Abstract
Resolvins are endogenous lipid mediators that actively regulate the resolution of acute inflammation. Resolvin E1 (RvE1; (5S,12R,18R)-trihydroxy-6Z,8E,10E,14Z,16E-eicosapentaenoic acid) is an endogenous anti-inflammatory and pro-resolving mediator derived from eicosapentaenoic acid that regulates leukocyte migration and enhances macrophage phagocytosis of apoptotic neutrophils to resolve inflammation. In the inflammatory milieu, RvE1 mediates counter-regulatory actions initiated via specific G protein-coupled receptors. Here, we have identified RvE1-specific signaling pathways initiated by the RvE1 receptor ChemR23. RvE1 stimulated phosphorylation of Akt that was both ligand- and receptor-dependent. RvE1 regulated Akt phosphorylation in a time (0–15 min)- and dose-dependent (0.01–100 nm) manner in human ChemR23-transfected Chinese hamster ovary cells. RvE1 stimulated phosphorylation of both Akt and a 30-kDa protein, a downstream target of Akt, identified using a phospho-Akt substrate antibody. The 30-kDa protein was identified as ribosomal protein S6, a translational regulator, and its phosphorylation was inhibited by a phosphatidylinositol 3-kinase (PI3K) inhibitor (wortmannin) and an ERK inhibitor (PD98059) but not by a p38-MAPK inhibitor (SB203580). Ribosomal protein S6 is a downstream target of the PI3K/Akt signaling pathway as well as the Raf/ERK pathway. In ChemR23-expressing differentiated HL60 cells, RvE1 also stimulated the phosphorylation of ribosomal protein S6. In addition, RvE1 enhanced phagocytosis of zymosan A by human macrophages, which are inhibited by PD98059 and rapamycin (mTOR inhibitor). These results indicate that RvE1 initiates direct activation of ChemR23 and signals receptor-dependent phosphorylation. These phosphorylation-signaling pathways identified for RvE1 receptor-ligand interactions underscore the importance of endogenous pro-resolving agonists in resolving acute inflammation.
Keywords: Diseases, Eicosanoids/Lipoxygenase Pathway, Immunology/Cellular Response, Lipid/Docosahexaenoic Acid/DHA, Lipid/Eicosapentaenoic Acid/EPA, Lipid/Fatty Acid, Receptors/Leukocyte/Lymphocyte
Introduction
The acute inflammatory response is a protective host reaction to foreign invaders or tissue injury. During acute inflammation, a cascade of events is well orchestrated at the molecular signaling level leading to the resolution phase, including cell migration, activation, proliferation, differentiation, and clearance (1–6). It is now clear that the resolution of acute inflammation is a biochemically active process that enables self-limited inflammatory challenge to return to homeostasis (1, 2, 5). A systems approach to the study of resolving exudates identified the first specialized chemical mediators, locally acting lipid mediators that play a pivotal role as endogenous agonists stimulating the resolution of acute inflammation (5, 7). Resolvin E1 (RvE1)3 is a potent local mediator biosynthesized from the omega-3 fatty acid, eicosapentaenoic acid (EPA), during resolution (5, 7). Systemic aspirin treatment enhances local exudate conversion of EPA to the potent, bioactive RvE1 (5). The complete stereochemical structure of RvE1 ((5S,12R,18R)-trihydroxy-6Z,8E,10E,14Z,16E-eicosapentaenoic acid) was determined using UV spectroscopy, liquid chromatography-tandem mass spectrometry (LC-MS/MS), and gas chromatography-mass spectrometry (5, 7), and its bioactivity and stereoselective actions were confirmed by total organic synthesis (8).
RvE1 stimulates the resolution of acute inflammation by reducing transmigration of polymorphonuclear leukocytes (PMN) (5, 7, 8), reducing NADPH oxidase activity in human PMN (9), limiting interleukin-12 production (8), and stimulating nonphlogistic macrophage phagocytosis (10). RvE1 is protective in rabbit periodontal disease (9) and murine colitis (11). In this setting, RvE1 significantly reduces tumor necrosis factor-α, interleukin-12 p40, the inducible nitric-oxide synthase, and COX-2 expression (11) (for recent review see Ref. 12).
RvE1 stimulates endogenous resolution mechanisms both in vivo in complex disease models and in vitro (12–14); in nanogram quantities RvE1 promotes resolution of acute inflammation by regulating leukocyte infiltration, increasing macrophage ingestion of apoptotic neutrophils, and enhancing clearance of phagocytes to lymph nodes and spleen (13). To validate RvE1 receptor candidates identified using a counter-regulatory screening system consisting of tumor necrosis factor-α signaling in an NFκB reporter system, [3H]RvE1 was prepared by total organic synthesis and found to specifically bind the identified recombinant human ChemR23 with high affinity (Kd = 11.3 ± 5.4 nm) (8). Also, recombinant human BLT1 (leukotriene B4 receptor 1) binds RvE1 with lower affinity (Kd = 48.3 nm) (15). These results indicate that RvE1 has at least two different specific receptors (ChemR23 and BLT1) differentially expressed by cells of the innate immune system that amplify its signal. Recently, Cash et al. (16) demonstrated that mice deficient in the ChemR23 receptor display an enhanced anti-inflammatory phenotype and identified synthetic peptides that are anti-inflammatory and act via ChemR23 signaling. Also, Campbell et al. (17) demonstrated that RvE1 and ChemR23 expressed on mucosal surfaces stimulate clearance of neutrophils during the resolution of mucosal inflammation. Thus, RvE1 is a potent endogenous agonist of anti-inflammation and pro-resolution pathways that are cell type-specific and stereoselective in their actions. Here, we report that RvE1 regulates protein signal transduction via activation of ChemR23 in macrophages and cell lines expressing the recombinant human receptor.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
For the detection of specific proteins, the following primary antibodies were used. Anti-ribosomal protein S6 (rS6) and anti-phospho-rS6 (Ser235/Ser236), anti-Akt and anti-phospho-Akt (Ser473), anti-phospho-ERKs (Thr202/Thr204), anti-phospho-p38-MAPK (Thr180/Thr182), and an antibody to the (R/K)X(R/K)XX(S*/T*) consensus-phosphorylated motif (phospho-Akt(S) Ab) were obtained from Cell Signaling Technology (Danvers, MA). Asterisks following the last S and T indicate the phosphorylated serine or threonine residues recognized by the phospho-Akt(S) antibody. A mouse anti-human ChemR23 monoclonal antibody (clone 84939) was purchased from R&D Systems (Minneapolis, MN) and labeled appropriately; class-matched mouse IgG3κ (BD Biosciences) was used as a negative control for ChemR23 staining. Anti-CD14 monoclonal Ab and isotype control IgG were purchased from eBioscience, Inc. (San Diego). Wortmannin (PI3K inhibitor), bisindolylmaleimide I (Bim I; protein kinase C inhibitor), SB203580 (p38-MAPK inhibitor), and PD98059 (ERK inhibitor) used as pharmacological inhibitors were purchased from Calbiochem. Rapamycin, an inhibitor of the mammalian target of rapamycin (mTOR), was purchased from Cell Signaling Technology. Synthetic RvE1 was prepared by total organic synthesis and handled as described previously (8). Chemerin COOH-terminal bioactive peptide (YHSFFFPGQFAFS), a ligand specific for the RvE1 receptor ChemR23, was synthesized as described elsewhere (8). LTB4 was obtained from Cayman Chemical (Ann Arbor, MI). CD14 Microbeads (Miltenyi Biotech Inc., Auburn, CA) and recombinant human granulocyte/monocyte colony-stimulating factor (R&D Systems) were used for human macrophage differentiation. Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich) was used for HL60 differentiation. Fluorescein isothiocyanate-serum-treated zymosan A (FITC-STZ) from Saccharomyces cerevisiae (Invitrogen) was used for the in vitro phagocytosis (10).
