Abstract
Extracellular acidosis often rapidly causes intracellular acidification, alters ion channel activities, and activates G protein-coupled receptors. In this report, we demonstrated a novel cellular response to acidosis: induction of the zymogen activation of matriptase. Acid-induced matriptase activation is ubiquitous among epithelial and carcinoma cells and is characterized by rapid onset, fast kinetics, and the magnitude of activation seen. Trace amounts of activated matriptase can be detected 1 min after cells are exposed to pH 6.0 buffer, and the vast majority of latent matriptase within the cells is converted to activated matriptase within 20 min. Matriptase activation may be a direct response to proton exposure because acid-induced matriptase activation also occurs in an in vitro, cell-free setting in which intracellular signaling molecules and ion channel activities are largely absent. Acid-induced matriptase activation takes place both on the cell surface and inside the cells, likely due to the parallel intracellular acidification that activates intracellular matriptase. Following matriptase activation, the active enzyme is immediately inhibited by binding to hepatocyte growth factor activator inhibitor 1, resulting in stable matriptase-hepatocyte growth factor activator inhibitor 1 complexes that are rapidly secreted. As an early response to acidosis, matriptase activation can also be induced by perturbation of intracellular pH homeostasis by 5-(N-methyl-N-isobutyl)-amiloride and 5-(N-ethyl-N-isopropyl)-amiloride, both of which inhibit Na+/H+ exchangers, and diisothiocyanostilbene-2,2′-disulfonic acid, which can inhibit other acid-base ion channels. This study uncovers a novel mechanism regulating proteolysis in epithelial and carcinoma cells, and also demonstrates that a likely function of matriptase is as an early response to acidosis.
Keywords: Cell/pH, Enzymes/Proteolytic, Membrane/Enzymes, Protease, Protease/Inhibitor, Proteases/Serine Protease, Protein/Post-translational Modification, Protein/Proenzyme Processing
Introduction
Matriptase, a type 2 transmembrane serine protease, is broadly expressed by epithelial and carcinoma cells (1–3), and plays essential roles in the maintenance of epithelial integrity, particularly for epidermal terminal differentiation and barrier function (4–6). A rare human inherited genetic disorder, autosomal recessive ichthyosis, is associated with two missense mutations of the human ST14 gene, which encodes matriptase (7, 8). In addition, the protease can exhibit potent oncogenic activity via ras-dependent and ras-independent pathways when it is even slightly overexpressed in the skin of transgenic mice (9). Several tumor xenograft models also provide further evidence that matriptase is strongly associated with cancer cell proliferation, invasion, and metastasis (10, 11). As is typical for classical serine proteases, matriptase is synthesized as a zymogen and must undergo activation by cleavage at Arg614 (R↓VVGG) to gain full proteolytic activity. It is the activity of the mature enzyme that is believed to be responsible for both the physiological and pathological functions of the enzyme identified by studies of animal models and human genetics (12–14).
In contrast to most serine proteases, however, which are typically activated through the action of other active proteases, the matriptase zymogen undergoes autoactivation to gain its full proteolytic capacity (15). This autoactivation process can only be initiated when the matriptase zymogen molecules are anchored to a lipid bilayer biomembrane (16). Other proteins, including hepatocyte growth factor activator inhibitor 1 (HAI-1),2 the cognate matriptase protease inhibitor, is also colocalized with matriptase and is intimately involved in matriptase autoactivation (17). The term “matriptase activation machinery” has been used to describe this complex matriptase activation system that must anchor on the cell membrane to perform two major biochemical events: 1) mediating the proteolytic cleavage of matriptase, which converts the single-chain matriptase zymogen into the two-chain active enzyme and 2) inhibition of the newly formed active matriptase through high affinity binding to HAI-1 resulting in the formation of a 120-kDa matriptase-HAI-1 complex (16). Many protease inhibitors are physically separated from their target proteases, and the colocalization of HAI-1 with matriptase is quite unusual (18, 19). The close spatial relationship of the two proteins, the presence of HAI-1 at significantly higher concentrations than matriptase, and the high affinity nature of the binding between active matriptase and HAI-1, results in HAI-1 binding and inhibition of matriptase proteolytic activity almost immediately after the enzyme is activated. The end product of matriptase activation is, therefore, paradoxically a functionally inactive 120-kDa matriptase-HAI-1 complex rather than free active matriptase. Further evidence for this unprecedented functional linkage between matriptase and HAI-1 is manifested by the rescue of the defects in placenta development and skin abnormality that are associated with HAI-1 (SPINT1) genetic ablation, by the simultaneous ablation of, or significant reduction in matriptase expression (20–23). The extremely short life of free, active matriptase underscores the importance of location at which the matriptase activation machinery is activated, and the stimuli responsible for its activation. In most polarized epithelial cells, matriptase activation and inhibition occurs at the basolateral plasma membrane (25). Interestingly, whereas a proportion of the matriptase·HAI-1 complex can be shed directly from the basolateral surface, most matriptase·HAI-1 complexes, particularly in vivo, are internalized, trafficked to the apical plasma membrane (transcytosis), and secreted into the lumen (25). Thus, the basolateral plasma membrane may be the subcellular site where matriptase acts on its substrates. In this context, the protease-activated receptor 2, a G protein coupled-receptor that is also located basolaterally, may be more likely to be activated by matriptase than other putative matriptase substrates (26, 27).
The matriptase activation machinery can be activated by several exogenous factors, some of which are specific for a particular cell-type or system. Sphingosine 1-phosphate, a blood-borne lysophospholipid, is capable of inducing matriptase activation through stimulation of sphingosine 1-phosphate membrane receptors (28); however, this mechanism only seems to occur in mammary epithelial cells. The androgen, dihydrotestosterone, can induce matriptase activation in LNCaP prostate cancer cells by androgen receptor-dependent transcriptional regulation (29). Breast cancer cells, however, respond to neither sphingosine 1-phosphate nor androgens for matriptase activation. Instead, breast cancer cells activate matriptase constitutively (30). Suramin, a sulfide-rich polyanionic compound, can stimulate matriptase activation in a variety of matriptase-expressing cells (19). These unrelated chemicals act through different mechanisms and induce different signal pathways, all of which converge at the matriptase activation machinery and induce matriptase activation and inhibition. Although the matriptase activation machinery is regulated by the divergent mechanisms, the activation machinery itself can be isolated from cells by simple homogenization of the cells followed by collection of the insoluble fractions of the homogenates (16). Matriptase, HAI-1, and the activation machinery all appear to be anchored to the internal membrane structures of the cell surrounding the nucleus. Within a narrow range of pH and ionic strength, the matriptase activation machinery can be turned on in vitro, under cell-free conditions. At pH 7.4 and above matriptase activation does not occur. As the buffer is made more acidic, matriptase activation increases with maximum activation occurring at pH 6.0. Buffer acidity, therefore, appears to serve as a switch, determining the degree to which matriptase activation takes place. In view of the rapid kinetics, the magnitude of the response, and the fact that this process is not dependent on upstream signaling pathways, led us to hypothesize that the matriptase activation machinery may serve as a pH sensor that responds directly to an acidic environment. In the current study, we evaluate this hypothesis using live cells to determine whether extracellular acidosis and perturbation of intracellular pH homeostasis can induce matriptase activation.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Alexa Fluor 488 goat anti-mouse IgG and SNARF-4F acetoxymethyl ester were obtained from Invitrogen. Benzamil was obtained from Alexis Biochemicals. Dulbecco's phosphate-buffered saline (DPBS) was obtained from Mediatech Inc. All other chemicals and reagents were obtained from Sigma, unless otherwise specified.
