Abstract
Loss of function mutations in the hERG (human ether-a-go-go related gene or KCNH2) potassium channel underlie the proarrhythmic cardiac long QT syndrome type 2. Most often this is a consequence of defective trafficking of hERG mutants to the cell surface, with channel retention and degradation at the endoplasmic reticulum. Here, we identify the Hsp40 type 1 chaperones DJA1 (DNAJA1/Hdj2) and DJA2 (DNAJA2) as key modulators of hERG degradation. Overexpression of the DJAs reduces hERG trafficking efficiency, an effect eliminated by the proteasomal inhibitor lactacystin or with DJA mutants lacking their J domains essential for Hsc70/Hsp70 activation. Both DJA1 and DJA2 cause a decrease in the amount of hERG complexed with Hsc70, indicating a preferential degradation of the complex. Similar effects were observed with the E3 ubiquitin ligase CHIP. Both the DJAs and CHIP reduce hERG stability and act differentially on folding intermediates of hERG and the disease-related trafficking mutant G601S. We propose a novel role for the DJA proteins in regulating degradation and suggest that they act at a critical point in secretory pathway quality control.
Keywords: Protein/Degradation, Chaperone/Chaperonin, Ion Channels, Protein Folding, Trafficking, Hsp40, hERG, Quality Control
Introduction
The KCNH2 or hERG (human ether-a-go-go related gene) gene encodes the Kv11.1 α-polypeptide, the transmembrane channel responsible for the rapidly activating delayed rectifier K+ current (IKr) in the heart (1–3). Loss of hERG function delays ventricular repolarization and is classified clinically as type 2 long QT syndrome (LQT2),3 characterized by a significantly increased risk of ventricular arrhythmia and sudden death (4). To date more than 200 naturally occurring LQT2 mutants have been discovered that result in either abnormal hERG channel function or a decrease in cell surface localization (5).
The most common mechanism for the loss of hERG function in LQT2 mutants is a defect in hERG trafficking from the site of synthesis at the endoplasmic reticulum (ER) to the active location at the cell surface (5). Importantly, functional rescue by restoring trafficking is possible in ∼70% of these mutants, making this phenotype a promising target for pharmacological intervention. These mutant channels, which are unable to productively fold or form active tetrameric channels, are retained at the ER and destroyed by proteasomal ER-associated degradation (ERAD) (6–9).
The mechanisms responsible for hERG folding and ERAD are still poorly characterized. By comparison, work with the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel has implicated the cytosolic heat shock protein (Hsp) chaperones Hsp70/Hsc70 and Hsp90, as well as several E3 ubiquitin ligases and membrane extraction factors, in its folding and degradation (10, 11). Like CFTR, much of the hERG polypeptide consists of structured domains in the cytosol, and many trafficking-defective mutations are found in these domains (5, 12, 13). Initial reports suggest that Hsc70 and Hsp90 also have key roles in hERG biogenesis. Both chaperones interact with the immature form of wild type (WT) hERG at the ER, most probably through its N- or C-terminal cytosolic domains. These interactions were enhanced for trafficking-defective mutants of hERG. Importantly, specific inhibition of Hsp90 activity with geldanamycin reduced the production of mature hERG and reduced the overall interaction between hERG and Hsp90 (14, 15).
Hsc70 and Hsp90 are ATP-dependent chaperones, and a range of co-chaperone proteins assist and direct their activity (16). Hsp40/DnaJ family co-chaperones stimulate the ATPase and polypeptide binding activities of Hsc70 and are essential for Hsc70 function (17–19). In the sequential chaperone pathway for the folding of progesterone and glucocorticoid steroid hormone receptors, binding of the Hsp40 protein DJA1 (DNAJA1/Hdj2) appears to be a first step, followed by Hsc70 binding (20–22). Another co-chaperone, Hop, is thought to promote substrate transfer from Hsc70 to Hsp90 by binding to both chaperones at once. Hsc70 and Hop dissociate and are replaced in Hsp90 complexes by other co-chaperones, such as peptidyl-prolyl isomerases. It is possible that the outline of this pathway also applies to other substrates of the chaperone system including hERG.
Recently, we undertook a proteomic screen to identify proteins important for hERG trafficking and found, in addition to Hsc70 and Hsp90, a number of associated co-chaperones. Among these was FKBP38 (FKBP8), a membrane-anchored peptidyl-prolyl isomerase that binds specifically to Hsp90. We determined that FKBP38 interacted and co-localized with immature hERG at the ER and that siRNA knockdown of endogenous FKBP38 reduced hERG trafficking efficiency (23). Because FKBP38 is the only known membrane-anchored ER co-chaperone, it may provide a direct link between the cytosolic chaperones and ER export mechanisms of hERG.
Other co-chaperones found to be associated with hERG were DJA1, the Bcl2-associated athanogene Bag2, and Hop (23). Bag2 is one of several nucleotide exchange factors for Hsc70 that reset the ATPase cycle of the chaperone after stimulation by Hsp40 (19). These co-chaperones were also found to be associated with CFTR or a trafficking-defective mutant of the protein (11). DJA1 binds to newly synthesized CFTR at the ER and was shown to assist Hsc70 in preventing aggregation (24). Bag2 was found to regulate degradation of CFTR through interactions with CHIP (C terminus of Hsc70-interacting protein), an E3 ubiquitin ligase that contacts Hsc70 and ubiquitylates chaperone-bound substrates (25–28). An increasingly recognized function of these co-chaperones and the chaperone system generally is to make “triage” decisions regarding whether substrates are folded or degraded (29–31). Interactions between Hsc70 and these co-chaperones are transient and sometimes competitive, for example, the Hsc70-binding domains in Hop and CHIP are homologous to each other and recognize the same site on Hsc70 (16, 32).
The roles of DJA1 and related Hsp40 proteins in this system of chaperone-mediated folding and degradation are still poorly understood. DJA1 is highly homologous to the co-chaperones DJA2 (DNAJA2/Hdj3/Rdj2) and the less abundant DJA4 (DNAJA4). All of these have a J domain that activates Hsc70 and a separate polypeptide-binding domain, although they diverge in the efficiencies of these features (33–35). We therefore tested the function of the Hsp40 proteins for hERG maturation in cells. Intriguingly, we found that overexpression of the DJA proteins reduces the levels of the mature form of hERG exported from the ER, indicating diminished hERG trafficking efficiency. This effect is due to an increase in proteasomal degradation of the immature form, most likely by increased ERAD through Hsc70 and the ubiquitin ligase CHIP. Our results provide the first demonstration of a degradation role for these Hsp40s and suggest that they act at a critical point in hERG quality control.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Unless otherwise stated, all of the chemical reagents were from Sigma Diagnostics Canada or BioShop Canada (Mississauga, Canada). Restriction enzymes and other recombinant DNA reagents were from New England Biolabs (Ipswich, MA), Invitrogen, and Stratagene (San Diego, CA). Geldanamycin was from LC Laboratories (Boston, MA). The following commercially available antibodies were used: anti-Myc (9E10) (Santa Cruz, CA), anti-hemagglutinin (HA.11) (Covance/BAbCO, Richmond, CA), anti-Hsc/Hsp70 (Stressgen, Victoria, Canada), anti-hERG (Chemicon, Temecula, CA), anti-FLAG (M2) (Sigma), anti-GFP (JL-8) (BD Living Colors; Clontech, Mountain View, CA), anti-DJA1 (Thermo Fisher Scientific), and anti-tubulin (Sigma). Additional antibodies specific for DJA1 and DJA2 were raised in rabbits against the synthetic peptides LVDFDPNQER and PEVPNIIGET, respectively, as described (34). Lactacystin was from the Cayman Chemical Company (Ann Arbor, MI). Protein G-Sepharose™ Fast Flow beads were from GE Healthcare. The Express Protein labeling mix, [35S], used for pulse-chase studies and in vitro translation experiments was from PerkinElmer Life Sciences. Untreated rabbit RL was from Green Hectares (Oregon, WI) and was desalted into buffer G (100 mm KOAc, 20 mm Hepes-KOH, pH 7.5, and 5 mm MgOAc2).
Plasmids
The generation of N-terminal hemagglutinin (HA)-tagged WT hERG has been described previously (12). The hERG mis-sense mutant G601S was engineered using the QuikChange XL site-directed mutagenesis kit (Stratagene) and a hERG-C cassette as the PCR template as described previously (12). Sequences of DJA1, DJA2, and DJA4 were inserted into pcDNA3.1 Myc-His C (Invitrogen) as described previously (34) as were DJA1- and DJA2-ΔJ (amino acids 98–397 and 100–412, respectively). hERG CT (amino acids 668–1159) and CL-CNBD (amino acids 668–870) were inserted into pGEM-11Z (Promega) with EcoRI and NotI. Myc-tagged CHIP was a kind gift from Jörg Höhfeld (25). HCN2 was in pCMV-Myc, and CFTR in pcDNA3-HA.
