Abstract
The effect of glycosylation on tissue factor (TF) activity was evaluated, and site-specific glycosylation of full-length recombinant TF (rTF) and that of natural TF from human placenta (pTF) were studied by liquid chromatography-tandem mass spectrometry. The amidolytic activity of the TF·factor VIIa (FVIIa) complex toward a fluorogenic substrate showed that the catalytic efficiency (Vmax) of the complex increased in the order rTF1–243 (Escherichia coli) < rTF1–263 (Sf9 insect cells) < pTF for the glycosylated and deglycosylated forms. Substrate hydrolysis was unaltered by deglycosylation. In FXase, the Km of FX for rTF1–263-FVIIa remained unchanged after deglycosylation, whereas the kcat decreased slightly. A pronounced decrease, 4-fold, in kcat was observed for pTF·FVIIa upon deglycosylation, whereas the Km was minimally altered. The parameters of FX activation by both rTF1–263D-FVIIa and pTFD-FVIIa were identical and similar to those for rTF1–243-FVIIa. In conclusion, carbohydrates significantly influence the activity of TF proteins. Carbohydrate analysis revealed glycosylation on asparagines 11, 124, and 137 in both rTF1–263 and pTF. The carbohydrates of rTF1–263 contain high mannose, hybrid, and fucosylated glycans. Natural pTF contains no high mannose glycans but is modified with hybrid, highly fucosylated, and sialylated sugars.
Keywords: Enzymes/Kinetics, Glycosylation, Carbohydrates, Extrinsic FXase, Fluorogenic Assay, Human Placenta, Recombinant, Tissue Factor Activity
Introduction
Tissue factor (TF)2 is an integral membrane glycoprotein, which upon injury or stimulus is exposed on cellular surfaces to circulating blood (1). TF, the nonenzymatic cofactor, is involved in the extrinsic factor Xase (FXase) formation with FVIIa, which initiates coagulation by activating FIX and FX to the corresponding serine proteases FIXa and FXa, respectively. The TF·FVIIa complex assembly accelerates the generation of FXa by FVIIa by 7 orders of magnitude (2). Serine proteases FIXa and FXa form the intrinsic FXase with FVIIIa and the prothrombinase complex with FVa, respectively. These membrane-bound complexes accelerate thrombin generation, which subsequently converts fibrinogen to fibrin and forms a stable clot (3).
TF is a 263-amino acid glycoprotein containing three domains: the extracellular domain (residues 1–219), the transmembrane domain (residues 220–242), and the cytoplasmic domain (residues 243–263) (4). The extracellular domain contributes to coagulation by binding FVIIa and thereby forming the extrinsic FXase (5). The interaction of TF with FVIIa is calcium-dependent, and the cleavage of both FIX and FX by the complex is membrane-dependent (6, 7). The transmembrane domain serves as an anchor to the cell membrane, whereas the cytoplasmic domain has been suggested to play a role in signal transduction (8, 9). Studies on the distribution of TF have found high activity levels in the brain, lung, heart, and placenta, and precise localization has been assigned to endothelial, monocytic, and vascular adventitia (10, 11). Although TF is expressed in many human tissues, the overall concentration of TF in vivo is extremely low. Surrogate rTFs from various expression systems have been widely used for functional studies; however, the structural and functional comparison of the natural protein with the recombinant forms has been lacking (12).
Posttranslational modifications of TF have also been partially described, and their potential role in activity of the protein has been analyzed (13, 14). Variations in posttranslational modifications of proteins have accentuated the fact that specific modifications, reflective of the production system, may play a role in the activity of proteins (15–17). The most pronounced diversity of posttranslational modification structures is found in Asn-linked (N-linked) and Ser/Thr (O-linked) glycosylation (18, 19). The carbohydrate moieties of some coagulation proteins have been described and shown to have influence on the activity of those proteins (20–24).
Although it is widely accepted that TF is the key initiator of blood coagulation, many controversies related to TF structure and function are continuously debated (25–29). Experimental data used for the understanding of TF structure and function are mostly acquired with rTF proteins. The cDNA deduced amino acid sequence of TF protein indicates that TF contains four potential sites for N-linked glycosylation based on the Asn-X-Ser/Thr motif (18). These sites include Asn-11, Asn-124, and Asn-137 in the extracellular domain and Asn-261 in the cytosolic domain. However, there is no consensus related to the influence of glycosylation on TF activities (12). Some reports state that TF glycosylation has no influence on TF procoagulant activity (30–35). Other observations suggest that glycosylation may be important (36–38).