Monoclonal Antibodies
Human matriptase was detected using either M32 or M24 mouse monoclonal antibodies (mAbs), which recognize both the latent and activated forms of matriptase. Activated matriptase was detected using the M69 mouse mAb, which recognizes an epitope present only on the activated form of the enzyme (28, 31). Human HAI-1 was detected using the HAI-1-specific mouse mAb M19 (32).
Cell Culture
184 A1N4 cells were cultured in a 50:50 mixture of Dulbecco's modified Eagle's/Ham's F-12 medium (DMEM-F12 50/50, Mediatech Inc.) supplemented with 0.5% fetal bovine serum (FBS) (Gemini Bio-Products), 5 μg/ml of recombinant human insulin (rh-insulin) (Invitrogen), 5 μg/ml of hydrocortisone (Sigma), 10 ng/ml of recombinant human epidermal growth factor (Promega), and 1% penicillin-streptomycin (Mediatech Inc.). MCF-10A cells were cultured in Dulbecco's modified Eagle's medium-F12 (50:50) supplemented with 5% horse serum (Gemini Bio-Products), 0.5 μg/ml of hydrocortisone, 10 μg/ml of rh-insulin, 20 ng/ml of recombinant human epidermal growth factor, and 1% penicillin-streptomycin. MTSV-1.1 B and MTSV-1.7 cells were cultured in modified Iscove's modified Dulbecco's modified Eagle's medium (Mediatech Inc.) supplemented with 10% FBS, 10 μg/ml of rh-insulin, 5 μg/ml of hydrocortisone, and 1% penicillin-streptomycin. MCF-7 cells were cultured in Eagle's minimum essential medium (Mediatech Inc.) supplemented with 10% FBS, 10 μg/ml of rh-insulin, and 1% penicillin-streptomycin. T-47D cells were cultured in RPMI 1640 medium (Mediatech Inc.) supplemented with 10% FBS, 45 μg/ml of rh-insulin, and 1% penicillin-streptomycin. MDA-MB-468 cells were cultured in Leibovitz L-15 medium (Mediatech Inc.) supplemented with 10% FBS, and 1% penicillin-streptomycin. RWPE-1 and RWPE-2 cells were cultured in keratinocyte serum-free medium (Invitrogen) supplemented with 50 μg/ml of bovine pituitary extract (Sigma) and 5 ng/ml of recombinant human epidermal growth factor, and 1% penicillin-streptomycin. LNCaP, OVCAR-3, OV2008, and IGROV-1 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, and 1% penicillin-streptomycin. DU 145 cells were cultured in Eagle's minimum essential medium (Mediatech Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin. PC-3 cells were cultured in F-12K medium (Mediatech Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were maintained at 37 °C in a humidified atmosphere with 5% CO2, except MDA-MB-468 cells, which were maintained without additional CO2.
Immunoblotting Assay
Cells were harvested by scraping and lysed in a buffer consisting of 0.5% Triton X-100, 1 mm 5,5′-dithiobis-(2-nitrobenzoic acid), and protease inhibitor mixture (Roche) in PBS. Samples of 50 μg of total protein per well were subjected to SDS-PAGE using a sample buffer that did not contain a reducing agent and without boiling the samples. Proteins resolved by 7.5% SDS-PAGE were transferred to BioTrace NT nitrocellulose membranes (Pall), and probed with monoclonal antibodies M32, M24, M69, and M19, as indicated. The binding of the primary antibody was detected using horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch), and visualized using the Western Lightening Chemiluminescence Reagent Plus (PerkinElmer Life Sciences) and x-ray film.
Immunofluorescence Staining
Cells were seeded on 18-mm diameter cover slides and incubated for 48 h. After the indicated treatments, the cells were fixed with 4% formaldehyde in PBS for 20 min at room temperature and then permeabilized using 0.1% Triton X-100 in PBS for another 20 min at room temperature. The fixed, permeabilized cells were then probed with mouse primary monoclonal antibodies at a concentration of 5 μg/ml, followed by labeling with 2 μg/ml of Alexa Fluor 488-conjugated goat anti-mouse IgG for visualization. Cellular nuclei were counterstained with 4′,6-diamidino-2-phenylindole, and images were recorded using an Olympus FluoView FV1000 confocal laser scanning microscope and edited with Olympus FV10-ASW software.
Intracellular pH Measurement
Intracellular pH (pHi) was determined by a SNARF-4F fluorescent pH indicator. For kinetics experiments (Fig. 4), cells were loaded with 10 μm SNARF-4F acetoxymethyl ester for 1 h and incubated in DPBS for 15 min to permit de-esterification before measurement. For transporter blocker experiments (Table 1), the cells were first treated with blockers for 2 h and the fluorescent dye was loaded 1 h before the end of blocker treatment. For the measurement, intracellular SNARF-4F was excited at 488 nm and fluorescence images at the dual emission wavelengths, 580 and 640 nm, were recorded simultaneously on a laser scanning confocal microscope (Zeiss LSM510). pHi at a given time point was determined from the ratio of background-corrected emission intensities at 580 and 640 nm, through the use of calibration parameters (33, 34). In situ pH calibration was performed in calibration buffers containing 130.5 mm potassium gluconate, 9.0 mm sodium gluconate, 1.5 mm sodium phosphate monobasic, 1.5 mm magnesium sulfate, 10 mm d-glucose, 2.0 mm calcium chloride, 10 mm HEPES, and 10 μm of the ionophore, nigercin, at either pH 5.0 or 8.0. Data analysis and layout were performed with SigmaPlot 2000 (SPSS Inc.)
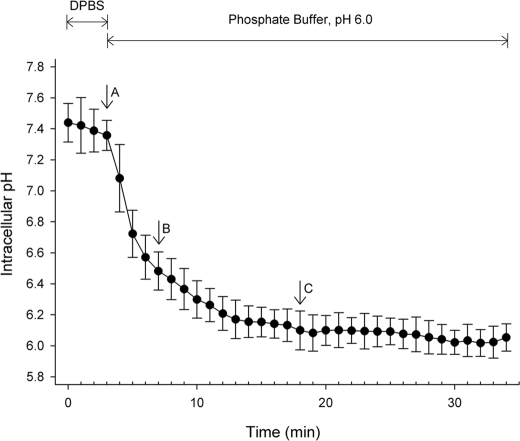
FIGURE 4.
The kinetics of intracellular acidification induced by extracellular acidosis. SNARF-4R-loaded 184 A1N4 cells were maintained in DPBS and then exposed to pH 6.0 buffer. Intracellular pH was recorded and computed as described under “Experimental Procedures.” The pHi value is presented as mean ± S.D. of 10 individual cells at each time point. When maintained in DPBS, the pHi was ∼7.4. After switching to pH 6.0 buffer, the pHi was rapidly decreased. At the indicated time points, matriptase activation was analyzed in two separated experiments by immunoblotting (Fig. 5) and immunofluorescence staining (Fig. 6).
TABLE 1.
Blockage of pH regulatory transporters by inhibitors leads to disturbance of intracellular pH homeostasis
184 A1N4 cells were incubated in basal media, or treated with 0.1% Me2SO, 50 μm EIPA, 50 μm MIBA, 50 μm DIDS, 500 μm DIDS, or 1 μm bafilomycin A1 for 2 h. To measure pHi, the cells were proloaded with SNARF-4R for 1 h. Intracellular pH of each treatment was recorded and computed as described under “Experimental Procedures.” The pHi value is presented as mean ± S.D. of 50 individual cells of each treatment. The Student's t tests were used in tests of significance.
| Treatment | Target transporter | pHi (mean ± S.D.) |
|---|---|---|
| Control | 7.30 ± 0.13 | |
| 0.1% Me2SO | 7.34 ± 0.12 | |
| 50 μm EIPA | NHEs | 6.49 ± 0.10a |
| 50 μm MIBA | NHEs | 6.95 ± 0.11a |
| 50 μm DIDS | NBCs | 7.10 ± 0.06a |
| 500 μm DIDS | NBC, NDCBE, AE | 6.94 ± 0.05a |
| 1 μm Bafilomycin A1 | V-ATPase | 7.38 ± 0.10 |
a Significant difference to the pHi of vehicle control (p < 0.05).