Cell Culture and Transfection
The GripTite™ 293 macrophage scavenger receptor cell line (Invitrogen) was used for all experiments except where indicated. This cell line is a genetically engineered HEK-293 cell line that expresses the human macrophage scavenger receptor type I, class A gene from the pCMV SPORT6 MSR.neo plasmid and strongly adheres to standard tissue culture plates (36). These cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 600 μg/ml geneticin (Invitrogen). The cells were cultured at 37 °C in 5% CO2. Plasmid transfections were carried out using Lipofectamine 2000 for all experiments as described by the manufacturer. The cells incubated at 26 °C were transfected as described and transferred to 26 °C for 24 h following transfection, where they remained for an additional 24 h, and then they were lysed. All of the siRNA duplexes were from Dharmacon/Thermo Fisher Scientific including the nonsilencing control (target sequence, 5′-UGGUUUACAUGUCGACUAA); the siRNA target sequences within DJA1 and DJA2 are in supplemental Table S1. 20 nm siRNA duplexes were transfected with Oligofectamine as described by the manufacturer, and the cells were passaged after transfection.
Stable Cell Line Generation
HeLa and GripTite 293 cell lines (ATCC) were generated that stably express WT hERG with HA (14) and Myc tags, respectively, inserted after residue 443 in the extracellular S1-S2 loop. A retroviral expression plasmid was generated by introducing the cDNA encoding the extracellular HA-tagged WT hERG into the HindIII/ApaI sites of the retroviral expression vector pTZV4-CMV-IRES-puro (Open Biosystems). For viral infection subconfluent HeLa cells in 6-well plates were incubated with 0.5 ml of viral expression plasmid in Opti-MEM (Invitrogen) for 12 h at 37 °C. The cells were cultured with DMEM supplemented with 10% fetal bovine serum and 1 μg/ml puromycin for 10–14 days to select the stable cells. 50–100 individual colonies were pooled and expanded for experiments.
In Vitro Binding Assay
His-tagged human DJA1, DJA2, and DJA4 were expressed in Rosetta 2 Escherichia coli cells (Novagen) and purified as described (34). Binding experiments of purified DJA1, DJA2, and DJA4 with hERG fragments were performed as described (35). Briefly, purified DJA1, DJA2, and DJA4 were prebound to nickel-Sepharose beads in buffer containing 500 mm NaCl, 20 mm Hepes-KOH, pH 7.5, and 5 mm MgOAc2 for 30 min at 4 °C. Cell-free translations of the hERG CT or CL-CNBD were performed with the TnT-coupled RL system with T7 polymerase (Promega) supplemented with [35S] methionine (GE Healthcare and PerkinElmer Life Sciences); diluted 1:20 into buffer G containing 20 mm imidazole, 0.1% Triton X-100, and 2 mg/ml ovalbumin; and added to the immobilized DJA proteins. The final reactions contained 5 μm DJA protein and 5% translation mixture. After 5 min at room temperature, the binding reactions were terminated by the addition of 0.1 unit/μl apyrase. The protein complexes were recovered at 4 °C for 30 min and washed with buffer G containing 20 mm imidazole and 0.1% Triton X-100 before being analyzed by SDS-PAGE and phosphorimaging quantification.
Immunoblot Analysis and Immunoprecipitation
Forty-eight hours post-transfection the cells were washed two times with cold phosphate-buffered saline and incubated in lysis buffer (0.5% Nonidet P-40, 75 mm NaCl, and 50 mm Tris, pH 8) plus a protease inhibitor mixture (Roche Applied Science) for a minimum of 15 min or, for Fig. 1 (D and E) and Fig. 2, in phosphate-buffered saline containing 1% Triton X-100 under identical conditions. The cells were homogenized by pipetting, harvested, and then left on ice for an additional 15 min with occasional vortexing. Detergent-insoluble material was sedimented at 16,000 × g for 20 min, after which the resulting supernatant was collected, and the protein concentration was determined with a detergent-compatible assay as per the manufacturer's instructions (Bio-Rad). For immunoprecipitation, samples of 0.5–1 mg of protein were incubated overnight at 4 °C in a volume of 0.5–1 ml with either monoclonal mouse anti-HA antibody (1:200) or monoclonal mouse anti-Myc antibody (1:100). For co-precipitation of hERG with endogenous DJA1 and DJA2, samples of 5 mg of protein were incubated overnight at 4 °C in a volume of 2 ml with either monoclonal mouse anti-HA antibody (1:100) or polyclonal rabbit anti-hERG antibody (1:250). The samples were incubated with protein G-Sepharose Fast Flow beads (GE Healthcare) for 2 h, after which the beads were washed extensively and then resuspended in 2× sample buffer (12% 0.5 m Tris, pH 6.8, 5% β mercaptoethanol, 20% glycerol, 20% SDS, bromphenol blue). The samples were resolved on a 7% polyacrylamide SDS gel and transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked for 1 h with 5% nonfat dry milk and 0.1% Tween 20 in phosphate-buffered saline and then incubated with the appropriate primary antibody for a minimum of 2 h at room temperature, washed extensively, and then incubated with goat anti-mouse/rabbit IgG conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories). After extensive washing the membranes were visualized on x-ray films using an ECL Plus detection kit (Amersham Biosciences).
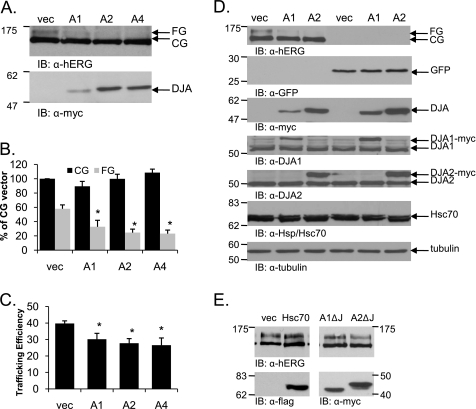
FIGURE 1.
Overexpression of the DJA proteins reduces the trafficking efficiency of hERG. A, HEK-293 (GripTite) cells were transiently transfected with 1 μg of HA-tagged hERG and 2 μg of Myc vector (vec), Myc-tagged DJA1 (A1), DJA2 (A2), or DJA4 (A4). Two days post-transfection, the cells were lysed, and protein expression was analyzed by Western blots with anti-hERG antibody for hERG visualization and anti-Myc antibody for DJA protein visualization. B and C, corresponding mean data (n = 8 experiments/condition). All of the values are normalized to the CG vector condition after background subtraction. Trafficking efficiency is calculated as FG/(CG+FG). *, p ≤ 0.01 versus vector CG. D, HEK-293 cells were transiently transfected with 1 μg of HA-tagged WT hERG or 2 μg of GFP and either Myc vector or Myc-tagged DJA1 or DJA2. Two days post-transfection the cells were analyzed as indicated in A. The membranes were blotted for hERG, GFP, tagged and endogenous DJA1 and DJA2, Hsc70, and tubulin as indicated. E, HEK-293 cells were transiently transfected with 1 μg of HA-tagged WT hERG and either Myc vector, FLAG-tagged Hsc70, or Myc-tagged DJA1 or DJA2 lacking their J domain (A1ΔJ and A2ΔJ) and analyzed as above. IB, immunoblot.
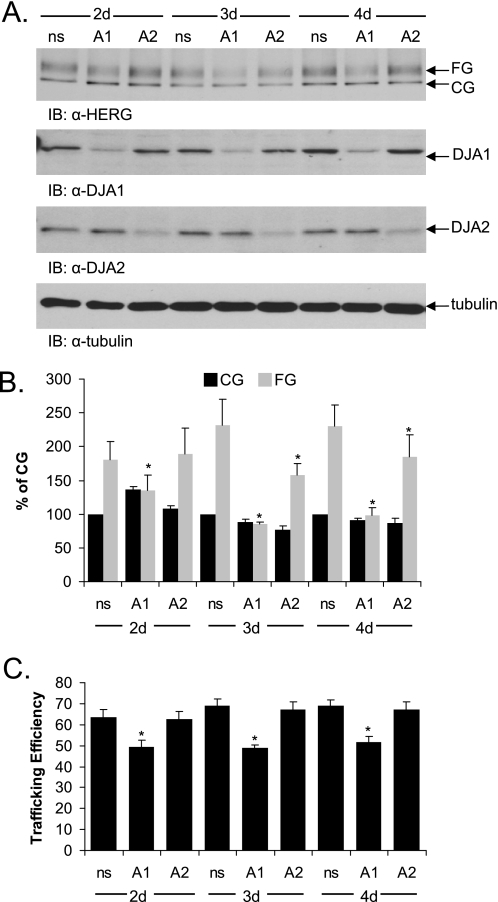
FIGURE 2.