Human ChemR23-transfected Chinese Hamster Ovary (CHO) Cells
CHO cells were cultured in Ham's F-12 (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen). CHO cells stably expressing human ChemR23 (CHO-hChemR23) or mock-transfected CHO (CHO-mock) cells were prepared (15) and maintained in the presence of 500 μg/ml G418 (Cellgro, Herndon, VA). Subconfluent cells were serum-depleted for 4 h preparatory to stimulation with RvE1.
Macrophage-like Differentiated HL60 Cells
The human leukemia cell line HL60 was obtained from American Type Culture Collection Technology (ATCC, Manassas, VA) and maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS. HL60 cells were differentiated to macrophage-like cells by incubating them for 48 h with PMA (16 nm) in RPMI supplemented with 10% FBS (16). Adherent PMA-differentiated cells (HL60-PMA) were then serum-depleted for 4 h preparatory to stimulation with RvE1. Differentiated cells were confirmed by flow cytometric analysis with anti-CD14 Ab.
Monocyte Isolation
Fresh venous blood was collected in 10 units/ml sodium heparin from de-identified healthy volunteers who denied taking any drugs for at least 2 weeks before the experiments. The protocol was approved by the Brigham and Women's Hospital Institutional Review Board (protocol 88-02642) and the Boston University Medical Center Institutional Review Board (protocol H-23425). The isolation of peripheral blood mononuclear cells was performed as described by Böyum (18). Briefly, peripheral blood mononuclear cells were separated by dextran sedimentation followed by Ficoll-Histopaque gradient centrifugation (400 × g, 30 min, 25 °C). Peripheral blood mononuclear cells were then incubated with CD14 Microbeads (20 μl/107 cells) (15 min, 4 °C) in calcium/magnesium-free Dulbecco's phosphate-buffered saline (DPBS, pH 7.45) containing 2 mm EDTA and 0.5% bovine serum albumin according to the manufacturer's protocol. Cells were subsequently washed and loaded onto a magnetic column (Miltenyi Biotech Inc.). Magnetically labeled CD14+ monocytes were eluted from the column and resuspended in the appropriate medium for differentiation (see below). Viability was >98% as evaluated by trypan blue and propidium iodide exclusion. The purity of the isolated fraction was routinely >95% as confirmed by fluorescence-activated cell sorting (FACS) analysis.
Monocyte Differentiation
Purified monocytes (1.0 × 106 cells/ml) were differentiated into macrophages in RPMI 1640 containing 10% heat-inactivated FBS and recombinant human granulocyte/monocyte colony-stimulating factor (10 ng/ml) at 37 °C in a 5% CO2 atmosphere. Complete fresh medium was added at day 3, and the cells were used for the experiments 7 days after isolation. Cells were then serum-depleted for 4 h prior to incubations with RvE1.
Flow Cytometric Analysis of Cell Surface Expression
Cell surface expression was assessed by FACS analysis on a FACSort (BD Bioscience). Cells (1.0 × 106 cells) were fixed with 3.7% formaldehyde in DPBS for 15 min at room temperature in the dark and washed three times for 5 min in DPBS. Cells were then incubated with anti-human ChemR23 Ab (1:100 ratio in FACS buffer) followed by incubation with FITC-labeled secondary Ab or phycoerythrin-conjugated anti-CD14 Ab (1:100) for 1 h at 4 °C. Appropriately labeled cross-matched isotype controls IgG were used as negative controls for ChemR23 and CD14 staining.
Immunofluorescence Microscopy Analysis of Cell Surface Expression
Cell surface expression of ChemR23 was assessed by immunofluorescence as well as flow cytometry. Cells (0.5 × 106 cells/well) incubated in 4-well chamber slides (Thermo Fisher Scientific, Rochester, NY) were fixed as described above and blocked with 2% goat serum (Sigma-Aldrich) in DPBS for 15 min followed by incubation with anti-ChemR23 Ab or isotype-matched control IgG3κ at a 1:100 ratio in 2% FBS/DPBS overnight at 4 °C. Cells were subsequently washed four times with DPBS and incubated with Alexa Fluor® 488-conjugated goat anti-mouse IgG (Invitrogen) at a 1:250 dilution in 2% FBS/DPBS for 1 h at room temperature in the dark. Following incubation, cells were washed five times and mounted using a Prolong® Gold antifade kit with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Digital images of stained cells were captured with an inverted microscope (Zeiss Axiovert 200, Carl Zeiss, Heidelberg, Germany) and a video camera (Sony DFW-X700, Sony Corp., Tokyo, Japan). The images were imported into image analysis software (MicroSuite, Olympus America Inc., Melville, NY) for analysis.
RvE1-specific Binding
[3H]RvE1 methyl ester was obtained by catalytic hydrogenation of synthetic 6,14-diacetylenic RvE1, which was supplied to American Radiolabeled Chemicals for custom labeling. The resulting mixture was obtained from American Radiolabeled Chemicals, and [3H]RvE1 was saponified to the corresponding carboxylic acid and isolated by reverse-phase high pressure liquid chromatography (8). The binding studies were carried out with the isolated tritiated RvE1 ([6,7,14,15-3H]RvE1; 100 Ci/mmol) as described previously (8). Briefly, aliquots of CHO-hChemR23 cells or CHO-mock cells (1.0 × 106 cells/0.5 ml) were incubated in binding mixtures at 4 °C for the indicated time periods. The binding mixtures contained cells and the indicated concentrations of [3H]RvE1 or unlabeled RvE1 as a competitor in DPBS with CaCl2 and MgCl2 (DPBS+/+). To determine ligand specificity by competition binding, cells were incubated with 10 nm [3H]RvE1 in the presence of the indicated concentrations of RvE1 (10−10 to 10−6 m) as a competitor for 1 h at 4 °C. To determine displacement of equilibrium-bound [3H]RvE1, cells were incubated at 4 °C in the presence of [3H]RvE1 (5 nm). After 10 min, unlabeled RvE1 was added to the incubations. For saturation binding, cells were incubated with increasing concentrations of [3H]RvE1 (0.5–10.0 nm) in the presence or absence of unlabeled RvE1 (2 μm) for 1 h at 4 °C. Bound and unbound radioligand was separated by filtration through Whatman GF/C glass microfiber filters (Fisher Scientific), and radioactivity was determined.
Cell Stimulation and Western Blotting
The signaling transduction induced by RvE1 was assessed by Western blotting as described previously (19). Concentrations of antagonists and stimulation times are presented in the legends to Figs. 2–6. Briefly, at the appropriate time intervals, cells (5.0 × 106/ml) were lysed rapidly by adding 60 μl of 5× SDS sample buffer to 240 μl of the reaction mixture and then boiling the samples for 5 min. The final composition of SDS sample buffer was 2% (w/v) SDS, 58.3 mm Tris-HCl, pH 6.8, 6% (v/v) glycerol, 5% (v/v) β-mercaptoethanol, 0.002% (w/v) bromphenol blue, 1% (v/v) protease inhibitor mixture (Sigma), and 1 mm phenylmethylsulfonyl fluoride. Aliquots of these samples were separated by SDS-PAGE (∼17 μg/lane) on 10 or 12% (v/v) polyacrylamide slab gels. The separated proteins were transferred immediately by electrophoresis to polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membranes were incubated with the appropriate primary antibodies overnight at 4 °C followed by incubation with the secondary antibodies (goat anti-mouse or anti-rabbit IgG-horseradish peroxidase conjugate, 1:3000 dilution; Cell Signaling) for 1 h at room temperature. The activity of horseradish peroxidase was visualized with a luminol-ECL detection system (Pierce) followed by autoradiography. To confirm that equal amounts of protein were present in each lane, the membranes were reblotted with anti-actin Ab (1:2000 dilution; Sigma-Aldrich). The band density was measured using an image densitometer (Bio-Rad) and normalized to the actin band.