In the present study, we provide experimental data showing the role of carbohydrates in the function of natural TF. We describe the activity of TF proteins from three different sources, including rTF1–243 expressed in Escherichia coli, rTF1–263 expressed in Sf9 insect cells, and, most importantly, affinity-purified natural human placental TF (pTF). We show that the activity of the natural TF protein is different relative to its recombinant full-length counterpart. In a previous report (39), we have shown that the mobility, by SDS-PAGE, of the recombinant protein is different from that of natural TF purified from human placenta and human monocytes.
We have also focused on the comparative analysis of the carbohydrate composition of the recombinant and natural TF. We describe the carbohydrate moieties of full-length rTF1–263 and natural pTF. The sites of glycosylation and the composition of glycans at each site are identified and described. The study revealed that the composition of carbohydrates at each site varies between the recombinant and natural human pTF. The activity of these proteins before and after deglycosylation revealed that glycosylation does alter TF activity.
EXPERIMENTAL PROCEDURES
Proteins
rTF1–243 (E. coli) was a gift from Drs. S. L. Liu and R. Lundblad (Baxter Healthcare Corp., Duarte, CA). rTF1–263, expressed in Sf9 insect cells, was a gift from Dr. R. Jenny (Haematologic Technologies, Inc.). pTF (Thromborel S) was a gift from Dr. D. Barrow (Behring Diagnostics Inc.). rFVIIa was provided as a gift by Dr. U. Hedner (Novo Nordisk). The concentration of rFVIIa was calculated using a molecular mass of 50,000 Da and an extinction coefficient (E280 nm0.1%) of 1.39 (40). Human FX was isolated from fresh frozen plasma using an anti-FX monoclonal antibody (mAb). The production and characterization of anti-TF-5 mAb and anti-TF-48 mAb was described elsewhere (39). Peptide-N-glycosidase F (PNGase F), native protein deglycosylation kit, proteomics grade trypsin, horseradish peroxidase-labeled goat anti-mouse Ig, and Glu-C enzyme were purchased from Sigma.
Materials
Synthetic phospholipids 1,2-dioleolyl-sn-glycero-3-phospho-l-serine and 1,2-dioleoyl-sn-glycero-3-phosphocholine were purchased from Avanti Polar Lipids Inc. (Alabaster, AL). Spectrozyme FXa was obtained from American Diagnostica Inc. (Greenwich, CT). Fluorogenic substrate 6-(d-FPR)amino-1-naphthalenebutylsulfonamide was synthesized in house (41). Sinapinic acid, dl-dithiothreitol, and iodoacetamide were purchased from Sigma. CHAPS, water, and acetonitrile were purchased from Fisher. Luminex microspheres were purchased from Luminex (Austin, TX). R-Phycoerythrin-streptavidin was purchased from Invitrogen. Formic acid was purchased from VWR (West Chester, PA).
Purification and Quantitation of TF
Human pTF was purified from the commercial thromboplastin, Thromborel S®, by a procedure described previously (39). The contents of 100 vials of Thromborel S were resuspended in Tris-buffered saline, pH 7.4, containing 0.2% Triton X-100 and affinity-purified on Sepharose-coupled anti-TF-5 mAb (2.5 mg of mAb/ml of resin). Proteins were quantitated by fluorescence-linked immunoassay (FLI) by capture on anti-TF-5 mAb-coupled beads, probed with biotinylated anti-TF-48 mAb, and detected with R-phycoerythrin-streptavidin (39). Each point in the FLI measurement is a mean of two experiments ± 1 S.D. The concentration of TF was confirmed by absorbance at 280 nm using an E280 nm0.1% value of 1.5 for rTF1–263 and 1.4 for rTF1–243 and binding stoichiometry to rFVIIa (42, 43).
Enzymatic Removal of Oligosaccharides from TF
For the functional studies, enzymatic deglycosylation of rTF1–263 and pTF was performed under non-denaturing conditions using PNGase F. One μl (1 unit) of the enzyme was added to 10 μg of TF protein, followed by a 4-h incubation at 37 °C. The efficiency of the oligosaccharide removal was assessed by immunoblotting. For carbohydrate analysis, enzymatic deglycosylation of rTF1–263 was performed under non-denaturing conditions using Endo F1, F2, and F3 from the native protein deglycosylation kit for rTF1–263 and PNGase F for pTF. One μl of each endoglycosidase (0.05 units of F1, 0.01 units of F2, and 0.01 units of F3) was added to 10 μg of rTF1–263 protein in the provided reaction buffer, followed by a 4-h incubation at 37 °C. The efficiency of the oligosaccharide removal was assessed by Coomassie Blue staining. The molecular weights of the glycosylated (G) and deglycosylated (D) forms of TF proteins were determined by mass spectrometry.