RESULTS
Exposure of Epithelial and Carcinoma Cells to a Mildly Acidic Extracellular Milieu Results in Robust Matriptase Activation
To determine whether the matriptase activation machinery could be activated in live cell exposure to a mildly acidic buffer, we transiently exposed a variety of matriptase-expressing epithelial and carcinoma cells to pH 6.0 buffer and evaluated matriptase activation. 184 A1N4 mammary epithelial cells were used as the model cell system for the initial characterization studies. Matriptase activation in intact cells was first compared with that seen in the in vitro, cell-free system (Fig. 1).
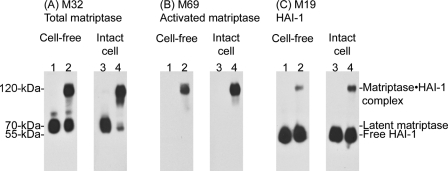
FIGURE 1.
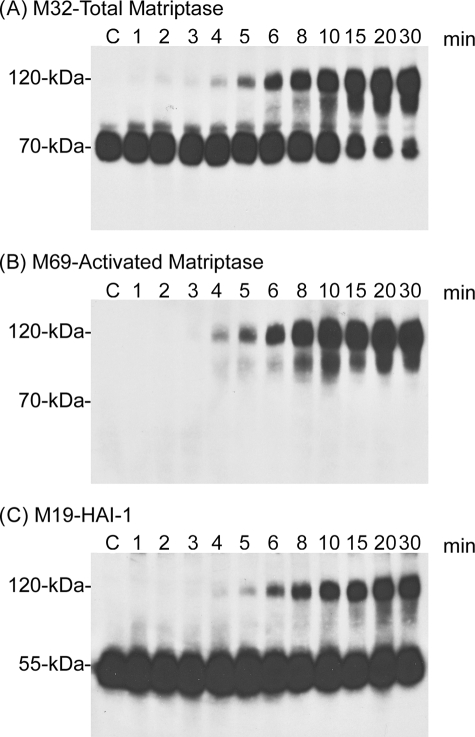
Cell-free versus intact cell matriptase activation. The insoluble fractions of cell homogenates (Cell-free, lanes 2) or intact cells (Intact cell, lanes 4) were exposed to pH 6.0 citric acid-phosphate buffer at room temperature for 20 min to induce matriptase activation. The non-activation control for the cell-free system was incubated at 4 °C (lanes 1) and the intact cell system was incubated in basal media (lanes 3). Lysates prepared from these samples were analyzed by immunoblotting for total matriptase using mAb M32, for activated matriptase using mAb M69, and for HAI-1 using mAb M19, as indicated.
After growth for 1 day in media supplemented with 0.5% serum, analysis of 184 A1N4 cells showed that the matriptase was present as the latent form of the enzyme, detected as a 70-kDa species using the matriptase mAb M32, in lysates prepared from either intact cells, or from the insoluble fractions of cell homogenates in a cell-free system (Fig. 1A, M32, lanes 1 and 3). Consistent with this observation, no protein bands were detected using the activated matriptase-specific mAb (Fig. 1B, M69, lanes 1 and 3) (28, 31). The inhibitor HAI-1 was detected in its uncomplexed form at 55 kDa, again consistent with the lack of active matriptase in these lysates (Fig. 1C, M19, lanes 1 and 3). Incubation of the cells and cell homogenates in neutral buffer at room temperature does not result in activation (data not shown). After exposure of the cell-free system to pH 6.0 citric acid-phosphate buffer for 20 min at room temperature, robust matriptase activation was observed consistent with our previous findings (16) (Fig. 1, lane 2). Interestingly, even more robust matriptase activation was seen when intact, live cells were exposed to pH 6.0 citric acid-phosphate buffer for a similar period (Fig. 1, lanes 4). Matriptase activation is evident due to the appearance of the activated matriptase·HAI-1 complex at 120-kDa, which is detected by the total matriptase mAb M32 (Fig. 1A, M32, lanes 2 and 4), by the activated matriptase-specific mAb M69 (Fig. 1B, M69, lanes 2 and 4), and by the HAI-1 mAb M19 (Fig. 1C, lanes 2 and 4). Concurrent with the emergence of the 120-kDa matriptase·HAI-1 complex, levels of 70-kDa latent matriptase are decreased (Fig. 1A, M32, lanes 2 and 4). HAI-1 is expressed at much higher levels than matriptase and so the formation of the matriptase·HAI-1 complex does not cause an obvious decrease in the levels of free, uncomplexed HAI-1 (Fig. 1C, M19, lanes 2 and 4). Comparing the ratio of complexed (activated) to free (latent) matriptase, the degree of matriptase activation in the intact cell system appears to be greater than that in the in vitro, cell-free activation system. These data suggest that the matriptase activation machinery can also be rapidly and robustly activated in live cells by exposure to a mildly acidic buffer. The essentially instantaneous binding of HAI-1 to the active matriptase after activation of the enzyme that is characteristic of the cell-free system appears to be well preserved in intact cell activation. Furthermore, given the fact that the vast majority of matriptase were detected in the matriptase·HAI-1 complex (Fig. 1A, lane 4) and that the complex only represents a small proportion of total HAI-1 (Fig. 1C, lane 4), 184 A1N4 cells express HAI-1 much higher than matriptase.
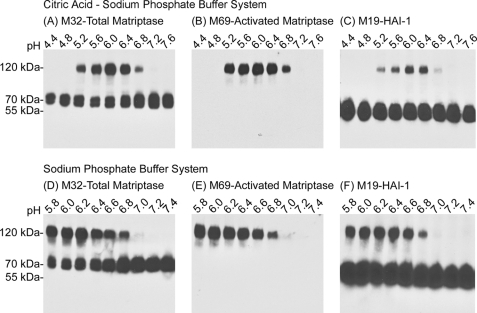
The extracellular pH range and optimum for the induction of matriptase activation in live cells is very similar to that seen with the in vitro, cell-free system (Fig. 2). Exposure of 184 A1N4 cells to citric acid-phosphate buffers (Fig. 2, upper panels) with pH values ranging from pH 4.4 to 7.6 showed that matriptase activation occurred within a narrow pH range between pH 5.2 and 6.8 with the optimum at pH 6.0, which is very similar to what has been observed in the in vitro, cell-free activation system (16). The intact-cell acid exposure study was repeated twice more using two different buffer systems, phosphate-based (Fig. 2B, lower panels) and citrate-based (not shown), with essentially identical results, confirming that the exposure to the low pH and not some buffer constituent was responsible for the induction of matriptase activation observed.
FIGURE 2.
The pH optimum for matriptase activation in the intact cell activation assay. 184 A1N4 cells were exposed to citric acid-phosphate-based (upper panels) or phosphate-based (lower panels) buffers with the pH indicated, at room temperature for 20 min. Cell lysates were then prepared and analyzed by immunoblotting for total matriptase using mAb M32, for activated matriptase using mAb M69, and for HAI-1 using mAb M19.