FIGURE 2.
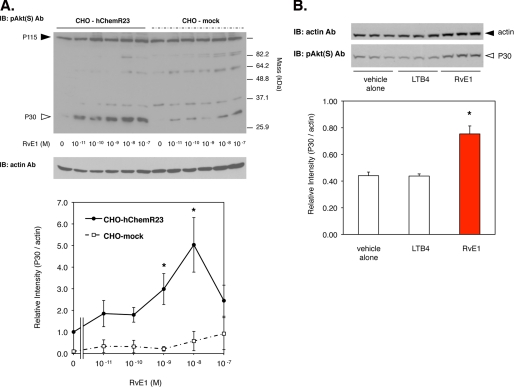
RvE1 regulates phosphorylation of a 30-kDa protein (p30) via human ChemR23. A, RvE1 regulates the phosphorylation of p30 in a concentration-dependent manner. Whole cell lysates were extracted from CHO-hChemR23 cells incubated with RvE1 (0.01–100.0 nm, 15 min, 37 °C) and analyzed by Western blotting with pAkt(S) Ab to determine the phosphoproteins as described under “Experimental Procedures.” The open arrowhead indicates the position of p30. The line graph summarizes the relative intensity of p30 phosphorylation in CHO cells treated with RvE1. Results represent the mean ± S.E. for four separate experiments (*, p < 0.05, when compared with vehicle). IB, immunoblot. B, RvE1 regulates p30 phosphorylation but not LTB4. Whole cell lysates were extracted from CHO-hChemR23 cells incubated with RvE1 (10 nm) or LTB4 (10 nm) at 37 °C for 15 min and analyzed by Western blotting with pAkt(S) Ab. The open arrowhead indicates the position of p30. The bar graph summarizes the relative intensity of p30 phosphorylation. Results represent the mean ± S.E. for three separate experiments (*, p < 0.05 when compared with vehicle).
FIGURE 3.
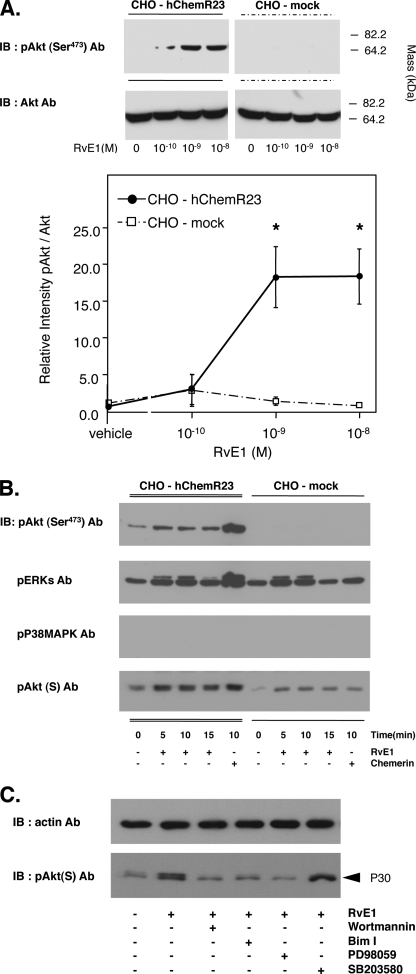
RvE1 regulates signal transduction via human ChemR23. A, RvE1 regulates Akt phosphorylation via ChemR23 in a concentration-dependent manner. Whole cell lysates were extracted from CHO-hChemR23 cells and CHO-mock cells incubated with the indicated concentrations of RvE1 (0.1–10.0 nm, 37 °C, 15 min) and analyzed by Western blotting with anti-pAkt (Ser473) Ab to determine the phosphorylation of Akt. The line graph summarizes the relative intensity of Akt phosphorylation. Results represent the mean ± S.E. for three separate data experiments (*, p < 0.05, when compared with vehicle in CHO-hChemR23 cells). IB, immunoblot. B, RvE1 regulates signal transduction in a time-dependent manner. Whole cell lysates were extracted from CHO-hChemR23 cells incubated with RvE1 (10 nm) or chemerin (100 nm) for the indicated time periods. Protein phosphorylation was assessed by Western blotting using phospho-Akt, ERK, p38-MAPK, and pAkt(S) antibodies as described under “Experimental Procedures.” C, reduction of RvE1-enhanced p30 phosphorylation by pharmacological inhibitors. Whole cell lysates were extracted from CHO-hChemR23 cells incubated with wortmannin (200 nm), PD98059 (50 μm), or SB203580 (10 μm) and Bim I (1 μm) for 15 min. Cells were exposed to RvE1 (10 nm, 37 °C, 15 min) and analyzed by Western blotting with pAkt(S) Ab.
FIGURE 4.
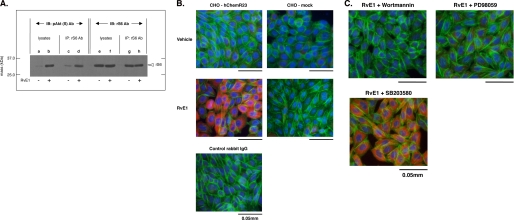
RvE1 regulates the phosphorylation of ribosomal protein S6. A, whole cell lysates were derived from CHO-hChemR23 cells treated with or without RvE1 (10 nm, 15 min, 37 °C) and pulled down with a mouse antibody to ribosomal protein S6 Ab. Immunoprecipitates (∼70 μg/lane) were blotted with the phospho-Akt substrate antibody (pAkt(S) Ab; lanes c and d). rS6 was detected with rS6 Ab (lanes g and h) to confirm protein loading. IB, immunoblot; IP, immunoprecipitation. B, immunofluorescence microscopy of rS6 phosphorylation. CHO-hChemR23 cells and CHO-mock cells were incubated with or without RvE1 (10 nm, 37 °C, 15 min). Cells were stained with anti-phospho-rS6 (Ser235/Ser236) Ab followed by Alexa Fluor® 568 secondary Ab (red) to determine the distribution of phosphorylated rS6 within the cells. Actin filaments were labeled with Alexa Fluor® 488-phalloidin (green), and nuclei were labeled with DAPI (blue) as described under “Experimental Procedures.” C, reduction of RvE1-enhanced p30 phosphorylation by pharmacological inhibitors. CHO-hChemR23 cells were incubated with wortmannin (200 nm), PD98059 (50 μm), or SB203580 (10 μm) for 15 min. Cells were then exposed to RvE1 (10 nm, 37 °C, 15 min) and stained with anti-phospho-rS6 (Ser235/Ser236) Ab using Alexa Fluor® 568 secondary Ab (red) to determine the phosphorylation of rS6. Actin filaments were labeled with Alexa Fluor® 488-phalloidin (green), and nuclei were labeled with DAPI (blue).
FIGURE 5.
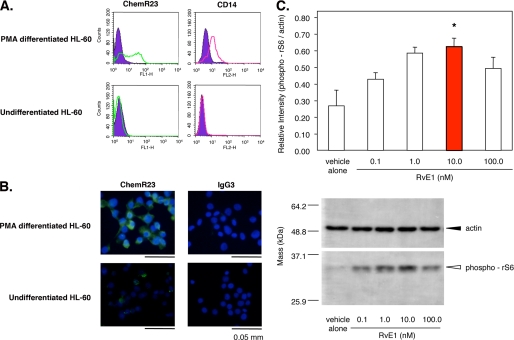
RvE1 regulates phosphorylation of ribosomal protein S6 in PMA-differentiated HL60 cells via ChemR23. A, flow cytometric analysis of ChemR23 and CD14 surface expression. PMA-differentiated HL60 cells were stained with anti-human ChemR23 Ab (left, open green histogram) or CD14 Ab (right, open red histogram) and FITC-labeled secondary antibody. The filled histogram indicates each isotype control IgG (purple). B, immunofluorescence microscopy analysis of ChemR23 surface expression. PMA-differentiated HL60 cells or undifferentiated HL60 cells were stained with anti-human ChemR23 antibody or IgG3 isotype control followed by labeling with Alexa Fluor® 488 secondary Ab (green). Nuclei were counterstained with DAPI (blue). C, RvE1 regulates phosphorylation of rS6 in PMA-differentiated HL60 cells. Whole cell lysates were extracted from PMA-differentiated HL60 cells incubated with the indicated concentration of RvE1 (0.1–100.0 nm, 37 °C, 15 min) and analyzed by Western blotting with phospho-rS6 (Ser235/Ser236) Ab. The bar graph summarizes the relative intensity of phospho-rS6. Results represent the mean values ± S.E. for three separate experiments (*, p < 0.05 when compared with vehicle).