Immunoblotting
Immunoblotting was performed as described previously (39). Three ng of TF was subjected to a 4–12% acrylamide SDS-PAGE gradient gel, transferred to nitrocellulose membrane, probed with a mix of anti-TF-5 mAb and anti-TF-48 mAb, and detected by horseradish peroxidase-goat anti-mouse Ig.
Molecular Weight Determination by Matrix-assisted Laser Desorption Ionization Time-of-flight (MALDI-TOF) Mass Spectrometry
Mass spectra of the G and D TFs were obtained on a Voyager-DETM PRO MALDI-TOF mass spectrometer (Perceptive Biosystems, Framingham, MA). The spectra were obtained in positive ion mode with delayed extraction equipped with a 337-nm nitrogen laser. A calibration mixture contained rabbit muscle aldolase [M + H]+ = 39,212.28 (average) and bovine serum albumin [M + H]+ = 66,430.09 (average). Ten pmol of G and D TF were spotted onto a stainless steel sample plate (catalog number V700666, Applied Biosystems). One drop of sinapinic acid in 100% methanol was applied to the plate, followed by sample application in sinapinic acid reconstituted in 60% acetonitrile and 0.03% trifluoroacetic acid in water. Experimentally obtained protein masses were compared with the masses calculated from amino acid composition. The mass difference between the experimental (mass spectrometry) and the calculated mass (amino acid composition) provided the mass of posttranslational modifications on the protein.
TF·FVIIa Amidolytic Activity in the Absence of Phospholipids
The previously described procedure consisted of a reaction mixture of 2.0 ml of 0.5 nm FVIIa, varying concentrations of TF (0.05–6.0 nm), and 50 μm SN-17c fluorogenic substrate (44). TF was preincubated with FVIIa for 10 min at 37 °C. Substrate was added, and the rate of substrate hydrolysis as the change in fluorescence intensity over time (5 min) was measured at an excitation wavelength of 350 nm and emission wavelength of 470 nm using a 450-nm cut-off filter in Jobin Yvan-Spex FluoroMax-2 (Instruments S.A. Inc., Edison, NJ). Each point is the mean of two independent experiments ± 1 S.D. Kinetic constants of substrate hydrolysis were determined using the non-linear fitting program GraphPad Prism4 and the Michaelis-Menten equation.
Relipidation of TF
Five nm TF was incubated with 10 μm PCPS (25% 1,2-dioleolyl-sn-glycero-3-phospho-l-serine and 75% 1,2-dioleoyl-sn-glycero-3-phosphocholine) in HBS and 5 mm CaCl2 for 30 min at 37 °C, followed by extensive dialysis in HBS and 5 mm CaCl2 to remove detergent (45). The relipidated TFs were quantitated by FLI.
Extrinsic FXase Assay
TF (0.1 nm) was incubated with 5 nm rFVIIa for 10 min at 37 °C in HBS, 5 mm CaCl2, pH 7.4, and varying concentrations of FX were added (0.01–8.0 μm). At selected time points (0–5 min), aliquots were quenched in 20 mm EDTA in HBS (pH 7.4) containing 0.2 mm Spectrozyme Xa, and substrate hydrolysis was monitored by the change in absorbance at 405 nm using a Molecular Devices (Menlo Park, CA) Vmax spectrophotometer. The FXa generation rate was calculated from a standard curve prepared by serial dilutions of FXa. Each point is the mean of two independent experiments ± 1 S.D. Michaelis-Menten kinetic constants were calculated using the GraphPad Prism4 program.
Enzymatic Digest of TF Proteins
Samples of G and D TF proteins were subjected to enzymatic digestion using trypsin. Three μg of rTF1–243 and rTF1–263 and 1.5 μg of pTF in 50 mm ammonium bicarbonate at pH 8.3 were reduced with 10 mm dl-dithiothreitol during 1 h of incubation at 57 °C. The protein was alkylated with 17 mm iodoacetamide in the dark at room temperature for 20 min. Trypsin was added to each protein solution at a 1:50 enzyme/protein ratio, and the digestion mixture was incubated at 37 °C overnight. Digestion was stopped upon the addition of formic acid to a final concentration of 1.25%. A second sample of G and D TF proteins was subjected to enzymatic digestion using GluC. Three μg of rTF1–243 and rTF1–263 and 1.5 μg of pTF were reduced and alkylated as described for the trypsin cleavage. The buffer pH was reduced to 4.0, and the proteins were incubated with Glu-C enzyme at 37 °C overnight.