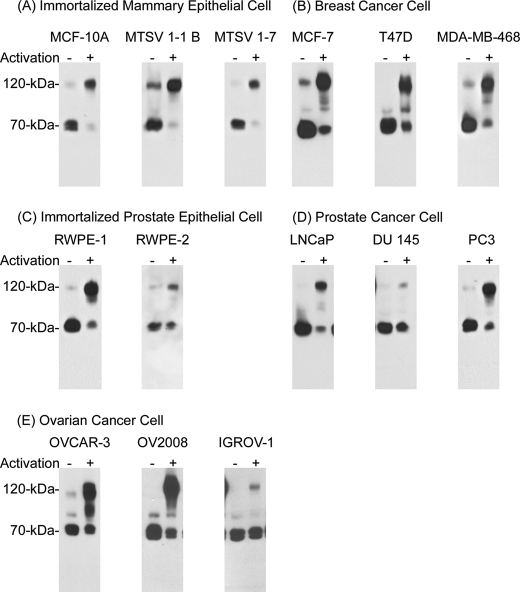
Matriptase is broadly expressed by a variety of epithelial and carcinoma cells of different origins, and so to determine whether the activation of matriptase in response to exposure to low pH is a general mechanism, 14 cell lines from several tissue origins were tested for their response to an acidic milieu. These lines included three immortal mammary epithelial cell lines, MCF-10A, MSTV-1-1 B, MSTV-1-7 (Fig. 3A), three breast cancer cell lines, MCF-7, T-47D, and MDA-MB-468 (Fig. 3B), two immortalized prostate epithelial cell lines, RWPE-1 and RWPE-2 (Fig. 3C), three prostate cancer cell lines, LNCaP, DU 145, and PC3 (Fig. 3D), and three ovarian cancer cell lines, OVCAR-3, OV2008, and IGROV-1 (Fig. 3E). Exposure of all of these cell lines to pH 6.0 buffer resulted in the induction of matriptase activation, although the level of activation in three of the lines was somewhat less than that seen in the 184 A1N4 mammary epithelial cells. These data suggested that extracellular acid-induced matriptase activation is likely a universal event among matriptase-expressing epithelial and carcinoma cells.
FIGURE 3.
Acid-induced matriptase activation in matriptase-expressing epithelial cells. Fourteen different cell lines were exposed to basal media (activation −) or pH 6.0 buffer to induce matriptase activation (activation +). Cell lysates were prepared and analyzed for total matriptase using the mAb M32. The cell lines used were three immortal mammary epithelial lines, MCF-10A, MSTV-1-1 B, and MSTV-1-7 (panel A), three breast cancer lines, MCF-7, T-47D, and MDA-MB-468 (panel B), two immortalized prostate epithelial lines, RWPE-1 and RWPE-2 (panel C), three prostate cancer lines, LNCaP, DU 145, and PC3 (panel D), and three ovarian cancer cell lines, OVCAR-3, OV2008, and IGROV-1 (panel E).
Intracellular Acidification and Matriptase Activation
Matriptase is known as an integral membrane protein that is localized at the plasma membrane, however, as we have shown before, the majority of matriptase resides inside the cell (18, 35). This intracellular pool of matriptase is anchored to the intracellular membranes surrounding the nucleus, and is the pool of enzyme that is assayed in the in vitro, cell-free activation system (16). The magnitude of matriptase activation seen in the intact cell activation system in response to acid exposure suggests that both the plasma membrane-localized matriptase and the internal matriptase pool have undergone activation. This suggests that the activation of internal matriptase is caused by intracellular acidification resulting from exposure of the cells to the extracellular acid milieu, thereby contributing to the high levels of matriptase activation. We, therefore, compared the kinetics of intracellular acidification (Fig. 4) with that of matriptase activation (Figs. 5 and 6) in response to extracellular acidic exposure. The kinetics of intracellular acidification was determined using the SNARF-4R pHi indicator. Before exposure to pH 6.0, the pHi of 184 A1N4 cells in DPBS was ∼7.4 and remained stable before exposure to pH 6.0 extracellularly (Fig. 4, before point A). Upon switching the extracellular pH to 6.0, pHi quickly decreased, reaching ∼6.5 after 4 min (Fig. 4, point B). Fifteen minutes after exposure of the cells to pH 6.0, pHi was stabilized at ∼6.1 (Fig. 4, point C).
FIGURE 5.
The kinetics of acid-induced matriptase activation. 184 AIN4 cells were incubated in pH 6.0 buffer at room temperature for the indicated times, after which cell lysates were prepared and analyzed by immunoblotting for total matriptase, activated matriptase, and HAI-1, as indicated.
FIGURE 6.
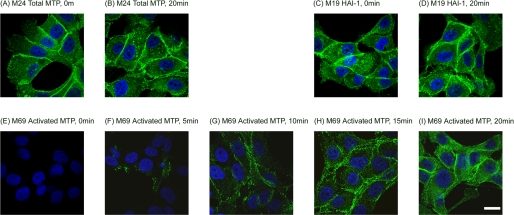
The kinetics and subcellular localization of acid-induced matriptase activation. 184 A1N4 cells were incubated in pH 6.0 buffer for the indicated times and then fixed, permeabilized, and analyzed by immunofluorescence staining for total matriptase (A and B), HAI-1 (C and D), and activated matriptase (E–I). The nuclei, shown in blue, were stained with 4′,6-diamidino-2-phenylindole. The scale bar represents 20 μm.
The kinetics of matriptase activation was determined both by immunoblotting of cell lysates (Fig. 5) and immunofluorescence staining of fixed cells (Fig. 6). When 184 A1N4 cells were exposed to pH 6.0 at room temperature, a trace amount of the 120-kDa matriptase·HAI-1 complex began to appear as soon as 1 min after exposure to the buffer (Fig. 5, 1 min), and was quite evident after around 4 min (Fig. 5, 4 min). At this time point, the intracellular pH was about 6.5 (Fig. 4, point B). Around half of the 70-kDa latent matriptase was converted into matriptase·HAI-1 complex 10 min after exposure to the buffer (Fig. 5, 1 min). The vast majority of the matriptase was detected in the 120-kDa complex form by 15 min after initiating the exposure of the cells to pH 6.0 buffer (Fig. 5, 15 min), at which point the intracellular pH was 6.1. These data show that intracellular acidification and matriptase activation have comparable kinetics.
The kinetics and subcellular localizations of matriptase activation were also studied by immunofluorescence staining using laser scanning confocal microscopy. Two days after growth of 184 A1N4 cells in media supplemented with low levels of serum, matriptase (Fig. 6A) and HAI-1 (Fig. 6C) were both detected on the cell surface, with more intense staining evident at the cell-cell contacts, and within the cells with a diffuse staining pattern. The subcellular locations of total matriptase and HAI-1 did not change notably during the course of matriptase activation (Fig. 6, A versus B and C versus D). As expected, staining with M69 demonstrated that the cells were negative for activated matriptase prior to the exposure of the cells to pH 6.0 (Fig. 5E). Activated matriptase began to be detectable, both on the cell surface and inside the cells, 5 min after exposure to pH 6.0, and gradually accumulated to much higher levels over the course of 20 min (Fig. 6, F to I). These data again demonstrate that the kinetics of intracellular acidification is consistent with the kinetics of matriptase activation, and suggest that, as we suspected, the matriptase activation that is induced in intact cells by exposure to an acidic milieu involves the cell surface, and more importantly the intracellular pools of the enzyme, and likely is the result of rapid intercellular acidification. These data suggest that both extracellular and intracellular pH environments may play important roles in regulation of matriptase activation.