FIGURE 6.
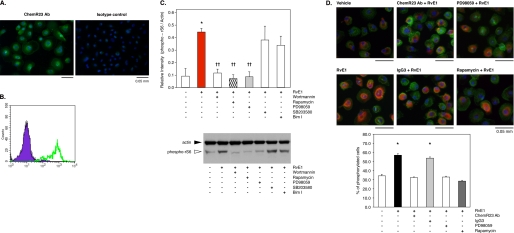
RvE1 regulates phosphorylation of ribosomal protein S6 in human macrophages via ChemR23. A, immunofluorescence microscopy analysis of ChemR23 surface expression. Human macrophages were stained with anti-human ChemR23 antibody or IgG3 isotype control followed by labeling with Alexa Fluor® 488 secondary Ab (green) as described under “Experimental Procedures.” Nuclei were counterstained with DAPI (blue). B, flow cytometric analysis of ChemR23 surface expression. Human macrophages were stained with anti-human ChemR23 antibody (green, open histogram) or isotype control mouse IgG3 (purple, filled histogram) and FITC-labeled secondary antibody. C, reduction of RvE1-enhanced rS6 phosphorylation by pharmacological inhibitors. Whole cell lysates were extracted from human macrophages incubated with wortmannin (200 nm), rapamycin (20 nm), PD98059 (50 μm), SB203580 (10 μm), or Bim I (1 μm) for 15 min followed by co-incubation with RvE1 (10 nm, 15 min, 37 °C) and then analyzed by Western blotting with phospho-rS6 (Ser 235/236) Ab. The bar graph summarizes the relative intensity of rS6 phosphorylation. Results represent the mean values ± S.E. for three separate experiments (*, p < 0.05 when compared with vehicle). D, immunofluorescence microscopy analysis of rS6 phosphorylation. Human macrophages were treated with human ChemR23 Ab (1:100 dilution), isotype control IgG3 (1:100), PD98059 (50 μm), or rapamycin (20 nm) followed by co-incubation with RvE1 (10 nm, 15 min, 37 °C). Cells were stained with anti-phospho-rS6 (Ser235/Ser236) Ab using Alexa Fluor® 568 secondary Ab (red) to determine the distribution of phosphorylation of rS6 within the cells. Actin filaments were labeled with Alexa Fluor® 488-phalloidin (green), and nuclei were labeled with DAPI (blue). The bar graph summarizes the percentage of rS6-phosphorylated cells. Results represent the mean values ± S.E. for three separate experiments (*, p < 0.05 when compared with vehicle).
Immunoprecipitation
Immunoprecipitations were performed to investigate the ability to detect rS6 phosphorylation by phospho-Akt(S) Ab, using a Seize® X-protein A immunoprecipitation kit (Pierce). Whole cell lysates were derived from CHO-hChemR23 cells (5.0 × 107 cells) in the presence or absence of RvE1 (10 nm, 15 min, 37 °C). Cells were lysed rapidly in the immunoprecipitation buffer and incubated on ice for 15 min followed by centrifugation (1000 × g, 10 min, 4 °C) to remove insoluble debris as described previously (18). Aliquots of the soluble proteome fraction (∼1 mg) were incubated with monoclonal rS6 immobilized antibody. Immunoprecipitates were eluted, denatured, loaded on 12% (v/v) polyacrylamide slab gels (≈70 μg/lane), and transferred onto polyvinylidene difluoride membrane.
The polyvinylidene difluoride membrane was blotted with the antibody to the (R/K)X(R/K)XX(S*/T*) consensus motif (phospho-Akt(S) Ab) to determine the phosphorylated rS6. Moreover, rS6 was detected from these samples using anti-rS6 Ab to confirm that the same amounts of protein were loaded.
Immunofluorescence Microscopy Analysis of rS6 Phosphorylation
Cells (0.5 × 106 cells/ml) were incubated in 4-well chamber slides with RvE1 (10 nm, 15 min, 37 °C). The wells were subsequently aspirated gently, and cells were washed with ice-cold DPBS. Cells were fixed with 3.7% formaldehyde/DPBS for 15 min at room temperature in the dark and washed with DPBS. Cells were subsequently permeabilized with 1% Triton X/DPBS for 10 min at room temperature. 5% goat serum/DPBS was used for blocking, and cells were stained with anti-phospho-rS6 (Ser235/Ser236) Ab or isotype-matched control IgG in a 1:100 dilution in 2% FBS/DPBS overnight at 4 °C. Following incubation, cells were washed and incubated with Alexa® Fluor 568-conjugated goat anti-rabbit IgG and Alexa® Fluor 488 conjugated with phalloidin. Nuclei were counterstained with DAPI. To evaluate ChemR23, PI3K, ERK, p38-MAPK, or mTOR involvement in RvE1-mediated phosphorylation by immunofluorescence microscopy, cells were treated with a monoclonal anti-human ChemR23 antibody, isotype-matched mouse IgG3κ control (1:100), PD98059 (50 μm), wortmannin (200 nm), SB203580 (10 μm), or rapamycin (20 nm) prior to incubation with RvE1.
Phagocytosis
The pro-resolving actions of RvE1 were assessed as described previously (10, 13). Briefly, for phagocytosis in vitro, human macrophages were plated onto 96-well plates (1.0 × 105 cells/well) and incubated with RvE1 (0.01–100.0 nm) for 15 min in DPBS+/+, pH 7.45. FITC-STZ (0.5 × 106 particles/well) was then added to macrophages for 30 min. The wells were subsequently aspirated, and extracellular fluorescence was quenched by briefly adding trypan blue. Cells were then washed, and intracellular fluorescence was measured (Excitation 485 nm/Absorption 535 nm) by a fluorescent plate reader (PerkinElmer Life Sciences). To evaluate ChemR23, ERK, or rapamycin involvement in RvE1-mediated phagocytosis, cells were treated with a monoclonal anti-ChemR23 Ab (1:100 dilution), isotype-matched control IgG3 (1:100), PD98059 (50 μm), or rapamycin (20 nm) prior to incubation with RvE1. To determine the distribution of rS6 phosphorylation and phagocytosis of FITC-STZ by immunofluorescence microscopy, cells were plated onto 4-well chamber slides (0.5 × 106 cells/wells) and treated with the same concentrations of antagonists as described above prior to incubation with RvE1 (10 nm) and FITC-STZ (2.5 × 105 particles/well). Cells were then stained with anti-phospho-rS6 (Ser235/Ser236) Ab using Alexa Fluor® 568 secondary Ab. Actin filaments were labeled with Alexa Fluor® 488-phalloidin, and nuclei were labeled with DAPI. Statistical analyses were carried out using Student's t test with p < 0.05 taken as significant.