LC-MS/MS Analysis of TF Proteolytic Peptides
Liquid chromatography/mass spectrometry (LC/MS) analysis of 1 pmol of TF peptides was carried out using a Shimadzu SIL-20AC autosampler with two LC-20AD pumps (Columbia, MD) and a Thermo LTQ mass spectrometer (San Jose, CA). The LC was connected to a 100-μm inner diameter by 150-mm length nanospray column that was pulled and packed in house using a stainless steel pressure bomb and Magic C18 reverse phase material (5-μm beads, Michrom, Auburn, CA). We used a slightly modified form of the two-split configuration described previously (46). Instead of capillaries, we used a 1 × 150-mm Atlantis C18 column (Waters, Milford, MA) to act as a counter restriction to the nanospray column. Total flow from the pumps was 80 μl/min during the loading period and 40 μl/min during the running period. The resulting flow through the nanospray column was 1 μl/min during the load and 500 nl/min during the running period. We used water (solvent A) and acetonitrile (solvent B) with 0.1% formic acid in both solvents to load and elute the sample from the nanospray column. The LC gradient consisted of the following: 97% A and 3% B for 20 min, 40% B at 57 min, and 60% B at 59 min. Total time for the sample analysis was 65 min. Due to the presence of CHAPS in the TF samples, each sample analysis was followed by a 5-μl injection of 50% isopropyl alcohol (Fisher) to “wash” the column. The isopropyl alcohol injection was carried out in 60% solvent B followed by a decrease to 3% in 5 min; total time for the column wash was 25 min. The LTQ mass spectrometer was tuned according to the manufacturer's recommended procedure. The nanospray column was placed in front of the mass spectrometer using an in-house built stage. The position of and voltage applied to the nanospray column were optimized to achieve a consistent spray. We applied 1.9 kV to the vent line on the tee at the head of the column. The transfer tube temperature was set to 150 °C. Our MS method consisted of a full MS scan from 250 to 2,000 Da in centroid mode, a zoom scan, and an MS/MS scan on the most intense ion from the full MS scan. We set the MS/MS parameters to the following: signal threshold = 6,000, isolation width = 2, normalized collision energy = 35, activation Q = 0.250, and activation time = 30. We chose the following data-dependent settings: recount = 2, repeat duration = 60 s, exclusion list size = 50, and exclusion duration = 100 s.
LC-MS/MS data files were viewed using Xcalibur Qualbrowser version 2.0. From Qualbrowser, we looked at the full MS in both the density map window and the spectrum window. The density map window allowed us to identify clusters of glycopeptides as described previously (47, 48). We also viewed the MS/MS spectra of the glycopeptides in Qualbrowser to identify glycan composition based on specific mass losses of different sugars as described previously (49–52).
Non-glycosylated and deglycosylated peptides were identified using the Sequest algorithm (53, 54). We made a reduced data base that only contained various forms of the TF protein: the TF protein predicted amino acid sequence and the TF protein where Asn was changed to Asp in NX(S/T) motifs. Sequest used this data base to identify TF peptides in the glycosylated and deglycosylated samples. We included the following possible modifications on amino acids in the Sequest search: carboxyamidomethylation of cysteine (57.05 Da); phosphorylation of serine, threonine, and tyrosine (79.97 Da); and the addition of a GlcNAc or a fucosylated (Fuc) N-acetyl glucosamine on Asn (203.20 and 349.34 Da, respectively). We used the following filters for high confidence identifications: ΔCN = 0.100; Xcorr versus charge state = 1.50, 2.00, and 2.50 for singly, doubly, and triply charged species, respectively; and peptide probability = 0.001. All Sequest-identified peptides were manually inspected to confirm correct b and y ion designations and to rule out possible false positive identifications.