Knockdown of DJA1 by siRNA reduces trafficking efficiency of hERG. A, HeLa cells stably expressing hERG were transfected with a nonsilencing (ns) siRNA or with siRNA pools against DJA1 (A1) or DJA2 (A2). 2–4 days post-transfection, protein expression was visualized by Western blots with anti-hERG, anti-DJA1, and anti-DJA2 antibodies. B and C, corresponding mean data for hERG (n = 3 experiments/condition for DJA1 and DJA2). All of the values are normalized to the CG nonsilencing condition. Trafficking efficiency is calculated as FG/(CG+FG). *, p < 0.01 versus nonsilencing where indicated. IB, immunoblot; d, days.
Proteasomal Inhibition
HEK-293 (GripTite) cells seeded in 6-well plates were transiently transfected with 1 μg of HA-tagged WT hERG and 2 μg of Myc vector or Myc-tagged DJA1, DJA2, or DJA4. Twenty-four hours post-transfection, the cells were incubated with DMEM containing Me2SO or 25 μm lactacystin (Cayman Chemical Company) for an additional 24 h, after which the cells were lysed as described above.
Pulse-Chase Metabolic Labeling
Twenty-four hours post-transfection, the cells were washed two times with phosphate-buffered saline and starved for 1 h in serum-free DMEM lacking methionine and cysteine in the presence of 0.25% bovine serum albumin and 2 mm glutamine. The cells were then metabolically labeled in the same medium containing 125–150 μCi/ml [35S]methionine/cysteine for 45 min, after which the medium was removed and replaced with DMEM containing 2 mm unlabeled methionine and cysteine and chased in the same medium for the indicated time intervals. The cells were lysed, immunoprecipitated as described, and subjected to SDS-PAGE, autoradiography for visualization, and phosphorimaging quantification.
Densitometry and Statistical Analysis
Densitometric analysis was carried out using the digital imagine program Image J (National Institutes of Health). For calculation of the relative protein expression levels, each band was quantified by a mean pixel value after subtraction of background. The values for CG and FG hERG bands were normalized to the value of the CG hERG band under control conditions (with Myc vector alone). hERG trafficking efficiency is represented by the ratio of the normalized FG hERG band to the total normalized HERG intensity (FG/(CG+FG)). Quantification of Hsc70 co-immunoprecipitated with hERG was performed by first normalizing to the amount of immunoprecipitated CG hERG and then reported relative to the amount of Hsc70 bound to CG hERG under control conditions. Paired t tests were used to compare groups of data to determine significance. The results are presented as the mean values ± standard error.
RESULTS
DJA Proteins Inhibit hERG Trafficking
To search for possible chaperones and co-chaperones that interact with hERG, we previously conducted proteomic screens of immunoprecipitated Myc-tagged hERG transfected into cardiac (HL-1) (37, 38) and noncardiac (HEK-293) cell lines (23). Several chaperones and co-chaperones were identified including DJA1, a member of the Hsp40/DnaJ family.
We first determined whether DJA1 and the closely related proteins DJA2 and DJA4 had an effect on hERG trafficking. As previously demonstrated, hERG maturation was observed in transiently transfected HEK-293 (GripTite) cells by Western blot detection of a 155-kDa form corresponding to hERG that had become FG during export through the Golgi, compared with the 135-kDa form representing CG hERG in the ER (Fig. 1A). Overexpression of Myc-tagged DJA1, DJA2, and DJA4 markedly reduced the abundance of mature FG hERG (p < 0.01 for all DJA proteins) without affecting the immature CG hERG, relative to control conditions (Fig. 1, A and B). When relative trafficking efficiency was calculated, a clearly diminished trafficking efficiency was observed in the presence of any of the three cytosolic Hsc70 co-chaperones (Fig. 1C; p < 0.01 for all DJA proteins).
To confirm the specificity of this effect, the expression of GFP in the presence of the DJA proteins was tested; spontaneous GFP refolding is robust in the absence of chaperones (39). As expected, GFP levels were not affected by overexpression of any of the DJAs in HEK-293 cells (Fig. 1D). Moreover, the abundance of other cellular proteins such as Hsc70 or tubulin remained unaffected (Fig. 1D). In previous work, overexpression of each of the DJAs increased Hsc70-dependent luciferase activity and mitochondrial import of the phosphate carrier (34), indicating that the transfected DJA proteins are functional in cells.
The DJA co-chaperones are all type I Hsp40 proteins with similar architecture: an N-terminal J domain that activates Hsc70, a central substrate-binding domain, and a C-terminal homodimerization domain. We hypothesized that the inhibition of hERG maturation might be mediated through the J domain because it is necessary for Hsc70 activation. To determine the effect of this domain deletion, mutants of the major Hsp40 proteins DJA1 and DJA2 were tested. Overexpression of mutant DJA1 and DJA2 proteins lacking the J domains (A1-ΔJ and A2-ΔJ) had no effect on hERG trafficking (Fig. 1E). Thus, the J domains of the co-chaperones were necessary for the inhibitory effect, implying that the interaction with Hsc70 was important. However, overexpression of Hsc70 itself had no effect on hERG maturation (Fig. 1E), suggesting that Hsc70 required activation by the DJAs to mediate inhibition.
We then asked what effect the reduction of DJA expression by siRNA knockdown would have on hERG trafficking. Because knockdown was much more efficient in HeLa cells than in HEK-293 cells, a HeLa cell line stably transfected with HA-tagged hERG was used. Transient transfection of DJA1 and DJA2 in these cells similarly impaired maturation of hERG. Interestingly, transfection of an siRNA duplex against DJA1 also partially diminished hERG maturation, compared with transfection of a nonsilencing control duplex (Fig. 2A). Similar siRNA knockdown of DJA2 had a minor effect on trafficking, and these observations were confirmed by quantification of the trafficking efficiency (Fig. 2, B and C; p < 0.01 for DJA1). It therefore appears that DJA1 can both promote and inhibit hERG maturation, depending on its expression level. The role of DJA2 may be more limited to inhibiting hERG trafficking, again depending on expression level, although a contribution to hERG folding is still possible. Because the individual knockdown of DJA1 or DJA2 did not increase hERG trafficking, it is likely that the endogenous level of either protein is sufficient to limit hERG trafficking to the level observed in control cells.
Effect of DJA Proteins on Other Channels
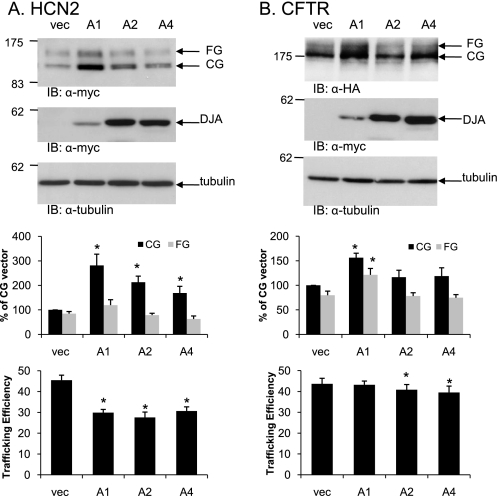
Next, the effect of the DJAs on two other ion channel proteins in the secretory pathway was tested. HCN2 is homologous to hERG in the transmembrane region and putative CNBD but divergent in other cytosolic regions. The CFTR chloride channel is unrelated to hERG but is a well studied model of folding, trafficking, and degradation at the ER. Both channels were transiently co-transfected with the DJAs in HEK-293 (GripTite) cells. Intriguingly, overexpression of DJA1 caused a strong accumulation of the CG form of HCN2 at the ER but no significant increase in the mature FG form. The overall effect was a marked inhibition of HCN2 trafficking, and a similar but lesser effect was observed with DJA2 and DJA4 (Fig. 3A). In contrast, DJA1 moderately increased the FG form of CFTR (Fig. 3B), consistent with an earlier report (24). However, the CG form was also increased, so the overall trafficking efficiency was unchanged. DJA2 and DJA4 had no apparent effect on CFTR (Fig. 3B). Thus, the DJAs may be more inhibitory of the trafficking of some types of membrane proteins than others.
FIGURE 3.