RESULTS
Specific Binding of [3H]RvE1 and Surface Expression of Human ChemR23
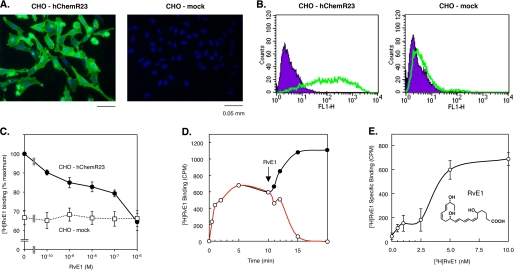
First, we established that the receptor was expressed on the surface of CHO cells transfected with hChemR23 (CHO-hChemR23) compared with wild type vectors (CHO-mock). Expression of human ChemR23 was only identified on CHO-hChemR23 cells but not on CHO-mock cells as assessed using immunofluorescence and flow cytometric analysis, respectively (Fig. 1, A and B).
FIGURE 1.
Surface expression of human ChemR23 in CHO cells and RvE1-specific binding. A, immunofluorescence microscopy analysis of ChemR23 cell surface expression. CHO-hChemR23 cells (left) or CHO-mock cells (right) were stained with anti-human ChemR23 Ab followed by secondary Ab labeled with Alexa Fluor® 488 (green), and nuclei were labeled with DAPI (blue) as described under “Experimental Procedures.” B, flow cytometric analysis of ChemR23 cell surface expression. CHO-hChemR23 cells (left) or CHO-mock cells (right) were stained with anti-human ChemR23 Ab (green, open histogram) or isotype control IgG3 (purple, filled histogram) and FITC-labeled secondary Ab as described under “Experimental Procedures.” C, competitive binding. CHO-hChemR23 cells (closed circles) or CHO-mock cells (open squares) were incubated at 4 °C with [3H]RvE1 (10 nm) in the presence of the indicated concentration of RvE1 (10−10 to 10−6 m) to determine the competition for specific [3H]RvE1 binding. Results represent the mean values ± S.E. for six separate experiments. D, displacement of equilibrium-bound [3H]RvE1 in CHO-hChemR23 cells. Cells (1.0 × 106 cells/0.5 ml) were incubated at 4 °C in the presence of [3H]RvE1 (5 nm). After 10 min (denoted by arrow), unlabeled RvE1 (2 μm; open circles) was added to incubations. In the absence of RvE1, the [3H]RvE1-specific binding did not decline for the time intervals indicated (closed circles). Results are representative of at least three separate experiments. E, [3H]RvE1-specific binding in CHO-hChemR23 cells. Cells were incubated in the indicated concentrations of [3H]RvE1 (0.5–10.0 nm) in the presence or absence of unlabeled RvE1 (2 μm) for 1 h at 4 °C to determine [3H]RvE1-specific binding. Results represent the mean values ± S.E. for five separate experiments.
We next examined competition binding of RvE1 with the radiolabeled homoligand RvE1 ([3H]RvE1) with CHO-hChemR23 and CHO-mock cells. ChemR23 is one of the high affinity G protein-coupled receptors that interacts with RvE1, demonstrating specific binding to ChemR23 using synthetic 3H-labeled RvE1 (8). Specific binding of [3H]RvE1 was displaced with increasing concentrations of the unlabeled homoligand and was completely reduced to levels of nonspecific binding following equilibrium by addition of a 3-log order excess of unlabeled RvE1. As shown in Fig. 1C, RvE1 displaced [3H]RvE1 binding to CHO-hChemR23, but not to CHO-mock cells, in a concentration-dependent manner. It is noteworthy that the amount of [3H]RvE1 binding to CHO-mock cells was ∼65% compared with the binding amount in CHO-hChemR23, and it was not displaced by unlabeled RvE1. These results gave the amount of nonspecific binding of [3H]RvE1 to CHO-mock cells. Displacement of [3H]RvE1 binding with excess RvE1 required ∼10 min to achieve 100% reversal of specific binding (Fig. 1D). In addition, [3H]RvE1 specific binding was saturated at 5 nm (Fig. 1E) after a 1-h incubation. The structure of RvE1 is provided in Fig. 1E. These results indicate that RvE1 binds specifically to CHO cells expressing the human ChemR23.
RvE1 Regulates Phosphorylation via ChemR23
CHO-hChemR23 cells stimulated with RvE1 exhibited intense phosphorylation of a 30-kDa protein (p30), monitored by Western blotting with pAkt(S) Ab, which recognizes the downstream target proteins of Akt containing a consensus-phosphorylated motif ((R/K)X(R/K)XX(S*/T*)) as described under “Experimental Procedure.” Treatment of CHO-hChemR23 cells with RvE1 (0.01–100.0 nm, 15 min, 37 °C) substantially increased phosphorylation of p30 (Fig. 2A, arrowhead; another example of the RvE1 time dependence is shown in Fig. 3B) in a concentration-dependent manner. In contrast, RvE1 did not appear to impact phosphorylation of a ∼115-kDa protein (Fig. 2A) in either CHO-hChemR23 or CHO-mock cells. The relative intensity of p30 phosphorylation in response to 10 nm RvE1 was higher in comparison with incubations in the absence of RvE1 (Fig. 2B). Phosphorylation of p30 was also obtained with CHO-hChemR23 cells incubated with 100 nm peptide derived from chemerin, a synthetic peptide fragment (YHSFFFPGQFAFS) ligand of ChemR23 (8, 20) (Fig. 3B), but not with 10 nm LTB4, a well known ligand for BLT1 (Fig. 2B). BLT1 is also a specific receptor for RvE1 that is present on neutrophils (15). These results indicate that ChemR23 stimulates phosphorylation of p30 in a RvE1-dependent manner.
Because pAkt(S) Ab recognizes the downstream target proteins of Akt containing a consensus-phosphorylated motif, p30 was next considered as a potential downstream target of Akt. RvE1 stimulated phosphorylation of Akt, ERK, and p30 (Fig. 3). RvE1-stimulated CHO-hChemR23 cells exhibited intense phosphorylation of Akt as detected by Western blotting with phospho-Akt (Ser473) Ab. At 1 nm, RvE1 significantly increased the intensity of Akt phosphorylation as did 10 nm RvE1. The relative intensity of Akt phosphorylation in RvE1-incubated cells was increased ∼20-fold in comparison with unstimulated cells (Fig. 3A). In contrast, RvE1 did not appear to stimulate consistent phosphorylation of Akt in CHO-mock cells. Thus, the actions of RvE1 were selective for the receptor ChemR23. The phosphorylation of Akt occurred by 5 min after exposure to RvE1 and remained level at 15 min. RvE1 also regulated phosphorylation of ERK and p30, but not p38-MAPK, in a time-dependent manner (Fig. 3B). Pharmacological inhibitors were used to determine which kinases activate the phosphorylation of p30 as determined by Western blotting (Fig. 3C). Reduction of RvE1-stimulated phosphorylation of p30 to essentially base line was obtained with wortmannin (200 nm; an inhibitor of PI3K) (21), Bim I (1 μm; an inhibitor of protein kinase C) (21), and PD98059 (50 μm; an inhibitor of ERK activation) (22). In contrast, SB203580 (10 μm); an inhibitor for activity of the α- and β-isoforms of p38-MAPK (22) did not affect the phosphorylation of p30.
RvE1 Regulates Phosphorylation of Ribosomal Protein S6
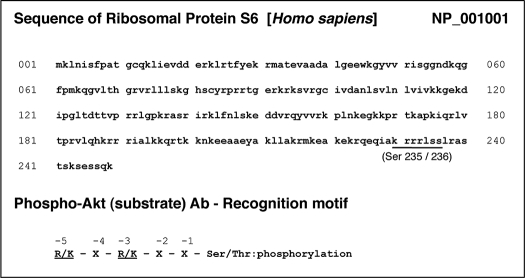
rS6, a part of the small ribosomal subunit involved in translation, is known as a phospho-protein that is stimulated with growth factors or hormones (21). It is also known that rS6 is a major substrate of ribosomal protein S6 kinase (p70S6K), a downstream of PI3K/Akt as well as Raf/ERK signal transductions. When rS6 is phosphorylated by p70S6K, the cells undergo growth (21). This is the same motif in which phospho-Akt(S) Ab recognizes rS6-phosphorylated sites (Ser235 and Ser236) (22). Here, we demonstrated p30 to be ribosomal protein S6 by immunoprecipitation (Fig. 4A) and by matching the protein sequence of rS6 with the motif from the pAkt(S) Ab (Table 1). Immunofluorescence microscopy was carried out to determine the distribution of rS6 phosphorylation within the cells. As shown in Fig. 4B, RvE1 phosphorylated rS6 (Ser235/236) in CHO-hChemR23 cells but not in CHO-mock cells.
