levy E. (2004) Neurobiol. Aging 25, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 9.Turk V., Stoka V., Turk D. (2008) Front. Biosci. 13, 5406–5420 [DOI] [PubMed] [Google Scholar]
- 10.Rawlings N. D., Tolle D. P., Barrett A. J. (2004) Nucleic Acids Res. 32, D160–D164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin J. R., Craven C. J., Jerala R., Kroon-Zitko L., Žerovnik E., Turk V., Waltho J. P. (1995) J. Mol. Biol. 246, 331–343 [DOI] [PubMed] [Google Scholar]
- 12.Engh R. A., Dieckmann T., Bode W., Auerswald E. A., Turk V., Huber R., Oschkinat H. (1993) J. Mol. Biol. 234, 1060–1069 [DOI] [PubMed] [Google Scholar]
- 13.Ekiel I., Abrahamson M., Fulton D. B., Lindahl P., Storer A. C., Levadoux W., Lafrance M., Labelle S., Pomerleau Y., Groleau D., LeSauteur L., Gehring K. (1997) J. Mol. Biol. 271, 266–277 [DOI] [PubMed] [Google Scholar]
- 14.Maruyama K., Kametani F., Ikeda S., Ishihara T., Yanagisawa N. (1992) Neurosci. Lett. 144, 38–42 [DOI] [PubMed] [Google Scholar]
- 15.Ii K., Ito H., Kominami E., Hirano A. (1993) Virchows Arch. A Pathol. Anat. Histopathol. 423, 185–194 [DOI] [PubMed] [Google Scholar]
- 16.Bernstein H. G., Rinne R., Kirschke H., Järvinen M., Knöfel B., Rinne A. (1994) Brain Res. Bull. 33, 477–481 [DOI] [PubMed] [Google Scholar]
- 17.Bernstein H. G., Kirschke H., Wiederanders B., Pollak K. H., Zipress A., Rinne A. (1996) Mol. Chem. Neuropathol. 27, 225–247 [DOI] [PubMed] [Google Scholar]
- 18.Benussi L., Binetti G., Ghidoni R. (2006) in Human Stefins and Cystatins ( Zerovnik E., Kopitar-Jerala N., Uversky V. N. eds) pp. 115–125, Nova Science Publishers, New York [Google Scholar]
- 19.Žerovnik E., Pompe-Novak M., Skarabot M., Ravnikar M., Musevic I., Turk V. (2002) Biochim. Biophys. Acta 1594, 1–5 [DOI] [PubMed] [Google Scholar]
- 20.Jenko S., Škarabot M., Kenig M., Gunčar G., Muševič I., Turk D., Žerovnik E. (2004) Proteins Struct. Funct. Bioinformat. 55, 417–425 [DOI] [PubMed] [Google Scholar]
- 21.Škerget K., Vilfan A., Pompe-Novak M., Turk V., Waltho J. P., Turk D., Žerovnik E. (2009) Proteins Struct. Funct. Bioinformat. 74, 425–436 [DOI] [PubMed] [Google Scholar]
- 22.Jerala R., Žerovnik E. (1999) J. Mol. Biol. 291, 1079–1089 [DOI] [PubMed] [Google Scholar]
- 23.Staniforth R. A., Giannini S., Higgins L. D., Conroy M. J., Hounslow A. M., Jerala R., Craven C. J., Waltho J. P. (2001) EMBO J. 20, 4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderluh G., Gutierrez-Aguirre I., Rabzelj S., Ceru S., Kopitar-Jerala N., Macek P., Turk V., Zerovnik E. (2005) FEBS J. 272, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 25.Ceru S., Kokalj S. J., Rabzelj S., Skarabot M., Gutierrez-Aguirre I., Kopitar-Jerala N., Anderluh G., Turk D., Turk V., Žerovnik E. (2008) Amyloid 15, 147–159 [DOI] [PubMed] [Google Scholar]
- 26.Rabzelj S., Viero S., Gutiérrez-Aguirre I., Turk V., Dalla Serra M., Anderluh G., Žerovnik E. (2008) FEBS J. 275, 2455–2466 [DOI] [PubMed] [Google Scholar]
- 27.Jerala R., Trstenjak M., Lenarcic B., Turk V. (1988) FEBS Lett. 239, 41–44 [DOI] [PubMed] [Google Scholar]
- 28.Rabzelj S., Turk V., Žerovnik E. (2005) Protein Sci. 14, 2713–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez R. G., Squazzo S. L., Koo E. H. (1996) J. Biol. Chem. 271, 9100–9107 [DOI] [PubMed] [Google Scholar]
- 30.Yoon I. S., Chen E., Busse T., Repetto E., Lakshmana M. K., Koo E. H., Kang D. E. (2007) FASEB J. 21, 2742–2752 [DOI] [PubMed] [Google Scholar]