Effect of DJA proteins on other channels. A, HEK-293 (GripTite) cells were transiently transfected as described for Fig. 1 but with Myc-tagged HCN2 in combination with Myc vector (vec), Myc-tagged DJA1 (A1), DJA2 (A2), or DJA4 (A4). HCN2 and the DJA proteins were visualized by Western blots using anti-Myc antibodies. B, the experiment was performed as above except with HA-tagged CFTR in combination with the DJA proteins, and the proteins were visualized using anti-HA and anti-Myc antibodies, respectively. The corresponding mean data are shown below (n = 5 experiments/condition for HCN2; n = 7 for CFTR). Trafficking efficiency is calculated as FG/(CG+FG). *, p < 0.01 versus vector where indicated. IB, immunoblot.
DJA Proteins Bind to hERG and Reduce the Hsc70-hERG Interaction
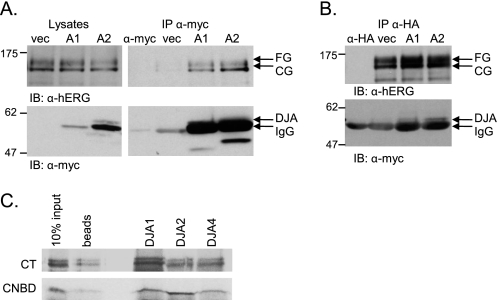
It is thought that Hsp40 proteins bind directly to substrate polypeptide and transfer them to Hsc70 after activation of the ATPase activity of that chaperone. Although the J domains of the DJAs were required for the effect on hERG trafficking, it is likely that the DJAs also interact with hERG through their substrate-binding domains. We therefore performed co-immunoprecipitation experiments from HeLa cells stably expressing HA-tagged hERG and transiently transfected with the vector control, DJA1, or DJA2. Tagged DJA1 and DJA2 were precipitated by the anti-Myc antibody, and hERG was clearly detected in the precipitate (Fig. 4A). hERG was not present in precipitates from cells transfected with the vector control or in mock precipitates containing only anti-Myc antibody (Fig. 4A). Notably, the immature CG form of hERG was primarily precipitated with the DJAs, as well as a small amount of the mature FG form.
FIGURE 4.
hERG binds DJA proteins. A, lysates of HeLa cells stably expressing HA-tagged hERG and transfected with Myc vector (vec), Myc-tagged DJA1 (A1), or DJA2 (A2) were immunoprecipitated with anti-Myc antibody, and the membranes were analyzed with anti-hERG or anti-Myc antibodies. Lysates alone are shown on the left side as input reference. IgG indicates the heavy chain of the anti-Myc antibody as also seen in a control lane loaded with 1 μg of anti-Myc alone (α-myc). B, lysates were treated as described above and immunoprecipitated with anti-HA antibody. Western blots were analyzed for both hERG and Myc-tagged DJA protein as indicated. IgG indicates the heavy chain of the anti-HA antibody as also seen in a control lane loaded with 0.5 μg of anti-HA alone (α-HA). C, DJA1, DJA2, and DJA4 were prebound to nickel-Sepharose beads and incubated with radiolabeled cell-free translations of the hERG CT (amino acids 668–1159) or CL-CNBD (amino acids 668–870). Final binding reactions containing 5 μm DJA protein and 5% translation mixture were terminated by the addition of 0.1 unit/μl apyrase. Protein complexes were recovered at 4 °C for 30 min and washed before being analyzed by SDS-PAGE and visualized by autoradiography. Nonspecific binding of translated hERG product to the nickel-Sepharose beads is shown in the lane labeled beads. IB, immunoblot; IP, immunoprecipitation.
As an alternate test, the above cells were immunoprecipitated with the anti-HA antibody, and HA-tagged hERG was detected in the precipitates. Both DJA1 and DJA2 were co-precipitated, visible as bands just above that of the immunoglobulin heavy chain (IgG) (Fig. 4B). No such band was detected in precipitates from control cells or mock precipitates with anti-Myc antibody alone. The interaction between mature FG hERG and the DJAs may represent a function of the co-chaperones in cell surface quality control, as has been suggested for CFTR (40), although this may be a secondary function of the DJAs.
The interactions between hERG and the endogenous co-chaperones were also confirmed. HEK-293 (GripTite) cells stably expressing HA-tagged hERG were treated with the proteasome inhibitor lactacystin to cause accumulation of hERG (discussed in more detail below; see Fig. 6) and immunoprecipitated with a polyclonal antibody specific for hERG. DJA1 was detected with a monoclonal antibody in the precipitate but not in precipitate from untransfected control cells (supplemental Fig. S1A). Furthermore, similarly treated cells were immunoprecipitated with the anti-HA monoclonal antibody, and both DJA1 and DJA2 were detected in the precipitates with the respective polyclonal antibodies (supplemental Fig. S1, B and C).
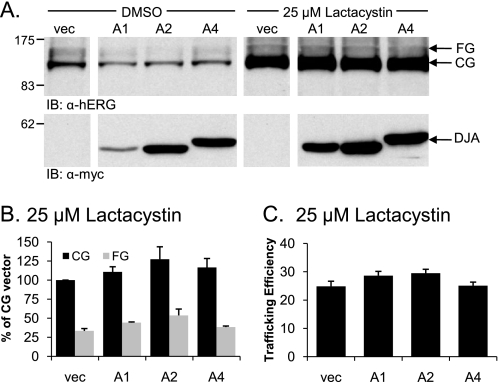
FIGURE 6.
Inhibiting the proteasome restores normal hERG trafficking in the presence of overexpressed DJA proteins. A, HEK-293 (GripTite) cells were transfected as indicated in Fig. 1, and 24 h post-transfection were incubated with DMEM containing dimethyl sulfoxide (DMSO) or 25 μm lactacystin for an additional 24 h. Cell lysates were prepared as described for Fig. 1, and protein expression was determined by Western blot as indicated. B and C, corresponding mean data after lactacystin treatment (n = 4 experiments/condition). All of the values are normalized to the CG vector (vec) condition after background subtraction. Trafficking efficiency is calculated as FG/(CG+FG). IB, immunoblot.
The CT cytosolic domain of hERG is known to be an important determinant of its trafficking, most probably corresponding to a folded structural unit including the putative CNBD (3, 13). To test whether this region of the polypeptide can interact with the DJA proteins, we performed in vitro binding studies using an established assay (34, 35). The entire CT domain (amino acids 668–1159) of hERG and a small fragment containing the CNBD (amino acids 668–870) were radiolabeled by cell-free translation and co-precipitated with purified His-tagged DJA1 and DJA2, as well as DJA4. Both fragments of hERG were bound by the DJA proteins, whereas only background binding was observed for control precipitates without His-tagged protein (Fig. 4C). Thus, the CNBD appears to provide at least one binding site for the DJA proteins, although this is likely not the only binding site.
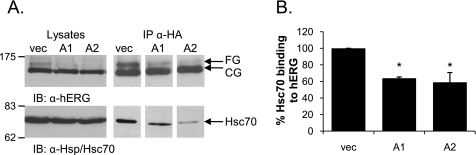
It is known that DnaJ proteins stimulate ATP hydrolysis by Hsc70 and consequently induce Hsc70-substrate binding (16). Given that Hsc70 interacts with hERG (14), we examined whether the overexpression of the DJA proteins influenced the interaction between hERG and Hsc70. To do this we conducted co-immunoprecipitation experiments in HEK-293 (GripTite) cells that were transiently co-transfected with HA-tagged hERG and Myc-tagged DJA1, DJA2, or control vector. When hERG was immunoprecipitated using the anti-HA antibody, endogenous Hsc70 was found in the precipitate. Strikingly, when DJA1 or DJA2 were overexpressed, the amount of Hsc70 that co-precipitates with hERG decreased substantially (Fig. 5). Quantification shows that Hsc70 binding to hERG was reduced to ∼60% of the control by both DJA1 and DJA2 (p = 0.01 for DJA1 and DJA2 versus control vector).
FIGURE 5.
Overexpression of DJA1 and DJA2 inhibits the interaction between hERG and Hsc70. A, lysates of HEK-293 (GripTite) cells transiently transfected with 1 μg of HA-tagged hERG and 2 μg of Myc or Myc-tagged DJA1 (A1) or DJA2 (A2) were immunoprecipitated with anti-HA antibody, and Western blots were analyzed for both hERG and Hsc70 protein as indicated. Lysates alone are shown on the left side as input reference. B, corresponding mean data (n = 3 experiments/condition for DJA1; n = 5 for DJA2 overexpression). Hsc70 binding to hERG is reported as a percentage of the amount of Hsc70 bound to hERG in the presence of Myc vector alone (vec) after first being normalized to the amount of CG hERG precipitated. *, p = 0.01 for A1 and A2 versus vector control. IB, immunoblot; IP, immunoprecipitation.