TABLE 1.
Ribosomal protein S6 sequence and phospho-Akt substrate Ab recognition motif
Again, the pharmacologic inhibitors were used to determine which kinases activated the phosphorylation of rS6 by immunofluorescence microscopy (Fig. 4C). Reduction of RvE1-stimulating phosphorylation of rS6 to base line was obtained with wortmannin (200 nm) and PD98059 (50 μm). In contrast, SB203580 (10 μm) did not affect the phosphorylation of rS6. In addition, the specific observation of rS6 phosphorylation by phospho-rS6 (Ser235/Ser236) Ab was confirmed using the isotype-matched control, rabbit IgG, as a negative control.
Macrophage-like HL60 Cells Express ChemR23, and RvE1 Regulates rS6 Phosphorylation via ChemR23
The expression of human ChemR23 was detected on adherent PMA-induced macrophage-like HL60 cells (PMA-HL60) by flow cytometry (Fig. 5A) and immunofluorescence microscopy analyses (Fig. 5B). In contrast, undifferentiated cells did not express ChemR23 on the surface. The differentiated cells were confirmed by observation of CD14 surface expression. RvE1-enhanced phosphorylation of rS6 protein was obtained by Western blotting with anti-phospho-rS6 (Ser235/Ser236) Ab. Treatment of PMA-HL60 cells with RvE1 (0.01–100.0 nm, 15 min, 37 °C) substantially increased rS6 phosphorylation (Fig. 5C, open arrowhead) in a concentration-dependent manner; the maximal level was at 10 nm. These results indicate that RvE1 regulates phosphorylation of rS6, which is a target protein of ChemR23 activation in human macrophage-like cells.
RvE1-mediated rS6 Phosphorylation Enhances Phagocytosis in Human Macrophages via ChemR23
Next, the expression of human ChemR23 was confirmed on human macrophages by immunofluorescence microscopy (Fig. 6A) and flow cytometric analyses (Fig. 6B). RvE1-exposed human macrophages exhibited intense rS6 phosphorylation by Western blotting using anti-phospho-rS6 (Ser235/Ser236) antibody (Fig. 6C). RvE1-enhanced rS6 phosphorylation was reduced with certain pharmacological inhibitors. As shown in Fig. 6C, wortmannin (200 nm; an inhibitor of PI3K), rapamycin (20 nm; a mTOR inhibitor), and PD98059 (50 μm; an ERK inhibitor) each substantially reduced phosphorylation of rS6. In contrast, SB203580 (10 μm; a p38-MAPK inhibitor) and Bim I (1 μm; a protein kinase C inhibitor) did not dramatically reduce the phosphorylation of rS6. We also tested the impact of inhibitors on RvE1-enhanced ERK phosphorylation (supplemental Fig. 1). Both PD98059 and wortmannin, but not rapamycin, SB203580, or Bim I, reduced ERK phosphorylation by RvE1.
RvE1-evoked rS6 phosphorylation in human macrophages was also observed by immunofluorescence microscopy using anti-phospho-rS6 (Ser235/Ser236) Ab (Fig. 6D). Here, ∼60% of the phosphorylated cells were observed following exposure to RvE1 (10 nm, 15 min, 37 °C). An increase in both the phosphorylated cell number and the intensity of phospho-rS6 was observed. Thus, we quantified the increases by determining the number of phosphorylated cells. Presently, there is no obvious explanation for this cellular pattern observed with RvE1 and ChemR23. In addition, anti-ChemR23 Ab (1:100 dilution; as a competitor for receptor binding), PD98059 (50 μm), and rapamycin (20 nm) each significantly reduced phosphorylation to the base-line value of ∼30%. In contrast, isotype control IgG3κ, used as a negative control for ChemR23 Ab, did not affect the phosphorylation.
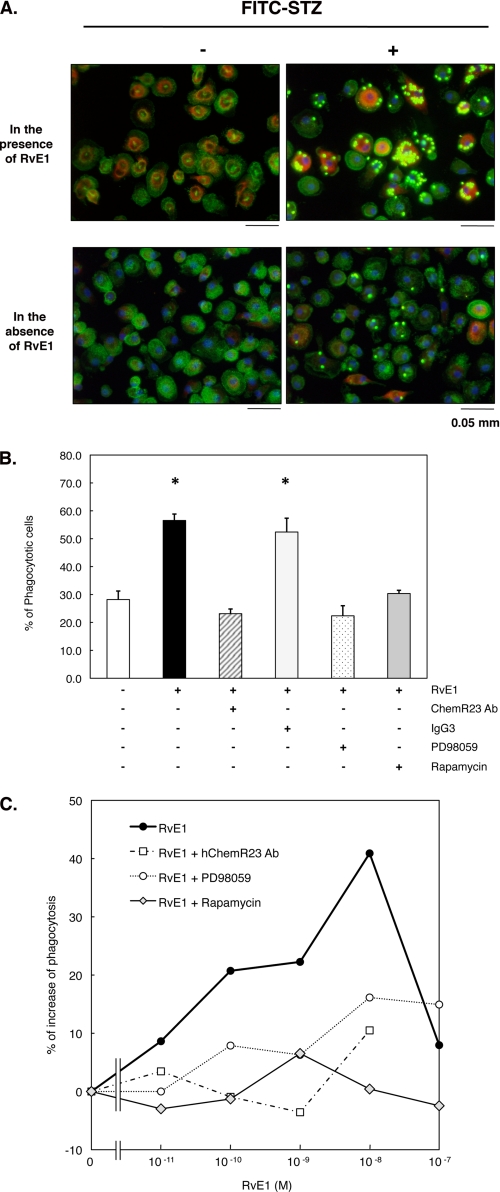
Because RvE1 was recently shown to enhance murine macrophage phagocytosis in vivo (13) and in vitro (10), we next tested RvE1 with human macrophages (Fig. 7). Phagocytosis was more than doubled by exposure to as little as 10 nm RvE1. Again, ChemR23 antibody (1:100 dilution) and the inhibitors PD98059 (50 μm) and rapamycin (20 nm) each significantly reduced phagocytizing cells to base line. This reduction was not observed with the IgG3κ antibody, which was used as a control for the ChemR23 antibody.
FIGURE 7.
RvE1 enhances phagocytosis of FITC-STZ in human macrophages. A, RvE1-induced FITC-STZ uptake and rS6 phosphorylation in human macrophages. Macrophages (4-well chamber slides, 0.5 × 106 cells/well) were treated with RvE1 (10 nm, 37 °C, 15 min) followed by co-incubation with or without FITC-STZ (green particles, 2.5 × 106 particles/well) for 30 min at 37 °C. Cells were stained with anti-phospho-rS6 (Ser235/Ser236) Ab using Alexa Fluor® 568 secondary Ab (red) to detect the phosphorylated form of rS6. Actin filaments were labeled with Alexa Fluor® 488-phalloidin (green), and nuclei were stained with DAPI (blue). B, reduction of agonist-enhanced phagocytosis of FITC-STZ by pharmacological inhibitors. Cells were treated with ChemR23 Ab (1:100 dilution), isotype control IgG3 (1:100), PD98059 (50 μm), or rapamycin (20 nm) for 15 min followed by RvE1 stimulus (10 nm, 37 °C, 15 min) and co-incubation with FITC-STZ (30 min, 37 °C). Phagocytosis was assessed by counting the percentage of cells ingesting FITC-STZ particles. Results represent the mean values ± S.E. for three separate experiments (*, p < 0.05 when compared with cells incubated with vehicle alone plus FITC-STZ). C, concentration-dependent increase in phagocytosis of FITC-STZ by human macrophages exposed to RvE1. Cells (96-well plate, 0.1 × 106 cells/well) were pretreated with ChemR23 Ab (1:100 dilution), PD98059 (50 μm), or rapamycin (20 nm) for 15 min. Cells were then treated with the indicated concentration (0.01–100.0 nm) at 37 °C for 15 min followed by co-incubation with FITC-STZ for 30 min at 37 °C. Phagocytosis was quantified using a fluorescent plate reader as described under “Experimental Procedures.” Results are representative of at least three separate experiments and are expressed as the percent of fluorescent intensity above cells incubated with vehicle alone plus FITC-STZ.