RESULTS
Enzymatic Removal of Oligosaccharides from TF
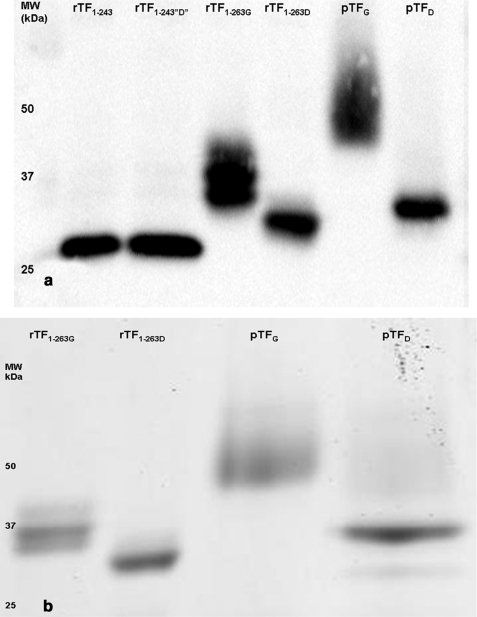
Fig. 1a shows the mobility of G and D TF proteins by immunoblotting. All TFs presented in this figure were treated with PNGase F and used in the functional assays. Fig. 1b shows the mobility of G and D forms stained with Coomassie Blue. In the experiments of this figure the rTF1–263 was deglycosylated using the Endo F enzymes and was used for MALDI-TOF and carbohydrate analysis by LC-MS/MS. The pTF in Fig. 1b was deglycosylated with PNGase F. Our first attempt at deglycosylation of both rTF1–263 and pTF was done using the native protein deglycosylation kit, which includes the Endo F1, F2, and F3 enzymes. The specificity of the endoglycosidases is as follows. Endo F1 cleaves Asn-linked high mannose (Man) and hybrid but not complex carbohydrates, and Endo F2 cleaves oligomannose and biantennary complex oligosaccharides, whereas Endo F3 is specific for the cleavage of core fucosylated biantennary and triantennary complex sugars. All three Endo F enzymes cleave between the two GlcNAc residues in the diacetylchitobiose core of the carbohydrate generating an Asn-linked GlcNAc-truncated sugar molecule (55). Fig. 1, a and b, shows rTF1–263G after deglycosylation (rTF1–263D) with PNGase F and the Endo F enzymes, respectively. The increased mobility of rTF1–263D in both figures suggests that deglycosylation took place.
FIGURE 1.
Immunoblot (a) and Coomassie Blue staining (b) of TF proteins. a, 2 ng of reduced TF proteins, rTF1–243, rTF1–243“D”, rTF1–263, rTF1–263D, pTF, and pTFD, were electrophoresed on a 4–12% linear gradient gel and immunoblotted with anti-TF-5 and anti-TF-48 mAb. The corresponding molecular mass (kDa) as determined from the molecular mass standard is indicated to the left of the immunoblot. All proteins were treated with PNGase F enzyme and used in the FVIIa·TF amidolytic activity assay and extrinsic FXase assay. b, 3 μg of reduced TF proteins, rTF1–263, rTF1–263D, pTF, and pTFD, were electrophoresed on 4–12% linear gradient gel and stained with Coomassie Blue. Both the immunoblot and staining gel demonstrate the increase in the apparent mobility of the rTF1–263D and pTFD relative to the glycosylated forms, rTF1–263G and pTFG.
Purification of pTF resulted in a protein with a higher apparent molecular weight and heterogeneity as compared with its full-length recombinant counterpart rTF1–263G (Fig. 1, a and b). The natural TF purified from human placenta is comparable in mobility and heterogeneity with that purified from human monocytes and is different from the recombinant forms (39). The mobility of pTF after treatment with Endo F enzymes remained the same even after prolonged exposure (data not shown). We therefore proceeded to treat pTF with PNGase F. PNGase F is specific for oligomannose, hybrid, and complex type glycans. The enzyme releases the Asn-linked carbohydrate, converting the Asn into Asp (56). Deglycosylation of pTFG with the enzyme (pTFD) resulted in a higher mobility and an increased homogeneity relative to the glycosylated form (Fig. 1, a and b). After deglycosylation, pTFD had a slightly lower mobility than rTF1–263D. The overall mobility of the proteins increased in the order rTF1–263D > pTFD > rTF1–263G > pTFG. The difference in mobility between rTF1–263D and pTFD may be attributed to other posttranslational modifications besides glycosylation (12).
Treatment of rTF1–243 with PNGase F, denoted as rTF1–243“D”, did not affect the mobility of the protein, consistent with the knowledge that rTF1–243 is not glycosylated as expected for a protein produced in E. coli (57). The presence and absence of carbohydrates on each protein, rTF1–243, rTF1–243“D”, rTF1–263G, rTF1–263D, pTFG, and pTFD, was also evaluated by carbohydrate staining, which showed positive staining for rTF1–263G and pTFG, and no staining was observed for rTF1–243, rTF1–243“D”, rTF1–263D, and pTFD (data not shown).
Quantitation of TF Proteins
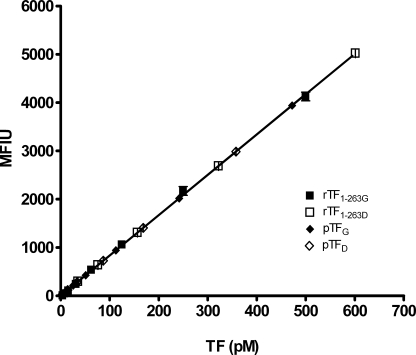
The concentration of the G and D proteins was measured by FLI (39), using rTF1–243 as a calibrator for the glycosidase-treated rTF1–243 and rTF1–263G as a calibrator for rTF1–263D, pTFG, and pTFD. The affinity of the anti-TF mAbs, used in the immunoassay, demonstrated equivalent recognition of the G and D forms of both rTF1–263 and pTF (Fig. 2).