Although these results may seem counter to the expected mechanism of Hsc70 substrate binding stimulated by the DJAs, we note that the co-immunoprecipitation experiments reflect a steady state situation in cells with a functional degradation system. The decrease in the amount of hERG complexed with Hsc70 could indicate a preferential degradation of Hsc70-bound channel. Indeed, Hsc70 is known to target substrate proteins for proteasomal degradation by its interaction with the E3 ubiquitin ligase CHIP (30, 41). We thus hypothesized that the DJA proteins inhibit hERG trafficking by targeting hERG to the proteasomal system for degradation through the Hsc70-CHIP pathway.
Reduced hERG Trafficking Caused by DJA Overexpression Is Proteasome-dependent
If the DJA proteins prevent hERG maturation by inducing degradation, inhibition of proteasomes should restore mature hERG. So HEK-293 (GripTite) cells co-transfected with hERG and the DJAs were treated with 25 μm of the specific proteasome inhibitor lactacystin for 24 h. Compared with cells treated with vehicle control (Me2SO), lactacystin increased the amount of both CG and FG hERG (Fig. 6A). Moreover, in the presence of lactacystin, the DJA proteins had no effect on the level of either CG or FG hERG relative to cells transfected with the vector control (Fig. 6A, right panel). The lack of effect of the DJAs was confirmed by quantification (Fig. 6B). Similarly, there was no effect of the DJAs on the trafficking efficiency of hERG in the presence of lactacystin (Fig. 6C). Therefore, the DJAs appear to promote the degradation of hERG by proteasomes, typical of other ERAD processes.
CHIP Reduces hERG Expression and Hsc70-hERG Binding
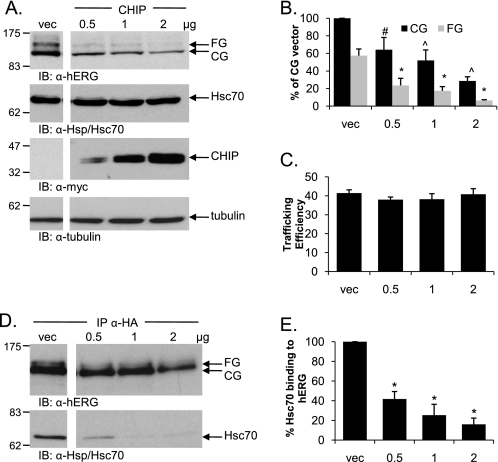
The E3 ubiquitin ligase CHIP was considered a prime candidate for the induction of hERG proteasomal degradation. Overexpression of Myc-tagged CHIP in HEK-293 (GripTite) cells co-transfected with HA-tagged hERG resulted in an equal reduction of both the CG and FG forms of hERG (Fig. 7, A and B). Given that both bands were reduced, there was no overall decrease in proportional trafficking efficiency (Fig. 7C); however, the reduced amount of FG hERG indicated that less was being trafficked to the cell surface, because an E3 ligase CHIP functions solely as a component of the degradation system and appeared to be very efficient at promoting the degradation of hERG, resulting in the reduction of the CG form as well as the FG form. In contrast, the DJA proteins as chaperones may not be as efficient at directly targeting hERG for degradation, and therefore there was no reduction in the CG hERG levels when they were overexpressed.
FIGURE 7.
Overexpression of CHIP reduces expression level of both hERG bands and decreases hERG-Hsc70 binding. A, HEK-293 (GripTite) cells were transiently transfected with 1 μg of HA-tagged hERG and either Myc alone or increasing amounts of Myc-tagged CHIP. Protein expression was analyzed for hERG, CHIP, Hsc70, and tubulin visualization as described for Fig. 1. B and C, corresponding mean data for A (n = 6 experiments/condition). All of the values are normalized to the CG vector (vec) condition after background subtraction. #, p < 0.05 versus vector CG; ^, p < 0.01 versus vector CG; *, p < 0.01 versus vector FG. Trafficking efficiency is calculated as FG/(CG+FG). D, lysates of HEK-293 (GripTite) cells transiently transfected as in A were immunoprecipitated with anti-HA antibody, and Western blots were analyzed for both hERG and Hsc70 as indicated. E, corresponding mean data for D (n = 4 experiments/condition). Quantification of Hsc70 binding is done as for Fig. 3. *, p < 0.01 for 0.5, 1.0, and 2.0 μg of CHIP versus vector control. IB, immunoblot.
Despite the difference in the efficiency of degradation induced by CHIP and the DJAs, they both dramatically reduced the interaction between hERG and Hsc70. As described above (Fig. 5), Hsc70 was co-immunoprecipitated with transiently transfected HA-tagged hERG. An incremental increase in the amount of co-transfected Myc-tagged CHIP led to a concomitant decrease in the amount of precipitated Hsc70 (Fig. 7D). Quantification showed that Hsc70 binding was diminished to ∼16% of the control at the highest expression level of CHIP tested (p < 0.01; Fig. 7E). These results are consistent with the DJAs acting through Hsc70 and CHIP to promote hERG degradation.
The DJA Proteins and CHIP Reduce hERG Stability
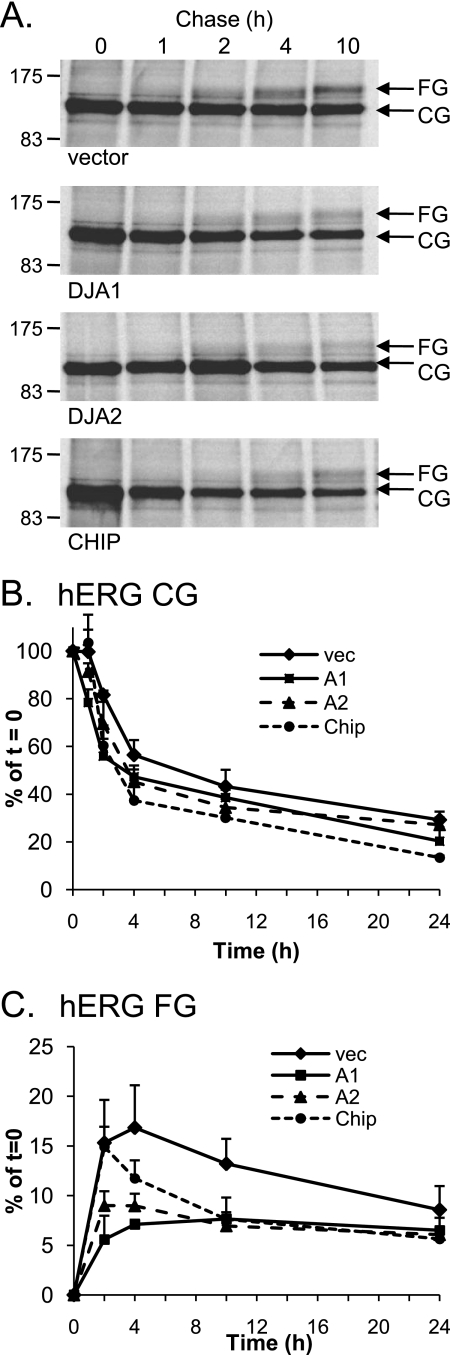
To compare the stability and turnover rate of hERG with or without the overexpression of DJA1, DJA2, or CHIP, we used pulse-chase analysis. In vector control transfected cells, hERG first appeared as the CG form, and a fraction subsequently shifted to the FG form after acquiring complex oligosaccharides ∼2 h after synthesis (Fig. 8A). This FG band was still visible after 24 h of chase time, confirming the relative stability of mature hERG. The abundant CG form progressively weakened, apparently because of degradation as well as conversion to the FG form at the cell surface.
FIGURE 8.
Overexpression of the DJA proteins and CHIP reduces the stability of hERG. A, lysates of HEK-293 (GripTite) cells transiently transfected with 1 μg of HA-tagged hERG and 2 μg of Myc (vector), Myc-tagged DJA1, DJA2, or CHIP were starved, metabolically labeled for 45 min, and then chased for the indicated time intervals. The cells were lysed and immunoprecipitated as described for Fig. 2 and analyzed by SDS-PAGE and autoradiography. B and C, corresponding mean data (n = 5 for vector, n = 3 for DJA1 (A1), n = 6 for DJA2 (A2), and n = 5 for CHIP) for core glycosylated (B) and fully glycosylated (C) hERG at the indicated time points. All of the values are normalized to the t = 0 value for their own condition.