DISCUSSION
Resolution of acute inflammation is an active process (1, 12) controlled in part by the temporal and spatially regulated formation of a new genus of pro-resolving lipid mediators (1, 2, 4, 5). Our recent results uncovered several new families of lipid mediators including resolvins (resolution phase interaction products) biosynthesized by evolving inflammatory exudates that are both anti-inflammatory and pro-resolving (1). Resolvin E1 is a potent anti-inflammatory and pro-resolving member of the E-series resolvins produced from EPA (5, 7, 8). The biosynthesis of RvE1 and related mediators with novel counter-regulatory and pro-resolving actions in inflammation as well as RvE1 receptor activation provides evidence for potent endogenous agonists of anti-inflammation (5). Hence, the pursuit of intracellular signaling pathways initiated by RvE1 can increase our understanding of the function of this mediator during resolution. In this report, we have demonstrated that RvE1 regulates phosphorylation of Akt (Ser473) via RvE1-specific interactions with ChemR23 on both human ChemR23-transfected CHO cells and human macrophages.
Macrophages are antigen-presenting cells that play key roles in both innate and adaptive immunity. ChemR23 is a G protein-coupled receptor related in nucleotide sequence to chemokine and eicosanoid receptors and is expressed in monocytes, macrophages, and dendritic cells (3, 8). The peptide chemerin, described originally as a chemoattractant ligand for ChemR23, has a role in both adaptive and innate immunity (20, 23).
Recently, Cash et al. (16) demonstrated that murine chemerin possesses potent anti-inflammatory components that are dependent on the proteolytic processing and peptide activation of ChemR23. A synthetic chemerin-derived peptide significantly reduces zymosan-induced peritonitis, and administration of an anti-chemerin antibody increases both PMN and monocyte recruitment (16). A key and important step in the resolution of acute inflammation is the uptake and clearance of apoptotic PMN by macrophages (1, 12). It is noteworthy that RvE1 enhances murine macrophage phagocytosis as a key pro-resolving action of this mediator (10), stimulates mucosal PMN clearance (17), and reduces airway inflammation (24, 25). As reported here, RvE1 regulates phosphorylation of ribosomal protein S6 (Fig. 6) and enhances phagocytosis by human macrophages (Fig. 7) via ChemR23.
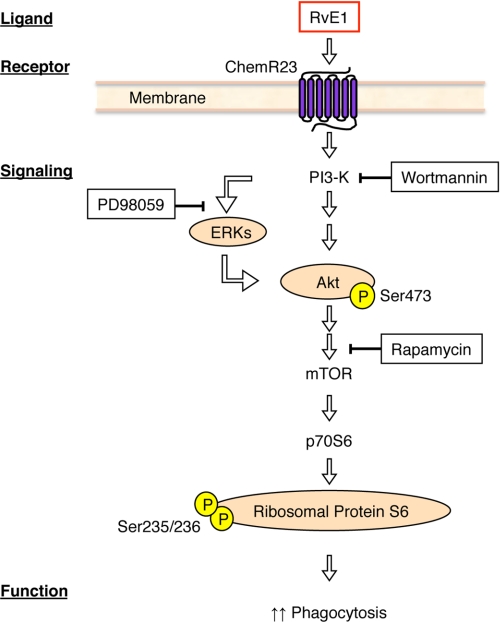
Ribosomal protein S6 is constitutively phosphorylated in a Bcr-Abl-dependent manner. mTOR acts as a nutrient and mitogen sensor to positively regulate translation and ribosome biogenesis (22). Upon activation by mitogens and amino acids, mTOR phosphorylates two key translational regulators: p70S6K and 4E-BP1. p70S6K activation leads to the phosphorylation of rS6, which is important in the translation of mRNAs bearing a 5′-terminal oligopolypyrimidine tract such as ribosomal proteins, elongation factors, and growth factors (26). The regulation of translation and the control of ribosome biogenesis are essential cellular processes that impact cell growth and proliferation. Also, protein biosynthesis on the ribosome requires accurate reading of genetic code in mRNA. It is noteworthy that the serine/threonine kinase Akt enhances FCγR-mediated phagocytosis through the activation of p70S6 kinase in vitro (26). These observations suggest that the phosphorylation of ribosomal S6 is regulated in an intracellular signaling pathway (Akt, mTOR, p70S6K, and rS6) that is enhanced during phagocytosis in macrophages (Fig. 8). The phosphorylation of ribosomal S6 is observed following differentiation of HL60 cells and with macrophages. PMA-differentiated HL60 cells express ChemR23 and regulate phosphorylation events when exposed to nanomolar levels of RvE1 (Fig. 5). Together, these results are consistent with RvE1-ChemR23-mediated Akt and rS6 phosphorylation acting to enhance phagocytosis.
FIGURE 8.
Hypothetical scheme for resolvin E1-ChemR23-dependent signaling in human macrophages. The scheme outlines the key phosphorylation-signaling components with RvE1 and the points of inhibitor action in this system. See under “Results” and legends to Figs. 2–6 for details.
Endogenous anti-inflammatory and pro-resolving lipid mediators are autacoids that act with high affinity (nm range) and stereoselectivity on structurally related receptors, as does aspirin-triggered lipoxin A4 generated from arachidonic acid, to enhance resolution by “stopping” PMN recruitment, reducing interleukin-12 production from antigen-presenting cells, and enhancing the uptake of apoptotic PMN by macrophages (reviewed in Refs. 12 and 27). RvE1 also blocks PMN transendothelial migration in vitro, inhibits migration and interleukin-12 production by antigen-presenting cells (8, 11), and regulates CD4+ T cells (28). Along these lines, mononuclear phagocytes are central to the control of local acute inflammation, as they produce many components that participate in and/or temporally regulate different enzyme systems and hence the mediators of the inflammatory response (14, 28). The nonphlogistic activation of ChemR23 by both RvE1 and anti-inflammatory peptides (16) is an additional example of the complexity and diversity of G protein-coupled receptor signaling (29), likely to conserve key essential functions as needed in the resolution of acute inflammation and the return to tissue homeostasis.
The potent anti-inflammatory and pro-resolving actions of RvE1 have recently been confirmed by others in airway inflammation (25) and collagen-induced arthritis in rats (30). Also, RvE1 was recently shown to act on bone marrow-derived dendritic cells and T cells (28). Because resolvin E1 interacts with at least two and possibly more distinct receptors, the relative contributions of these receptors to RvE1 signaling may depend on the local milieu of RvE1 formation. Whether additional receptors are involved in RvE1-demonstrated pro-resolving actions remains of interest. Nonetheless, the results of the present report clearly indicate that RvE1-ChemR23 interactions stimulate phosphorylation of signaling proteins, i.e. Akt and ribosomal S6 (see Fig. 8), which are central to signal transduction in many systems. This stimulation of phosphorylation with RvE1 contrasts with the actions of lipoxin A4, another endogenous anti-inflammatory and pro-resolving mediator, biosynthesized from arachidonic acid (12), that inhibits tyrosine phosphorylation in eosinophilic granulocytes (31). In summary, we have demonstrated that the small molecule local mediator RvE1, which stimulates the resolution of inflammation (4, 8, 11), acts in part via ChemR23 to regulate Akt phosphorylation events. These findings also suggest an endogenous mechanism that may underlie some of the beneficial actions of the RvE1 precursor omega-3 EPA in regulating local inflammation.