FIGURE 2.
Fluorescence-linked immunoassay of TF proteins. Varying concentrations of rTF1–263G, rTF1–263D, pTFG, and pTFD were captured on anti-TF-5 mAb-coated beads and probed with biotinylated anti-TF-48 mAb and R-phycoerythrin-streptavidine. The immunoassay detects both the G and D forms of TF proteins. Each point is recorded as a mean fluorescence intensity unit (MFIU) and is a mean of two experiments ± 1 S.D.
Determination of Molecular Weights by MALDI-TOF
Table 1 presents a comparison of the molecular masses of TF proteins determined by SDS-PAGE and MALDI-TOF. The backbone molecular masses calculated from the amino acid composition are also shown. The molecular masses of all three forms of TF based on mass spectrometry do not coincide with their mass calculated by amino acid composition and vary from 377 to 6,605 Da. The mass of rTF1–243 is unaltered before and after treatment with PNGase F with a difference of 377 Da from that calculated by amino acid composition. Treatment of rTF1–263 with Endo F enzymes reduced the mass by 2,994 Da, which differs from the calculated mass of 29,600 Da by 610 Da. This mass difference possibly accounts for the GlcNAc residues that remain on the Asn after deglycosylation with the Endo F enzymes (55). The mass of the natural human pTFG is 36,179 Da; however, we were not able to determine the mass of the rTF1–263 or the mass of pTF after deglycosylation with the PNGase F enzyme. Any other deviations between the calculated masses and masses of glycosidase-treated proteins obtained by MALDI-TOF are probably related to glycation or other posttranslational modifications, some of which were described previously (12).
TABLE 1.
Molecular masses (Da) of TF proteins
| TF species | Source | Calculateda | MALDI-TOF | Differenceb | SDS-PAGE |
|---|---|---|---|---|---|
| Da | Da | Da | Da | ||
| rTF1–243 | E. coli | 27,423 | 27,800 | 377 | 31,000 |
| rTF1–243“D” | E. coli | 27,423 | 27,800 | 377 | 31,000 |
| rTF1–263G | Sf9 | 29,592 | 33,196 | 3,604 | 37,000 |
| rTF1–263D | Sf9 | 29,592 | 30,202 | 610c | 33,000 |
| pTFG | Human placenta | 29,592 | 36,179 | 6,605 | 45,000 |
| pTFD | Human placenta | 29,592 | ND | ND | 34,000 |
a Calculated from amino acid composition.
b Difference between calculated mass by amino acid composition and that obtained by MALDI-TOF.
c This mass was determined on rTF1–263 deglycosylated with EndoF enzymes.
TF·FVIIa Amidolytic Activity
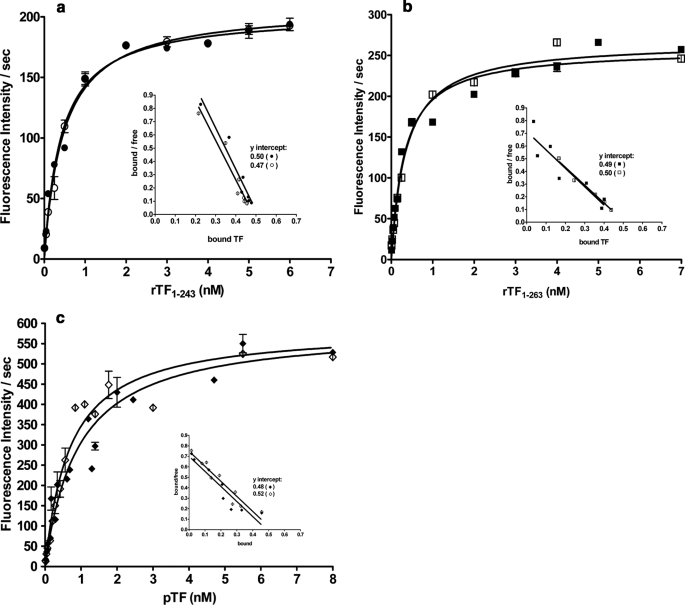
The affinities of the TF proteins for FVIIa in the absence of phospholipids were evaluated in the fluorogenic assay (Fig. 3) and are summarized in Table 2. All Kd(app) values are in the range of those previously reported (43). Treatment of rTF1–243 with PNGase F (Fig. 3a) does not alter the Kd(app) for FVIIa or the maximum rate of substrate hydrolysis by the complex. Similarly, deglycosylation of rTF1–263G (Fig. 3b) does not affect the affinity of the deglycosylated protein for FVIIa or the Vmax of the complex. Similarly, after deglycosylation, pTF does not change in either the Kd(app) or Vmax (Fig. 3c). All TF forms exhibit a functional 1:1 stoichiometry with FVIIa (Fig. 3, a–c, insets, and Table 2). Thus, functional concentrations are consistent with the mass concentrations of TF proteins determined by the immunoassay. The catalytic efficiency of various TF forms increases in the order rTF1–243 < rTF1–263 < pTF and is not changed by deglycosylation.