Measured immediately after radiolabeling (t = 0 h), similar quantities of CG hERG were synthesized under control conditions or in the presence of DJA1, DJA2, or CHIP (Fig. 8A). In control cells, the CG form was reduced to ∼30% after 24 h of chase. DJA1 and DJA2 induced a moderate decrease in CG levels relative to control conditions, whereas CHIP caused the largest decrease in CG hERG at 24 h of chase (Fig. 6B; CHIP, p < 0.02). The greater effect of CHIP compared with DJA1 or DJA2 corresponds with the results obtained under steady state conditions. In those experiments, it was only the overexpression of CHIP that resulted in a reduction of the CG hERG species (Fig. 7B), whereas the DJA proteins had no effect relative to the vector control (Fig. 1B). The FG form of hERG was most apparent in the control cells, peaking at 4 h of chase at ∼16% of the amount of CG hERG present at t = 0 (Fig. 8C), whereas transfection of DJA1 and DJA2 caused a reduction of peak FG hERG levels to ∼7 and 9% of initial CG hERG after 4 h of chase. The effect of CHIP on FG hERG was not evident until the 4-h chase point and was most noticeable at 10 h of chase. Again, these results correlate with those obtained under steady state conditions where overexpression of all three proteins reduced the expression of the mature FG form of hERG. Overall, these results suggest that the major effect of the DJAs was to prevent the formation of mature hERG and to increase degradation of the polypeptide consistent with involvement of CHIP.
These ideas were further confirmed by examining the population of polyubiquitylated hERG. HeLa cells stably expressing HA-tagged hERG were treated with lactacystin to prevent degradation of the channel, and hERG was immunoprecipitated with antibody against the tag. High molecular weight species were detected in the precipitate with antibodies against ubiquitin (supplemental Fig. S2), corresponding to hERG modified with polyubiquitin. Consistent with our proposed model, overexpression of DJA1 and DJA2 as well as CHIP caused a clear increase in the amount of polyubiquitylated hERG.
DJA2 and CHIP Reduce the Trafficking of a LQT2 Mutant under Permissive Conditions
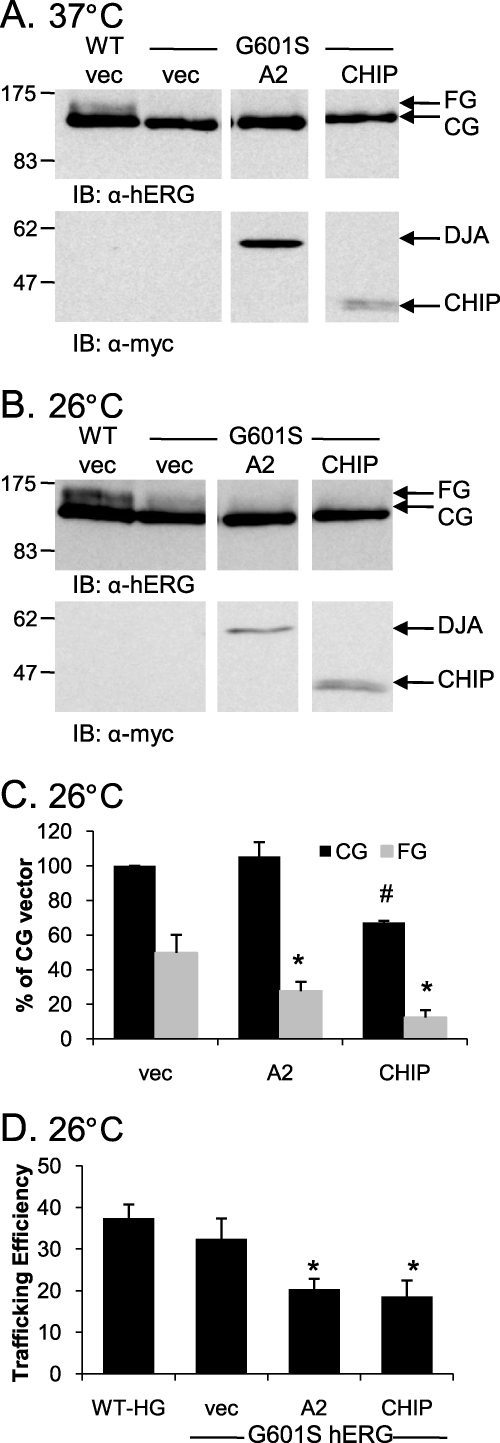
Although wild type hERG can progress to the mature form, clinical LQT2 trafficking mutants of hERG are in many cases unable to do so. It is possible that the degradation promoted by the DJAs and CHIP contributes to the trafficking defect of such mutants. To address this question, we expressed an established hERG trafficking mutant, G601S (42–45), in HEK-293 (GripTite) cells. G601S hERG is unable to traffic to the cell surface in cells at 37 °C, but incubation at 26 °C allows some trafficking of the mutant. HA-tagged G601S hERG was therefore co-transfected with DJA2, which had the greatest effect on WT hERG maturation among the DJAs or with CHIP at 37 or 26 °C.
As expected, there was no trafficking of G601S hERG at 37 °C under any of the conditions tested (Fig. 9A). However, at the trafficking-permissive temperature of 26 °C several interesting results emerged. First, the trafficking of G601S hERG was restored to levels close to those achieved for WT hERG at 37 °C as monitored by the appearance of the FG form (Fig. 9, A and B). Second, the overexpression of DJA2 caused a striking inhibition of G601S hERG maturation at 26 °C, comparable with the effect on WT hERG at 37 °C. Specifically, overexpression of DJA2 resulted in a reduction of the rescued FG G601S hERG band (p < 0.05 relative to vector control) but had no effect on the CG band (Fig. 9C), resulting in a reduced trafficking efficiency relative to control conditions (p = 0.02; Fig. 9D).
FIGURE 9.
hERG G601S in trafficked less efficiently at 26 °C with the overexpression of DJA2 or CHIP. A, HEK-293 (GripTite) cells were transiently transfected with 1 μg of HA-tagged WT hERG or HA-tagged G601S hERG and 2 μg of Myc, Myc-tagged DJA2, or CHIP. The cells were lysed, and protein expression was analyzed as described for Fig. 1. B, cells were treated identically to those in A, except that 24 h post-transfection they were incubated at 26 °C for 24 h prior to lysis. C and D, corresponding mean data (n = 3 experiments/condition). All of the values are normalized to the CG vector (vec) condition after background subtraction. C, #, p = 0.03 versus CG vec; *, p < 0.05 versus FG vector. D, trafficking efficiency is calculated as FG/(CG+FG). *, p < 0.02 for DJA2 and CHIP versus vector G601S. IB, immunoblot.
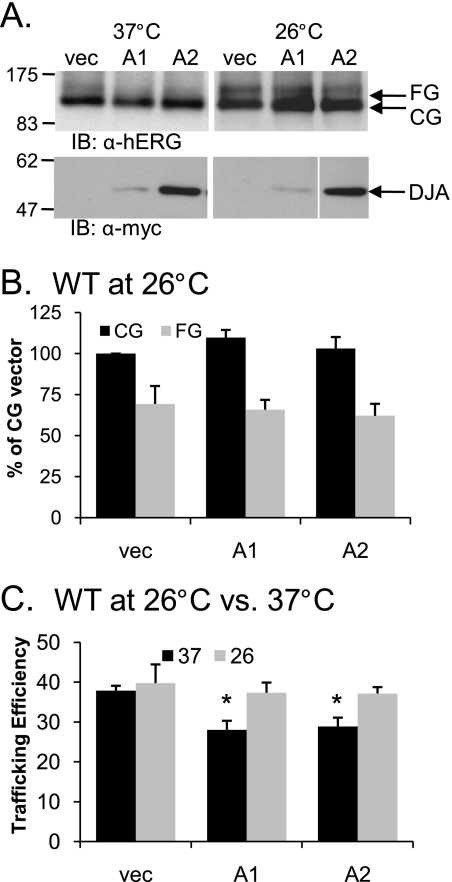
On the other hand, the overexpression of CHIP at 26 °C resulted in a greater reduction of the G601S hERG FG band (p < 0.05) relative to the reduction of the CG band (Fig. 9, B and C). The overall trafficking efficiency of G601S hERG at 26 °C was thus reduced by CHIP (p = 0.02; Fig. 9D), unlike its effect on WT hERG at 37 °C (Fig. 9C). This result suggests that there is differential processing of mutant and WT hERG by CHIP. How this difference is recognized by CHIP, perhaps through Hsc70, is not currently understood, although it seems reasonable that chaperones may differentially recognize the folding intermediates of WT or mutant hERG. Analysis of WT hERG expression at 26 °C in the presence of overexpressed DJA1 and DJA2 produced the unexpected observation that unlike at 37 °C, they had no effect on WT hERG expression or trafficking efficiency (Fig. 10). This observation again indicates that these chaperones along with CHIP recognize their substrates differently under different conditions.
FIGURE 10.