Supplementary Material
Acknowledgments
We thank Mary H. Small for expert assistance with manuscript preparation and Gabrielle Fredman and Dr. Tomoyuki Iwata for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DE-016191 (to C. N. S. and T. E.V. D.), DE-015566 (to T. E.V. D.), and GM38765 (to C. N. S.). Brigham and Women's Hospital and Boston University are assigned patents on resolvins that are licensed for clinical development and are subject to consultant agreements for Drs. Charles N. Serhan and Thomas E. Van Dyke.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- RvE1
- resolvin E1 ((5S,12R,18R)-trihydroxy-6Z,8E,10E,14Z,16E-eicosapentaenoic acid)
- CHO
- Chinese hamster ovary
- DAPI
- 4′,6-diamidino-2-phenylindole
- EPA
- eicosapentaenoic acid
- ERK
- extracellular signal-regulated kinase
- FBS
- fetal bovine serum
- LTB4
- leukotriene B4
- MAPK
- mitogen-activated protein kinase
- mTOR
- mammalian target of rapamycin
- p70S6K
- ribosomal protein S6 kinase
- rS6
- ribosomal protein S6
- PI3K
- phosphatidylinositol 3-kinase
- PMN
- polymorphonuclear leukocyte
- Bim I
- bisindolylmaleimide I
- PMA
- phorbol 12-myristate 13-acetate
- FITC
- fluorescein isothiocyanate
- STZ
- serum-treated zymosan A
- Ab
- antibody
- DPBS
- Dulbecco's phosphate-buffered saline
- FACS
- fluorescence-activated cell sorter
- hChemR23
- human ChemR23
- pAkt(S)
- phospho-Akt substrate.
REFERENCES
- 1.Serhan C. N., Savill J. (2005) Nat. Immunol. 6, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 2.Gilroy D. W., Lawrence T., Perretti M., Rossi A. G. (2004) Nat. Rev. Drug. Discov. 3, 401–416 [DOI] [PubMed] [Google Scholar]
- 3.Shimizu T. (2009) Annu. Rev. Pharmacol. Toxicol. 49, 123–150 [DOI] [PubMed] [Google Scholar]
- 4.Serhan C. N., Brain S. D., Buckley C. D., Gilroy D. W., Haslett C., O'Neill L. A., Perretti M., Rossi A. G., Wallace J. L. (2007) FASEB J. 21, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serhan C. N., Clish C. B., Brannon J., Colgan S. P., Chiang N., Gronert K. (2000) J. Exp. Med. 192, 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gronert K. (2008) Mol. Interv. 8, 28–35 [DOI] [PubMed] [Google Scholar]
- 7.Serhan C. N., Hong S., Gronert K., Colgan S. P., Devchand P. R., Mirick G., Moussignac R. L. (2002) J. Exp. Med. 196, 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arita M., Bianchini F., Aliberti J., Sher A., Chiang N., Hong S., Yang R., Petasis N. A., Serhan C. N. (2005) J. Exp. Med. 201, 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasturk H., Kantarci A., Ohira T., Arita M., Ebrahimi N., Chiang N., Petasis N. A., Levy B. D., Serhan C. N., Van Dyke T. E. (2006) FASEB J. 20, 401–403 [DOI] [PubMed] [Google Scholar]
- 10.Hong S., Porter T. F., Lu Y., Oh S. F., Pillai P. S., Serhan C. N. (2008) J. Immunol. 180, 3512–3519 [DOI] [PubMed] [Google Scholar]
- 11.Arita M., Yoshida M., Hong S., Tjonahen E., Glickman J. N., Petasis N. A., Blumberg R. S., Serhan C. N. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7671–7676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serhan C. N. (2007) Annu. Rev. Immunol. 25, 101–137 [DOI] [PubMed] [Google Scholar]
- 13.Schwab J. M., Chiang N., Arita M., Serhan C. N. (2007) Nature 447, 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor K. M., SanGiovanni J. P., Lofqvist C., Aderman C. M., Chen J., Higuchi A., Hong S., Pravda E. A., Majchrzak S., Carper D., Hellstrom A., Kang J. X., Chew E. Y., Salem N., Jr., Serhan C. N., Smith L. E. (2007) Nat. Med. 13, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arita M., Ohira T., Sun Y. P., Elangovan S., Chiang N., Serhan C. N. (2007) J. Immunol. 178, 3912–3917 [DOI] [PubMed] [Google Scholar]
- 16.Cash J. L., Hart R., Russ A., Dixon J. P., Colledge W. H., Doran J., Hendrick A. G., Carlton M. B., Greaves D. R. (2008) J. Exp. Med. 205, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell E. L., Louis N. A., Tomassetti S. E., Canny G. O., Arita M., Serhan C. N., Colgan S. P. (2007) FASEB J. 21, 3162–3170 [DOI] [PubMed] [Google Scholar]
- 18.Böyum A. (1968) Scand. J. Clin. Lab. Invest. Suppl.21, 77–89 [PubMed] [Google Scholar]
- 19.Ohira T., Zhan Q., Ge T., Van Dyke T., Badwey J. A. (2003) J. Immunol. Methods 281, 79–94 [DOI] [PubMed] [Google Scholar]
- 20.Zabel B. A., Zuniga L., Ohyama T., Allen S. J., Cichy J., Handel T. M., Butcher E. C. (2006) Exp. Hematol. 34, 1021–1032 [DOI] [PubMed] [Google Scholar]
- 21.Alessi D. R., Caudwell F. B., Andjelkovic M., Hemmings B. A., Cohen P. (1996) FEBS Lett. 399, 333–338 [DOI] [PubMed] [Google Scholar]
- 22.Ly C., Arechiga A. F., Melo J. V., Walsh C. M., Ong S. T. (2003) Cancer Res. 63, 5716–5722 [PubMed] [Google Scholar]
- 23.Yoshimura T., Oppenheim J. J. (2008) J. Exp. Med. 205, 2187–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haworth O., Cernadas M., Yang R., Serhan C. N., Levy B. D. (2008) Nat. Immunol. 9, 873–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aoki H., Hisada T., Ishizuka T., Utsugi M., Kawata T., Shimizu Y., Okajima F., Dobashi K., Mori M. (2008) Biochem. Biophys. Res. Commun. 367, 509–515 [DOI] [PubMed] [Google Scholar]
- 26.Ganesan L. P., Wei G., Pengal R. A., Moldovan L., Moldovan N., Ostrowski M. C., Tridandapani S. (2004) J. Biol. Chem. 279, 54416–54425 [DOI] [PubMed] [Google Scholar]
- 27.Maderna P., Godson C. (2003) Biochim. Biophys. Acta 1639, 141–151 [DOI] [PubMed] [Google Scholar]
- 28.Vassiliou E. K., Kesler O. M., Tadros J. H., Ganea D. (2008) J. Immunol. 181, 4534–4544 [DOI] [PubMed] [Google Scholar]
- 29.Maudsley S., Martin B., Luttrell L. M. (2005) J. Pharmacol. Exp. Ther. 314, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savinainen A., Qin S., Crandall T., Dubrovskiy A., Trocha M., Schwartz E., Gjorstrup P., Wu L. (2007) 10th International Conference on Bioactive Lipids in Cancer, Inflammation and Related Diseases, Montreal, September 16–19, 2007 (abstract) [Google Scholar]
- 31.Starosta V., Pazdrak K., Boldogh I., Svider T., Kurosky A. (2008) J. Immunol. 181, 8688–8699 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.