The Matriptase·HAI-1 Complex Is Rapidly Secreted
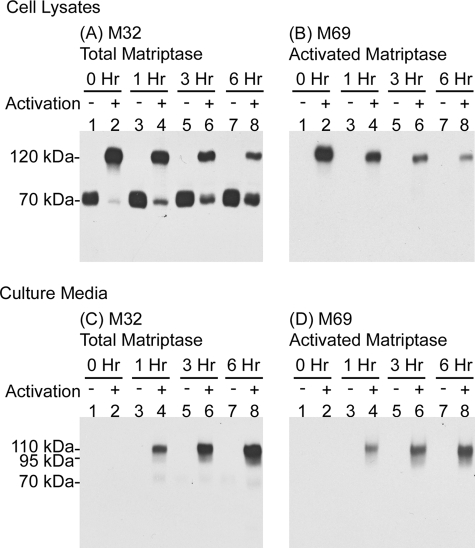
One of the remarkable features of matriptase activation is that the 120-kDa matriptase·HAI-1 complex is rapidly shed into the extracellular milieu (25). The effective shedding of matriptase·HAI-1 complexes results in the presence of very low levels of matriptase·HAI-1 complex in cells and the abundance of such complexes in body fluids, such as milk, urine, and semen, as well as the conditioned media of cultured cells. The rapid shedding of matriptase·HAI-1 complexes appears also to occur after extracellular acidosis-induced matriptase activation. During the course of acid exposure, matriptase·HAI-1 complexes were not efficiently shed into the extracellular milieu (data not shown), however, as soon as the pH 6.0 buffer was removed and the cells returned to basal media, matriptase·HAI-1 complexes were rapidly shed into the media. One hour after the cells were returned to basal media, levels of the 120-kDa cellular matriptase·HAI-1 complex detectable in cell lysates had dropped significantly (Fig. 7, A and B, comparing lane 4 with lane 2) and increasing levels of matriptase were detectable in the conditioned media as 95- and 110-kDa complexes that represent the cleaved products of the 120-kDa cellular matriptase·HAI-1 complexes (Fig. 7, C and D, lane 4). Most of the matriptase·HAI-1 complex was released into media within 3–6 h (Fig. 6). In addition, intracellular 70-kDa latent matriptase levels appeared to be rapidly replenished (Fig. 7A, lanes 2, 4, 6, and 8), and in contrast to activated matriptase species, essentially none of the 70-kDa latent matriptase was shed during the 6-h time course (Fig. 7C, lanes 1, 3, 5, and 7), suggesting that the cells selectively and rapidly secrete matriptase·HAI-1 complex following matriptase activation.
FIGURE 7.
Matriptase·HAI-1 complexes are shed into extracellular milieu following acid-induced activation. 184 A1N4 cells were preincubated with basal media (activation −) or pH 6.0 buffer to induce matriptase activation (activation +). After 20 min the cells were washed and then incubated in basal media for the indicated periods after which cell lysates and conditioned media were collected and analyzed by immunoblotting for total matriptase and activated matriptase, as indicated.
Disturbance of Intracellular pH Homeostasis Causes Matriptase Activation and Shedding
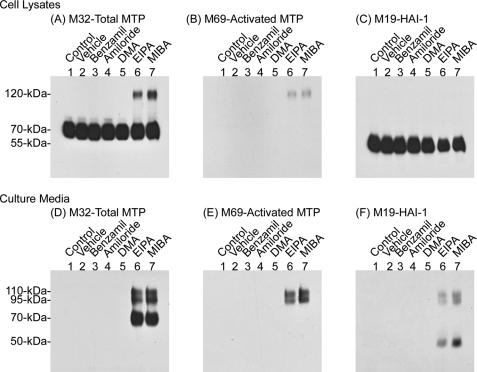
The cell-free and intact cell matriptase activation systems clearly show that both extracellular and intracellular acidity are prominent factors that can stimulate matriptase activation. Matriptase activation may, therefore, serve as an important and early response to intracellular and extracellular acidosis. pHi is tightly regulated to maintain a narrow pH range that is critical for cellular processes. pHi can be affected by passive H+ fluxes, metabolism, and fluxes of ionized forms of weak acids and bases. Extrusion of excessive H+ through several ion transporters is the major and essential mechanism of pHi homeostasis. Several types of acid-base ion transporters are involved in the regulation of pHi homeostasis in mammalian tissues, including Na+/H+ exchangers (NHEs), Na+/HCO3− co-transporters (NBCs), Na+-dependent Cl−/HCO3− exchangers (NDCBEs), Na+-independent Cl−/HCO3− or anion exchangers (AEs) (36, 37). Blockage of these pHi regulatory transporters reduces proton efflux as well as the alkalinity by extruding anions resulting in an increase in intracellular acidity (38–40). Among these transporters, the NHEs are the most prominent in the regulation of proton efflux and play a crucial role in maintaining pHi. Thus, we first tested whether perturbation of pHi by inhibition of NHE activity could increase matriptase activation. After a 2-h treatment of 184 A1N4 cells with two selective amiloride-derived NHE inhibitors, 5-(N-methyl-N-isobutyl)-amiloride (MIBA) and 5-(N-ethyl-N-isopropyl)-amiloride (EIPA), we observed acidic pHi in a large proportion of the treated cells, and the average pHi was determined to be around 6.95 for MIBA-treated cells and 6.49 for EIPA-treated cells (Table 1). Accordingly, accumulation of activated matriptase was clearly seen in the cell lysates (Fig. 8, A–C, lanes 6 and 7) as well as in the culture media of the MIBA- and EIPA-treated cells (Fig. 8, D–F, lanes 6 and 7). Furthermore, matriptase activation induced by MIBA-mediated NHE blockage can be negated by trimethylamine (Fig. 9), which can cause intracellular alkalinization without a change in the pH of the media (41). Determination of the average pHi for the cells co-treated with MIBA and trimethylamine was not feasible due to the fact that pHi in one-fifth of the cell population was higher than pH 8.0, and so beyond the detection range of SNARF-4R. Nevertheless, we have observed acidic pHi in one-fifth of the cells. The low levels of activated matriptase detected may have resulted from these cells with acidic pHi. These data suggest that intracellular acidosis alone can quickly induce the matriptase activation machinery, resulting in matriptase activation, HAI-1 inhibition, and shedding (Fig. 8). In contrast to MIBA and EIPA, treatments with other amiloride analogs, benzamil, amiloride, and dimethyl amiloride, all of which potently inhibit other types of Na+ channels, but have less inhibitory activity against the NHEs (42), caused no increase in matriptase activation and shedding (Fig. 8, lanes 3–5).
FIGURE 8.
NHE inhibition induces matriptase activation and shedding. 184 A1N4 cells were incubated in basal media (lane 1) or treated with 0.1% Me2SO (lane 2), 50 μm benzamil (lane 3), 50 μm amiloride (lane 4), 50 μm dimethyl amiloride (DMA) (lane 5), 50 μm EIPA (lane 6), or 50 μm MIBA (lane 7) for 2 h. Cell lysates and culture medium were collected and analyzed for total matriptase, activated matriptase, and HAI-1, as indicated.