Overexpression of DJA proteins has no effect on WT hERG at 26 °C. A, HEK-293 (GripTite) cells were transiently transfected with 1 μg of HA-tagged WT hERG and 2 μg of Myc alone or Myc-tagged DJA1 or DJA2. The cells were incubated at 37 °C for 24 h post-transfection and were then left at 37 °C or transferred to 26 °C for an additional 24 h. The cells were lysed, and protein expression was analyzed as described for Fig. 1. B and C, corresponding mean data (n = 4 experiments/condition). All of the values are normalized to the CG vector (vec) condition after background subtraction. Trafficking efficiency is calculated as FG/(CG+FG). *, p < 0.05 versus vector at 37 °C. IB, immunoblot.
DISCUSSION
This study identifies the Hsp40/DnaJ type I co-chaperones as key modulators of the degradation pathway of hERG as demonstrated by their ability to reduce hERG trafficking efficiency in a proteasomal-dependent manner. The three human co-chaperones DJA1, DJA2, and DJA4 were similar in their effect, with DJA2 perhaps slightly more effective than DJA1 at promoting hERG degradation. This targeting of hERG for degradation is most probably mediated by preferential degradation of Hsc70-bound polypeptide because of the action of the CHIP E3 ubiquitin ligase. Increased degradation of Hsc70-bound hERG leads to an overall reduction in Hsc70 complexes with hERG and premature destruction of hERG, which otherwise would be exported from the ER to the cell surface (Fig. 9). The DJA proteins as well as CHIP increased the turnover rate of hERG, and moreover, they act differentially on folding intermediates of WT hERG and the LQT2 trafficking mutant G601S. We propose a novel role for the DJA proteins in hERG degradation and suggest that they act at a critical point in quality control.
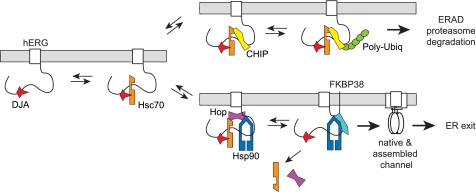
It is likely that polypeptide folding, and not degradation, is the major cellular function of the DJA and Hsc70 chaperone system. However, we propose that hERG folding involves several chaperone-mediated steps (Fig. 11) with Hsc70 acting at an early stage when either folding or degradation is a possible outcome. The first step in this pathway may be the binding of hERG to DJA proteins followed by the recruitment of Hsc70 to the complex as has been demonstrated for the progesterone receptor (20–22) and CFTR (24). The reduced maturation of hERG upon knockdown of DJA1 suggests that this first step is important for hERG folding. Both CHIP, which favors degradation, and Hop, which favors substrate transfer to Hsp90, compete for binding to the same C-terminal site of Hsc70 (27, 46). The outcome of this competition likely dictates which of the two eventually irreversible fates of hERG, ER export or degradation, is favored. Hop binding would recruit Hsp90 to the complex and cause Hsc70 to dissociate. Hop may then be replaced by FKBP38, because these co-chaperones are thought to compete for Hsp90 binding (46, 47). The activities of Hsp90 and FKBP38, which are known to promote proper hERG folding (23), may constitute a final chaperone-mediated folding step before export. However, other later steps such as substrate sorting into vesicles may also be limiting for export.
FIGURE 11.
Model of chaperone-mediated hERG folding and degradation. DJA proteins (DJA1, DJA2, and DJA4) bind hERG and activate binding by Hsc70. CHIP ubiquitylates Hsc70-bound hERG and targets it for degradation. Hop competes with CHIP and recruits Hsp90, which acts with FKBP38 to complete hERG folding.
In contrast, if CHIP binds to Hsc70-hERG complexes, it will ubiquitylate the channel and target it for degradation. We propose that the DJAs promote Hsc70 complexes with hERG folding intermediates, maintaining the availability of hERG for ubiquitylation by CHIP. Because CHIP does not recognize misfolded polypeptide directly, substrates must be presented to it by Hsc70 for ubiquitylation. By extension, co-chaperones such as the DJAs, which regulate polypeptide binding by Hsc70, would be expected to affect degradation by Hsc70-CHIP complexes. Indeed, an in vitro study found that DJA1 and Hsc70 were required for maximal activity of CHIP on CFTR (28). Consistent with this, we show that the Hsc70-activating J domains were required for the DJAs to promote degradation. Moreover, by promoting Hsc70 binding to hERG, the DJA proteins may also restrict the flow of hERG to Hsp90, further shifting the CHIP-Hop balance in favor of CHIP. We expect that this mechanism will apply to other polypeptide substrates in addition to hERG.
Growing evidence suggests that the cytosolic chaperone network maintains a balance between folding and degradation of its various substrates as a general mechanism with CHIP acting as a major link between the two systems (30, 41, 48). For CFTR, which has been extensively studied, not just CHIP but a number of membrane-anchored E3 ubiquitin ligases have been implicated in ERAD, as well as the derlin1/2 and Bap31 extraction complex and cytosolic p97/VCP (49–51). Interaction of the DJAs with one of these components for hERG degradation cannot be ruled out. The Hsc70 nucleotide exchange factors Bag1, Bag2, and HspBP1 have been shown to influence the degradation of polypeptides bound by the chaperone. Bag1 can link Hsc70 directly to the proteasome through its ubiquitin-like domain; Bag2 and HspBP1 appear to bind directly to CHIP and inhibit its activity (25, 26, 52, 53). As a next step in the current research, it will be interesting to investigate the involvement of nucleotide exchange factors or other ubiquitin ligases in hERG processing. Furthermore, the DJAs caused accumulation of HCN2 at the ER, but DJA1 promoted CFTR maturation, so the contribution of other co-chaperones and ubiquitin ligases to the control of trafficking will likely vary between proteins or families of proteins.
Interestingly, the DJA proteins had no effect on the expression level or trafficking efficiency of WT hERG at 26 °C, whereas they had a negative effect on the rescued trafficking of the LQT2 mutant G601S. As mentioned, it seems likely that the DJA proteins act on mutant hERG by altering the chaperone balance to favor degradation as they appear to do for WT hERG at 37 °C. However, the lack of DJA effect on WT hERG at 26 °C may suggest that other factors also influence degradation. It is possible that the folding intermediates of WT and G601S hERG at 26 °C may be structurally different, albeit subtly, and therefore may differentially bind to the DJA proteins. Alternatively, given that both WT and mutant hERG fold more efficiently at 26 °C compared with at 37 °C, but WT more so than G601S (Fig. 9B), some mechanism other than the DJA-Hsc70 system may be the main determinant of degradation and folding of WT hERG at 26 °C. One or more of the membrane-associated ERAD factors may be possible candidates. Also, it is possible that WT hERG is somehow capable of bypassing the quality control system, including recognition by the DJA proteins, at 26 °C, whereas the G601S mutant cannot.
Our results have implications for addressing hERG synthesis and mechanisms of LQT2 at the molecular level by revealing the complexity of the cellular chaperone and degradation pathways. The proposed involvement of the DnaJ family of chaperones on protein aggregation in CAG trinucleotide repeat diseases is a good example of this complexity. Although it was initially proposed that DJA1 overexpression could repress aggregate formation (54), it was later determined that in different cell types DJA1 overexpression in fact increased aggregate formation (55). Simple approaches such as increasing chaperone levels or activities can thus lead to effects that are unexpected and even unwanted. In the case of hERG, this effect was an actual reduction in cell surface expression. Future experiments are needed to explore the mechanisms responsible for such effects in an attempt to better understand hERG folding, trafficking, and degradation. In conclusion, we have revealed the Hsp40/DnaJ family of proteins as important agents in hERG quality control that could potentially be targeted for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Gergely Lukacs and Tsukasa Okiyoneda for the stably transfected cell lines and David Thomas for the CFTR plasmid.
This work was supported by Canadian Institutes of Health Research Grants MOP-86589 and MOP-68825 and by the Canadian Foundation for Innovation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- LQT2
- long QT syndrome type 2
- ER
- endoplasmic reticulum
- ERAD
- ER associated degradation
- CFTR
- cystic fibrosis transmembrane conductance regulator
- Hsc70
- 70-kDa heat shock cognate protein
- Hsp90
- 90-kDa heat shock protein
- WT
- wild type
- FKBP38
- 38-kDa FK506-binding protein
- FG
- fully glycosylated
- CG
- core glycosylated
- GFP
- green fluorescent protein
- CT
- C terminus
- CNBD
- cyclic nucleotide-binding domain
- E3
- ubiquitin-protein isopeptide ligase
- HA
- hemagglutinin
- DMEM
- Dulbecco's modified Eagle's medium
- siRNA
- small interfering RNA
- HCN
- hyperpolarization-activated cyclic nucleotide-gated potassium channel.