FIGURE 3.
FVIIa·TF amidolytic activity. Varying concentrations of rTF1–243 (●) and rTF1–243“D” (○) in a, rTF1–263G (■) and rTF1–263D (□) in b, and pTF (♦) and pTFD (◇) in c were incubated with FVIIa (0.5 nm) in HBS, 0.1% polyethylene glycol, 2 mm CaCl2, pH 7.4, for 10 min. Fluorogenic substrate was added (50 μm), and the rate of substrate hydrolysis was recorded. The inset in each graph represents the calculated 1:1 stoichiometry of the TF·FVIIa complex. Each point is a mean of two experiments ± 1 S.D.
TABLE 2.
Binding of TF to FVIIa in a fluorogenic assay
| TF species | Kd(app) | Vmaxa | Stoichiometry (TF/FVIIa) |
|---|---|---|---|
| nm | pm/s | ||
| rTF1–243 | 0.41 ± 0.40 | 59.7 ± 1.2 | 1.1:1.0 |
| rTF1–243“D” | 0.47 ± 0.03 | 61.2 ± 1.0 | 0.9:1.0 |
| rTF1–263G | 0.31 ± 0.02 | 75.8 ± 1.6 | 0.9:1.0 |
| rTF1–263D | 0.35 ± 0.02 | 78.3 ± 1.4 | 1.0:1.0 |
| pTFG | 0.92 ± 0.15 | 173.1 ± 8.4 | 0.9:1.0 |
| pTFD | 0.69 ± 0.09 | 172.8 ± 10.0 | 1.0:1.0 |
a Maximum rate of substrate fluorogenic hydrolysis at 0.5 nm FVIIa and saturating TF.
Factor Xa Generation
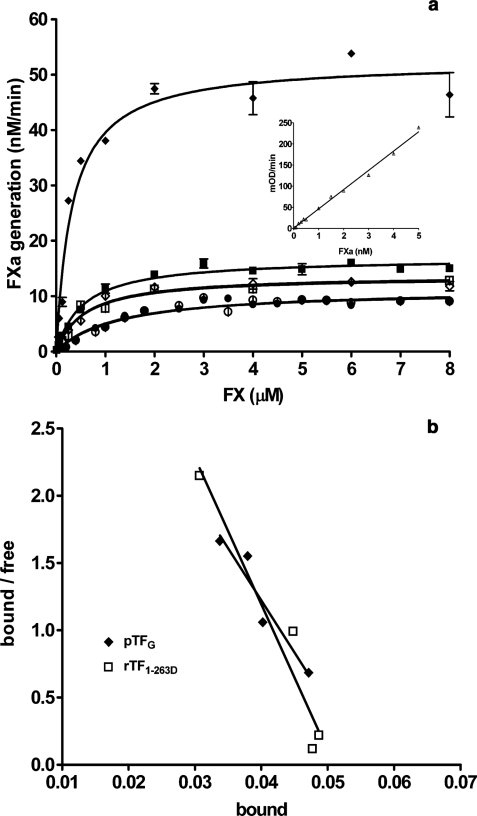
As demonstrated in Fig. 4a, natural human pTF is (5-fold) more active than any rTF. Comparison of rTF1–243 and rTF1–243“D” shows that treatment of the protein with the glycosidase does not alter the affinity (Km) or the catalytic efficiency (kcat) of the complex (Table 3). Deglycosylation of rTF1–263 does not alter the Km but results in a slight decrease in the kcat (from 2.8 to 2.2 s−1). A slight increase in Km was observed when pTF was deglycosylated (from 0.32 to 0.57 μm). In contrast, deglycosylation caused a pronounced reduction in the kcat (from 8.7 to 2.3 s−1). This decrease in the kcat and Km of the pTFD-FVIIa complex resulted in ∼6-fold reduction in the second order rate constant (kcat/Km) of the complex. The stoichiometry of TF·FVIIa was unchanged (Table 3). When the kinetic constants of the three TF proteins were compared, an overall increase in Km of ∼3-fold was observed in the order pTFG < rTF1–263G < rTF1–243. The catalytic efficiency (kcat) increased in the reversed order rTF1–243 < rTF1–263 < pTF, with pTF being 5-fold more active than the most active recombinant counterpart (Table 3). These differences lead to the 18-fold difference in the second order rate constant between rTF1–243 and pTFG. After deglycosylation, the parameters of FX activation by both rTF1–263D and pTFD were almost identical (Km = 0.57 and 0.57 μm, and kcat = 2.2 and 2.3 s−1, respectively). The stoichiometry of the G and D forms of relipidated TFs was confirmed to be 1:1, as presented in Fig. 4b for pTFG and rTF1–263D.