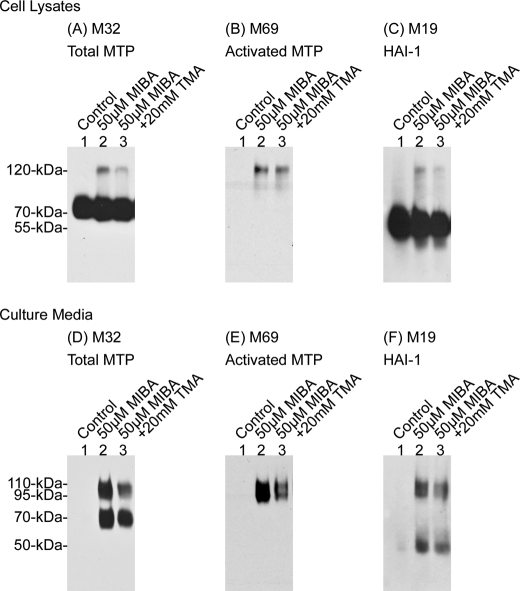
FIGURE 9.
Intracellular alkalinization attenuates matriptase activation caused by the NHE blockage. 184 A1N4 cells were treated with 20 mm trimethylamine (TMA) along with 50 μm MIBA (lane 3), a NHE potent inhibitor, for 2 h. Untreated cells (lane 1) and cells treated with 50 μm MIBA alone (lane 2) were used as experimental controls. Cell lysates (upper panels) and culture media (lower panel) were collected and analyzed by immunoblotting for total matriptase, activated matriptase, and HAI-1.
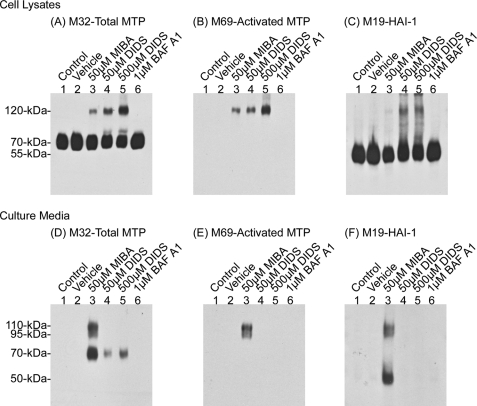
Inhibition of the NBCs, NDCBEs, and AEs by DIDS, a broad spectrum inhibitor of all three types of transporters, also lowered pHi and activated matriptase in a dose-dependent manner (Table 1 and Fig. 10, upper panels, lanes 4 and 5) (37, 43, 44). Interestingly, whereas higher levels of matriptase·HAI-1 complex were detected in the lysates of DIDS-treated cells versus MIBA-treated cells (Fig. 10, upper panels, comparing lanes 4 and 5 with lane 3), much higher levels of matriptase· HAI-1 complexes were shed in the culture media of MIBA-treated than DIDS-treated cells (Fig. 10, lower panels, comparing lane 3 with lanes 4 and 5). DIDS treatment apparently hindered the shedding of matriptase, resulting in the accumulation of matriptase·HAI-1 complex within the cells. In contrast to the inhibition of proton extrusion by EIPA, MIBA, and DIDS, prevention of proton transport from the cytosol to cellular organelles by bafilomycin A1, a potent inhibitor of the vacuolar H+-ATPase (V-ATPase), apparently does not reduce cytosolic pH sufficiently for inducing matriptase activation (Table 1 and Fig. 10, lane 6) (45). In summary, these data suggest that manipulation of pHi by inhibition of proton extrusion through the known acid-base ion transporters can activate the matriptase activation machinery.
FIGURE 10.
DIDS causes matriptase activation. 184 A1N4 cells were incubated in basal media (lane 1), or treated with 0.1% Me2SO (lane 2), 50 μm MIBA (lane 3), 50 μm DIDS (lane 4), 500 μm DIDS (lane 5), or 1 μm bafilomycin A1 (lane 6) for 2 h. Cell lysates and culture media were collected and analyzed for total matriptase, activated matriptase, and HAI-1, as indicated.
DISCUSSION
Acidosis is a common phenomenon in a variety of physiological and pathological processes. Under physiological conditions, urine pH in the distal tubules and bladder can fall below 5.5 due to the reabsorption of bicarbonate and secretion of hydrogen ions (46). In the skin, acidity plays essential roles in stratum corneum integrity and cohesion, epidermal permeability barrier and antimicrobial barrier function (47–49); in the epididymis, luminal acidification is essential for sperm maturation and storage (50). In pathological contexts, severe tissue ischemia causes extracellular acidosis (51), inflammation could induce interstitial acidification resulting in decreased extracellular pH values (52, 53), and tumor-associated microenvironments are more acidic than normal tissues due to hypoxia, increased glucose metabolism, and ineffective removal of the acidic metabolites. The acidic conditions in the tumor milieu have been thought to be a driving force for the selection of more aggressive cancer subclones (54, 55). Matriptase is almost ubiquitously expressed in all normal and malignant epithelial tissues. The identification of acidosis as a ubiquitous and potent factor that induces matriptase activation in epithelial and carcinoma cells suggests that matriptase activation may indeed be regulated by acidosis in these epithelial tissues and cancer. For example, matriptase is expressed by the kidney collecting ducts and epididymis, and activated matriptase in HAI-1 complexes is detected in human urine and semen, suggesting that matriptase activation machinery is induced in these tissues (2, 25). We have also demonstrated that breast cancer cells constitutively activate matriptase and significant amounts of matriptase·HAI-1 are secreted into conditioned media (30).
Acidosis-induced matriptase activation is characterized by: 1) dependence on the chemical environment but not cellular events; 2) rapid initiation, rapid kinetics, and the magnitude of the activation seen; 3) occurrence at both the cell surface and within the cell; and 4) the immediate HAI-1-mediated inhibition of the newly activated matriptase. The first three characteristics may result in the matriptase system being one of the few mechanisms by which cells can directly respond to acute extracellular and cytoplasmic acidosis. Other mechanisms in cells that respond directly include the proton-induced G-protein coupled receptors and acid-sensitive ion channels (24, 56–58). Although the functional consequences of matriptase activation and inhibition remain to be determined, the kinetics and magnitude of the matriptase activation seen demonstrate the readiness of the matriptase activation machinery to respond instantly to acidosis. The rapid kinetics and extent of matriptase activation induced by acidosis may be a function of the unique autoactivation mechanism involved. Matriptase is synthesized as a zymogen which, despite containing a pre-formed active site triad, exhibits only very weak proteolytic activity due to the lack of a well formed substrate binding pocket. To gain its full proteolytic activity, an irreversible cleavage at the canonical activation motif has to be made. The autoactivation of matriptase has been proposed to depend on interactions between two matriptase zymogen molecules and other proteins (15). These protein-protein interactions are thought to facilitate the activation cleavage carried out by the intrinsic, weak proteolytic activity of the matriptase zymogen. Anchorage of these proteins on the cell membrane is a prerequisite for matriptase activation, which is probably because such membrane attachment facilitates efficient protein-protein interactions (16). The involvement of biomembranes in matriptase activation underscores the importance of the physical environment for the process. Once anchored on the cell membrane, matriptase activation and HAI-1 inhibition can spontaneously and simultaneously take place in buffers with the appropriate pH and ionic strength. Thus, chemical environments, such as a mildly acidic pH, allow matriptase zymogens to interact with one another for the activation cleavage. The dependence on the appropriate physical and chemical environments, rather than on the presence of an upstream protease activator, may allow the activation of multiple matriptase zymogen molecules to occur simultaneously, with the result that the vast majority of cellular matriptase can be converted to the activated enzyme within 20 min. It is unlikely that this magnitude of activation could be achieved as rapidly through the action of another active proteases, the classic mechanism for protease activation. Further evidence for this non-conventional mechanism is provided by the atypical biochemical features of matriptase activation observed in our cell-free, in vitro activation system (16), in which matriptase activation is very sensitive to minor changes in pH, ionic strength, and temperature, and the presence of detergents. This susceptibility again seems to suggest a process that involves important protein-protein interactions facilitated by the microenvironment rather than one involving classical enzymatic action. One might expect that if matriptase activation involves the action of other active proteases the effects on the levels of matriptase activation by changes in pH, ionic strength, and temperature and by the presence of detergents would be in line with the effects on the activity of the presumed protease activators. Drastic changes in the proteolytic activity of the presumed matriptase activator would not be expected to be produced by minor changes in the chemical environment. It is, therefore, the autoactivation mechanism that allows cellular matriptase pools to be rapidly and almost completely activated in response to the changes in extracellular environments or the presence of activation inducers, such as sphingosine 1-phosphate, androgens, or suramin.