REFERENCES
- 1.Sanguinetti M. C., Jiang C., Curran M. E., Keating M. T. (1995) Cell 81, 299–307 [DOI] [PubMed] [Google Scholar]
- 2.Trudeau M. C., Warmke J. W., Ganetzky B., Robertson G. A. (1995) Science 269, 92–95 [DOI] [PubMed] [Google Scholar]
- 3.Warmke J. W., Ganetzky B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curran M. E., Splawski I., Timothy K. W., Vincent G. M., Green E. D., Keating M. T. (1995) Cell 80, 795–803 [DOI] [PubMed] [Google Scholar]
- 5.Anderson C. L., Delisle B. P., Anson B. D., Kilby J. A., Will M. L., Tester D. J., Gong Q., Zhou Z., Ackerman M. J., January C. T. (2006) Circulation 113, 365–373 [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z., Gong Q., Epstein M. L., January C. T. (1998) J. Biol. Chem. 273, 21061–21066 [DOI] [PubMed] [Google Scholar]
- 7.Sanguinetti M. C., Curran M. E., Spector P. S., Keating M. T. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2208–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ficker E., Dennis A. T., Obejero-Paz C. A., Castaldo P., Taglialatela M., Brown A. M. (2000) J. Mol. Cell Cardiol. 32, 2327–2337 [DOI] [PubMed] [Google Scholar]
- 9.Gong Q., Keeney D. R., Molinari M., Zhou Z. (2005) J. Biol. Chem. 280, 19419–19425 [DOI] [PubMed] [Google Scholar]
- 10.Riordan J. R. (2005) Annu. Rev. Physiol. 67, 701–718 [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Venable J., LaPointe P., Hutt D. M., Koulov A. V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., Riordan J. R., Kelly J. W., Yates J. R., 3rd, Balch W. E. (2006) Cell 127, 803–815 [DOI] [PubMed] [Google Scholar]
- 12.Akhavan A., Atanasiu R., Shrier A. (2003) J. Biol. Chem. 278, 40105–40112 [DOI] [PubMed] [Google Scholar]
- 13.Akhavan A., Atanasiu R., Noguchi T., Han W., Holder N., Shrier A. (2005) J. Cell Sci. 118, 2803–2812 [DOI] [PubMed] [Google Scholar]
- 14.Ficker E., Dennis A. T., Wang L., Brown A. M. (2003) Circ. Res. 92, e87–100 [DOI] [PubMed] [Google Scholar]
- 15.Ficker E., Kuryshev Y. A., Dennis A. T., Obejero-Paz C., Wang L., Hawryluk P., Wible B. A., Brown A. M. (2004) Mol. Pharmacol. 66, 33–44 [DOI] [PubMed] [Google Scholar]
- 16.Young J. C., Agashe V. R., Siegers K., Hartl F. U. (2004) Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
- 17.Qiu X. B., Shao Y. M., Miao S., Wang L. (2006) Cell. Mol. Life Sci. 63, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cyr D. M., Langer T., Douglas M. G. (1994) Trends Biochem. Sci. 19, 176–181 [DOI] [PubMed] [Google Scholar]
- 19.Mayer M. P., Bukau B. (2005) Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernández M. P., Chadli A., Toft D. O. (2002) J. Biol. Chem. 277, 11873–11881 [DOI] [PubMed] [Google Scholar]
- 21.Pratt W. B., Toft D. O. (2003) Exp. Biol. Med. 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 22.Cintron N. S., Toft D. (2006) J. Biol. Chem. 281, 26235–26244 [DOI] [PubMed] [Google Scholar]
- 23.Walker V. E., Atanasiu R., Lam H., Shrier A. (2007) J. Biol. Chem. 282, 23509–23516 [DOI] [PubMed] [Google Scholar]
- 24.Meacham G. C., Lu Z., King S., Sorscher E., Tousson A., Cyr D. M. (1999) EMBO J. 18, 1492–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arndt V., Daniel C., Nastainczyk W., Alberti S., Höhfeld J. (2005) Mol. Biol. Cell 16, 5891–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai Q., Qian S. B., Li H. H., McDonough H., Borchers C., Huang D., Takayama S., Younger J. M., Ren H. Y., Cyr D. M., Patterson C. (2005) J. Biol. Chem. 280, 38673–38681 [DOI] [PubMed] [Google Scholar]
- 27.Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 28.Younger J. M., Ren H. Y., Chen L., Fan C. Y., Fields A., Patterson C., Cyr D. M. (2004) J. Cell Biol. 167, 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickner S., Maurizi M. R., Gottesman S. (1999) Science 286, 1888–1893 [DOI] [PubMed] [Google Scholar]
- 30.McDonough H., Patterson C. (2003) Cell Stress Chaperones 8, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Höhfeld J., Cyr D. M., Patterson C. (2001) EMBO Rep. 2, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearl L. H., Prodromou C. (2006) Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 33.Terada K., Mori M. (2000) J. Biol. Chem. 275, 24728–24734 [DOI] [PubMed] [Google Scholar]
- 34.Bhangoo M. K., Tzankov S., Fan A. C., Dejgaard K., Thomas D. Y., Young J. C. (2007) Mol. Biol. Cell 18, 3414–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzankov S., Wong M. J., Shi K., Nassif C., Young J. C. (2008) J. Biol. Chem. 283, 27100–27109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto A., Naito M., Itakura H., Ikemoto S., Asaoka H., Hayakawa I., Kanamori H., Aburatani H., Takaku F., Suzuki H., Kobari Y., Miyai T., Takahashi K., Cohen E. H., Wydro R., Housman D. E., Kodama T. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9133–9137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claycomb W. C., Lanson N. A., Jr., Stallworth B. S., Egeland D. B., Delcarpio J. B., Bahinski A., Izzo N. J., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2979–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White S. M., Constantin P. E., Claycomb W. C. (2004) Am. J. Physiol. Heart Circ. Physiol. 286, H823–H829 [DOI] [PubMed] [Google Scholar]
- 39.Enoki S., Saeki K., Maki K., Kuwajima K. (2004) Biochemistry 43, 14238–14248 [DOI] [PubMed] [Google Scholar]
- 40.Sharma M., Pampinella F., Nemes C., Benharouga M., So J., Du K., Bache K. G., Papsin B., Zerangue N., Stenmark H., Lukacs G. L. (2004) J. Cell Biol. 164, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marques C., Guo W., Pereira P., Taylor A., Patterson C., Evans P. C., Shang F. (2006) FASEB J. 20, 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furutani M., Trudeau M. C., Hagiwara N., Seki A., Gong Q., Zhou Z., Imamura S., Nagashima H., Kasanuki H., Takao A., Momma K., January C. T., Robertson G. A., Matsuoka R. (1999) Circulation 99, 2290–2294 [DOI] [PubMed] [Google Scholar]
- 43.Ficker E., Obejero-Paz C. A., Zhao S., Brown A. M. (2002) J. Biol. Chem. 277, 4989–4998 [DOI] [PubMed] [Google Scholar]
- 44.Rajamani S., Anderson C. L., Anson B. D., January C. T. (2002) Circulation 105, 2830–2835 [DOI] [PubMed] [Google Scholar]
- 45.Delisle B. P., Anderson C. L., Balijepalli R. C., Anson B. D., Kamp T. J., January C. T. (2003) J. Biol. Chem. 278, 35749–35754 [DOI] [PubMed] [Google Scholar]
- 46.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 47.Edlich F., Erdmann F., Jarczowski F., Moutty M. C., Weiwad M., Fischer G. (2007) J. Biol. Chem. 282, 15341–15348 [DOI] [PubMed] [Google Scholar]
- 48.Demand J., Alberti S., Patterson C., Höhfeld J. (2001) Curr. Biol. 11, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 49.Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Koulov A. V., Kellner W. A., Riordan J. R., Balch W. E. (2008) Traffic 9, 1878–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riordan J. R. (2008) Annu. Rev. Biochem. 77, 701–726 [DOI] [PubMed] [Google Scholar]
- 52.Lüders J., Demand J., Höhfeld J. (2000) J. Biol. Chem. 275, 4613–4617 [DOI] [PubMed] [Google Scholar]
- 53.Alberti S., Böhse K., Arndt V., Schmitz A., Höhfeld J. (2004) Mol. Biol. Cell 15, 4003–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stenoien D. L., Cummings C. J., Adams H. P., Mancini M. G., Patel K., DeMartino G. N., Marcelli M., Weigel N. L., Mancini M. A. (1999) Hum. Mol. Genet. 8, 731–741 [DOI] [PubMed] [Google Scholar]
- 55.Wyttenbach A., Carmichael J., Swartz J., Furlong R. A., Narain Y., Rankin J., Rubinsztein D. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.