- 31.Kopitar-Jerala N., Curin-Serbec V., Jerala R., Krizaj I., Gubensek F., Turk V. (1993) Biochim. Biophys. Acta 1164, 75–80 [DOI] [PubMed] [Google Scholar]
- 32.Bolte S., Cordelières F. P. (2006) J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 33.Ritonja A, Machleidt W., Barrett A. J. (1985) Biochem. Biophys. Res. Commun. 131, 1187–1192 [DOI] [PubMed] [Google Scholar]
- 34.Stubbs M. T., Laber B., Bode W., Huber R., Jerala R., Lenarcic B., Turk V. (1990) EMBO J. 9, 1939–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Žerovnik E., Rabzelj S., Kenig M., Turk V. (2005) in Amyloid and Amyloidosis ( Grateau G., Kyle R. A., Skinner M. eds) pp. 21–23, CRC Press, Boca Raton, FL [Google Scholar]
- 36.Žerovnik E., Jerala R., Kroon-Zitko L., Turk V., Lohner K. (1997) Eur. J. Biochem. 245, 364–372 [DOI] [PubMed] [Google Scholar]
- 37.Jenko Kokalj S., Guncar G., Stern I., Morgan G., Rabzelj S., Kenig M., Staniforth R. A., Waltho J. P., Žerovnik E., Turk D. (2007) J. Mol. Biol. 366, 1569–1579 [DOI] [PubMed] [Google Scholar]
- 38.Bückig A., Tikkanen R., Herzog V., Schmitz A. (2002) Histochem. Cell Biol. 118, 353–360 [DOI] [PubMed] [Google Scholar]
- 39.Janowski R., Kozak M., Jankowska E., Grzonka Z., Grubb A., Abrahamson M., Jaskolski M. (2001) Nat. Struct. Biol. 8, 316–320 [DOI] [PubMed] [Google Scholar]
- 40.Mi W., Pawlik M., Sastre M., Jung S. S., Radvinsky D. S., Klein A. M., Sommer J., Schmidt S. D., Nixon R. A., Mathews P. M., Levy E. (2007) Nat. Genet. 39, 1440–1442 [DOI] [PubMed] [Google Scholar]
- 41.Iverfeldt K., Walaas S. I., Greengard P. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4146–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Di Giamo R., Riccio M., Santi S., Galeotti C., Ambrosetti D. C., Melli M. (2002) Hum. Mol. Genet. 11, 2941–2950 [DOI] [PubMed] [Google Scholar]
- 43.Cipollini E., Riccio M., Di Giaimo R., Dal Piaz F., Pulice G., Catania S., Caldarelli I., Dembic M., Santi S., Melli M. (2008) Biochim. Biophys. Acta 1783, 312–322 [DOI] [PubMed] [Google Scholar]
- 44.Morgan G. J., Giannini S., Hounslow A. M., Craven C. J., Žerovnik E., Turk V., Waltho J. P., Staniforth R. A. (2008) J. Mol. Biol. 375, 487–498 [DOI] [PubMed] [Google Scholar]
- 45.Saibil H. R. (2008) Curr. Opin. Struct. Biol. 18, 35–42 [DOI] [PubMed] [Google Scholar]
- 46.Raman B., Ban T., Sakai M., Pasta S. Y., Ramakrishna T., Naiki H., Goto Y., Rao Ch. M. (2005) Biochem. J. 392, 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rekas A., Adda C. G., Andrew Aquilina J., Barnham K. J., Sunde M., Galatis D., Williamson N. A., Masters C. L., Anders R. F., Robinson C. V., Cappai R., Carver J. A. (2004) J. Mol. Biol. 340, 1167–1183 [DOI] [PubMed] [Google Scholar]
- 48.Joensuu T., Lehesjoki A. E., Kopra O. (2008) Epilepsia 49, 557–563 [DOI] [PubMed] [Google Scholar]
- 49.Pennacchio L. A., Lehesjoki A. E., Stone N. E., Willour V. L., Virtaneva K., Miao J., D'Amato E., Ramirez L., Faham M., Koskiniemi M., Warrington J. A., Norio R., de la Chapelle A., Cox D. R., Myers R. M. (1996) Science 271, 1731–1734 [DOI] [PubMed] [Google Scholar]
- 50.Lehtinen M. K., Tegelberg S., Schipper H., Su H., Zukor H., Manninen O., Kopra O., Joensuu T., Hakala P., Bonni A., Lehesjoki A. E. (2009) J. Neurosci. 29, 5910–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.