Despite the rapid and robust activation of matriptase in response to acidosis and other inducers, the immediate consequence of the induced matriptase activation remains elusive. The challenge of defining the key matriptase substrates has been hampered by the surprising subsequent events associated with matriptase activation and locations at which matriptase activation takes place. Matriptase activation is tightly coupled to its inhibition by HAI-1 (19). The prompt inhibition by HAI-1 of active matriptase right after its activation implies that the active matriptase has only a very short period of time to act on its substrates before being inhibited by HAI-1. Any relevant in vivo matriptase substrates must, therefore, be present in very close proximity to the matriptase zymogen. This proximity would allow the active matriptase to have direct access to and be able to activate these substrates prior to inhibition by HAI-1 binding. Identification of the subcellular location at which matriptase activation takes place is, therefore, of paramount importance to the identification of relevant matriptase substrates and the consequence of matriptase activation. In polarized epithelial cells, matriptase is targeted to the basolateral plasma membrane where matriptase activation and inhibition take place (25). Important matriptase substrates are, therefore, likely also present at the basolateral plasma membrane. The loss of epithelial polarity in carcinoma cells may allow matriptase to have access to other substrates that may not otherwise be processed by matriptase in normal epithelial cells. In addition to the cell surface, matriptase activation also occurs in the secretory granules of gastric chief cells, suggesting that matriptase may also act on substrates present in the secretory granules en route to the plasma membrane (25).
In summary, we have established that matriptase activation is an early response to acidosis. Sudden drops in extracellular pH, blockage of proton extrusion, or disruption of anion influx rapidly causes intracellular acidosis. Extracellular and intracellular acidification appears to act directly on the matriptase activation machinery anchored on the cell membrane and facilitates protein-protein interactions, leading to activational cleavage. This newly generated active matriptase, however, only has a very short time to activate its substrates, which are probably located within or directly proximal to the activation machinery. HAI-1 rapidly inhibits the active matriptase by forming a very stable complex that is subsequently secreted into extracellular milieu. These studies not only reveal a novel mechanism governing proteolysis in epithelial and carcinoma cells, but also demonstrate that a likely function of matriptase is to act as an early response to cellular acidosis.
Acknowledgment
We thank Dr. Amy Fulton for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA096851 and R01CA104944 (to C. Y. L.) and the Maryland Cigarette Restitution Fund Program.
- HAI-1
- hepatocyte growth factor activator inhibitor 1
- pHi
- intracellular pH
- NHEs
- Na+/H+ exchangers
- NBCs
- Na+/HCO3− co-transporters
- NDCBEs
- Na+-dependent Cl−/HCO3− exchangers
- AEs
- Na+-independent Cl−/HCO3− or anion exchangers
- MIBA
- 5-(N-methyl-N-isobutyl)-amiloride
- EIPA
- 5-(N-ethyl-N-isopropyl)-amiloride
- DIDS
- 4,4′-diisothiocyanostilbene-2,2′-disulfonic acid
- DPBS
- Dulbecco's phosphate-buffered saline
- mAb
- monoclonal antibody
- FBS
- fetal bovine serum
- rh
- recombinant human.
REFERENCES
- 1.Oberst M., Anders J., Xie B., Singh B., Ossandon M., Johnson M., Dickson R. B., Lin C. Y. (2001) Am. J. Pathol. 158, 1301–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oberst M. D., Singh B., Ozdemirli M., Dickson R. B., Johnson M. D., Lin C. Y. (2003) J. Histochem. Cytochem. 51, 1017–1025 [DOI] [PubMed] [Google Scholar]
- 3.List K., Hobson J. P., Molinolo A., Bugge T. H. (2007) J. Cell. Physiol. 213, 237–245 [DOI] [PubMed] [Google Scholar]
- 4.List K., Haudenschild C. C., Szabo R., Chen W., Wahl S. M., Swaim W., Engelholm L. H., Behrendt N., Bugge T. H. (2002) Oncogene 21, 3765–3779 [DOI] [PubMed] [Google Scholar]
- 5.List K., Szabo R., Wertz P. W., Segre J., Haudenschild C. C., Kim S. Y., Bugge T. H. (2003) J. Cell Biol. 163, 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.List K., Kosa P., Szabo R., Bey A. L., Wang C. B., Molinolo A., Bugge T. H. (2009) Am. J. Pathol. 175, 1453–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basel-Vanagaite L., Attia R., Ishida-Yamamoto A., Rainshtein L., Ben Amitai D., Lurie R., Pasmanik-Chor M., Indelman M., Zvulunov A., Saban S., Magal N., Sprecher E., Shohat M. (2007) Am. J. Hum. Genet. 80, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avrahami L., Maas S., Pasmanik-Chor M., Rainshtein L., Magal N., Smitt J., van Marle J., Shohat M., Basel-Vanagaite L. (2008) Clin. Genet. 74, 47–53 [DOI] [PubMed] [Google Scholar]
- 9.List K., Szabo R., Molinolo A., Sriuranpong V., Redeye V., Murdock T., Burke B., Nielsen B. S., Gutkind J. S., Bugge T. H. (2005) Genes Dev. 19, 1934–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ihara S., Miyoshi E., Ko J. H., Murata K., Nakahara S., Honke K., Dickson R. B., Lin C. Y., Taniguchi N. (2002) J. Biol. Chem. 277, 16960–16967 [DOI] [PubMed] [Google Scholar]
- 11.Suzuki M., Kobayashi H., Kanayama N., Saga Y., Suzuki M., Lin C. Y., Dickson R. B., Terao T. (2004) J. Biol. Chem. 279, 14899–14908 [DOI] [PubMed] [Google Scholar]
- 12.Lin C. Y., Anders J., Johnson M., Sang Q. A., Dickson R. B. (1999) J. Biol. Chem. 274, 18231–18236 [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi T., Shuman M. A., Craik C. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11054–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M. G., Chen C., Lyu M. S., Cho E. G., Park D., Kozak C., Schwartz R. H. (1999) Immunogenetics 49, 420–428 [DOI] [PubMed] [Google Scholar]
- 15.Oberst M. D., Williams C. A., Dickson R. B., Johnson M. D., Lin C. Y. (2003) J. Biol. Chem. 278, 26773–26779 [DOI] [PubMed] [Google Scholar]
- 16.Lee M. S., Tseng I. C., Wang Y., Kiyomiya K., Johnson M. D., Dickson R. B., Lin C. Y. (2007) Am. J. Physiol. Cell Physiol. 293, C95–C105 [DOI] [PubMed] [Google Scholar]
- 17.Oberst M. D., Chen L. Y., Kiyomiya K., Williams C. A., Lee M. S., Johnson M. D., Dickson R. B., Lin C. Y. (2005) Am. J. Physiol. Cell Physiol. 289, C462–C470 [DOI] [PubMed] [Google Scholar]
- 18.Hung R. J., Hsu IaW., Dreiling J. L., Lee M. J., Williams C. A., Oberst M. D., Dickson R. B., Lin C. Y. (2004) Am. J. Physiol. Cell Physiol. 286, C1159–C1169 [DOI] [PubMed] [Google Scholar]
- 19.Lee M. S., Kiyomiya K., Benaud C., Dickson R. B., Lin C. Y. (2005) Am. J. Physiol. Cell Physiol. 288, C932–C941 [DOI] [PubMed] [Google Scholar]
- 20.Szabo R., Kosa P., List K., Bugge T. H. (2009) Am. J. Pathol. 174, 2015–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szabo R., Molinolo A., List K., Bugge T. H. (2007) Oncogene 26, 1546–1556 [DOI] [PubMed] [Google Scholar]
- 22.Carney T. J., von der Hardt S., Sonntag C., Amsterdam A., Topczewski J., Hopkins N., Hammerschmidt M. (2007) Development 134, 3461–3471 [DOI] [PubMed] [Google Scholar]
- 23.Mathias J. R., Dodd M. E., Walters K. B., Rhodes J., Kanki J. P., Look A. T., Huttenlocher A. (2007) J. Cell Sci. 120, 3372–3383 [DOI] [PubMed] [Google Scholar]
- 24.Drummond H. A., Jernigan N. L., Grifoni S. C. (2008) Hypertension 51, 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J. K., Lee M. S., Tseng I. C., Chou F. P., Chen Y. W., Fulton A., Lee H. S., Chen C. J., Johnson M. D., Lin C. Y. (2009) Am. J. Physiol. Cell Physiol. 297, C459–C470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi T., Harris J. L., Huang W., Yan K. W., Coughlin S. R., Craik C. S. (2000) J. Biol. Chem. 275, 26333–26342 [DOI] [PubMed] [Google Scholar]
- 27.Bertog M., Letz B., Kong W., Steinhoff M., Higgins M. A., Bielfeld-Ackermann A., Frömter E., Bunnett N. W., Korbmacher C. (1999) J. Physiol. 521, 3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benaud C., Oberst M., Hobson J. P., Spiegel S., Dickson R. B., Lin C. Y. (2002) J. Biol. Chem. 277, 10539–10546 [DOI] [PubMed] [Google Scholar]
- 29.Kiyomiya K., Lee M. S., Tseng I. C., Zuo H., Barndt R. J., Johnson M. D., Dickson R. B., Lin C. Y. (2006) Am. J. Physiol. Cell Physiol. 291, C40–C49 [DOI] [PubMed] [Google Scholar]
- 30.Benaud C. M., Oberst M., Dickson R. B., Lin C. Y. (2002) Clin. Exp. Metastasis 19, 639–649 [DOI] [PubMed] [Google Scholar]
- 31.Benaud C., Dickson R. B., Lin C. Y. (2001) Eur. J. Biochem. 268, 1439–1447 [DOI] [PubMed] [Google Scholar]
- 32.Lin C. Y., Anders J., Johnson M., Dickson R. B. (1999) J. Biol. Chem. 274, 18237–18242 [DOI] [PubMed] [Google Scholar]
- 33.Marcotte N., Brouwer A. M. (2005) J. Phys. Chem. B 109, 11819–11828 [DOI] [PubMed] [Google Scholar]
- 34.Hunter R. C., Beveridge T. J. (2005) Appl. Environ. Microbiol. 71, 2501–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin C. Y., Wang J. K., Torri J., Dou L., Sang Q. A., Dickson R. B. (1997) J. Biol. Chem. 272, 9147–9152 [PubMed] [Google Scholar]
- 36.Bernardo A. A., Kear F. T., Santos A. V., Ma J., Steplock D., Robey R. B., Weinman E. J. (1999) J. Clin. Invest. 104, 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor C. J., Nicola P. A., Wang S., Barrand M. A., Hladky S. B. (2006) J. Physiol. 576, 769–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masereel B., Pochet L., Laeckmann D. (2003) Eur. J. Med. Chem. 38, 547–554 [DOI] [PubMed] [Google Scholar]
- 39.Soleimani M., Burnham C. E. (2001) J. Membr. Biol. 183, 71–84 [DOI] [PubMed] [Google Scholar]
- 40.Wang C. Z., Yano H., Nagashima K., Seino S. (2000) J. Biol. Chem. 275, 35486–35490 [DOI] [PubMed] [Google Scholar]
- 41.Taggart M., Austin C., Wray S. (1994) J. Physiol. 475, 285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cox T. (1997) J. Exp. Biol. 200, 3055–3065 [DOI] [PubMed] [Google Scholar]
- 43.Romero M. F., Hediger M. A., Boulpaep E. L., Boron W. F. (1997) Nature 387, 409–413 [DOI] [PubMed] [Google Scholar]
- 44.Muallem S., Loessberg P. A. (1990) J. Biol. Chem. 265, 12813–12819 [PubMed] [Google Scholar]
- 45.Furuchi T., Aikawa K., Arai H., Inoue K. (1993) J. Biol. Chem. 268, 27345–27348 [PubMed] [Google Scholar]
- 46.Chernew I., Braude A. I. (1962) J. Clin. Invest. 41, 1945–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hachem J. P., Crumrine D., Fluhr J., Brown B. E., Feingold K. R., Elias P. M. (2003) J. Invest. Dermatol. 121, 345–353 [DOI] [PubMed] [Google Scholar]
- 48.Mauro T., Holleran W. M., Grayson S., Gao W. N., Man M. Q., Kriehuber E., Behne M., Feingold K. R., Elias P. M. (1998) Arch. Dermatol. Res. 290, 215–222 [DOI] [PubMed] [Google Scholar]
- 49.Korting H. C., Hübner K., Greiner K., Hamm G., Braun-Falco O. (1990) Acta Derm. Venereol. 70, 429–431 [PubMed] [Google Scholar]
- 50.Shum W. W., Da Silva N., Brown D., Breton S. (2009) J. Exp. Biol. 212, 1753–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichihara K., Haga N., Abiko Y. (1984) Am. J. Physiol. Heart Circ. Physiol. 246, H652–H657 [DOI] [PubMed] [Google Scholar]
- 52.Simmen H. P., Battaglia H., Giovanoli P., Blaser J. (1994) Infection 22, 386–389 [DOI] [PubMed] [Google Scholar]
- 53.Press A. G., Hauptmann I. A., Hauptmann L., Fuchs B., Fuchs M., Ewe K., Ramadori G. (1998) Aliment. Pharmacol. Ther. 12, 673–678 [DOI] [PubMed] [Google Scholar]
- 54.Gillies R. J., Raghunand N., Garcia-Martin M. L., Gatenby R. A. (2004) IEEE Eng. Med. Biol. Mag. 23, 57–64 [DOI] [PubMed] [Google Scholar]
- 55.Gatenby R. A., Gawlinski E. T., Gmitro A. F., Kaylor B., Gillies R. J. (2006) Cancer Res. 66, 5216–5223 [DOI] [PubMed] [Google Scholar]
- 56.Ludwig M. G., Vanek M., Guerini D., Gasser J. A., Jones C. E., Junker U., Hofstetter H., Wolf R. M., Seuwen K. (2003) Nature 425, 93–98 [DOI] [PubMed] [Google Scholar]
- 57.Murakami N., Yokomizo T., Okuno T., Shimizu T. (2004) J. Biol. Chem. 279, 42484–42491 [DOI] [PubMed] [Google Scholar]
- 58.Stockand J. D., Staruschenko A., Pochynyuk O., Booth R. E., Silverthorn D. U. (2008) IUBMB Life 60, 620–628 [DOI] [PubMed] [Google Scholar]