FIGURE 4.
Extrinsic FXase assay (a) and stoichiometry of relipidated TF (b). a, varying concentrations of FX were incubated with FVIIa (5 nm) in complex with rTF1–243 (●), rTF1–243“D” (○), rTF1–263G (■), rTF1–263D (□), pTF (♦), and pTFD (◇) (0.1 nm). FXa generation was measured and calculated from the FXa standard curve (inset). b, the stoichiometry of relipidated TF was analyzed in the TF·FVIIa amidolytic assay. Varying concentrations of rTF1–263D (□) and pTFG (♦) were incubated with FVIIa (0.05 nm) in HBS, 0.1% polyethylene glycol, 2 mm CaCl2, pH 7.4, for 10 min. Fluorogenic substrate was added (50 μm), and the rate of substrate hydrolysis was recorded. The graph represents the calculated 1:1 stoichiometry of the TF·FVIIa complex. Each point is a mean of two experiments ± 1 S.D.
TABLE 3.
Activity of TF·FVIIa complex in the extrinsic FXase assay
| TF species | Km | kcat | kcat/Km | Relative activity | Stoichiometry (TF/FVIIa) |
|---|---|---|---|---|---|
| μm | s−1 | μm−1s−1 | |||
| rTF1–243 | 1.19 ± 0.22 | 1.8 | 1.5 | 1.0 | 1.0: 1.0 |
| rTF1–243“D” | 1.31 ± 0.26 | 1.9 | 1.4 | 0.9 | 1.0: 1.0 |
| rTF1–263G | 0.54 ± 0.05 | 2.8 | 5.2 | 3.5 | 1.0: 1.0 |
| rTF1–263D | 0.57 ± 0.17 | 2.2 | 3.8 | 2.5 | 0.9: 1.0 |
| pTFG | 0.32 ± 0.04 | 8.7 | 26.8 | 17.9 | 0.9: 1.0 |
| pTFD | 0.57 ± 0.07 | 2.3 | 4.0 | 2.7 | 0.9: 1.0 |
MS Analyses of TF Peptides
Digestion of full-length TF with trypsin should generate 30 fragments (supplemental Table 1). Included in the table are fragments generated by a Glu-C digest of fragment 10 (marked with an asterisk). These smaller peptides obtained from Glu-C digestion allowed for the increased recovery and identification of fragment 10. According to the previously determined amino acid sequence of a mature TF protein (4), the first 10 residues of the intact N terminus are Ser, Gly, Thr, Thr, Asn, Thr, Val, Ala, Ala, and Tyr. Amino acid sequence analysis of the rTF1–263 by Edman degradation confirms this sequence.3 The Edman degradation analysis also shows that the majority of the recombinant protein exists in a truncated form (∼80%) with the first two residues missing, resulting in the following sequence: Thr, Thr, Asn, Thr, Val, Ala, Ala, Tyr. Similarly, our MS analysis revealed that 75% exists as the truncated form. The natural pTF exists mainly in the full-length form (69%). The truncated form of TF was previously observed and reported (4, 31). No evidence of leader peptides or fragments were identified by Edman degradation or by MS analysis. The N- and C-terminal peptides of rTF1–263 and pTF were recovered and identified by MS. Sequest search result data identify the recovery of the N- and C-terminal peptides with high confidence, suggestive of the correct identity of the two terminals. Manual inspection of peptides also confirms the absence of any adducts. Likewise, the N terminus of rTF1–243 was recovered and identified. The truncated form was identified by MS in ratios similar to those of rTF1–263. The C terminus of rTF1–243, which constitutes the transmembrane domain of TF, was not recovered. The recovery of this peptide was not anticipated, based on our hypothesis that this very hydrophobic peptide would not elute from the column under our conditions or would elute with CHAPS, which would greatly diminish the ionization and identification of this peptide. rTF1–243 was constructed and purified as described previously (8, 31). Supplemental Table 1 presents the cleavage sites, peptides generated, and hypothetical masses of the peptides (acquired from the ExPASy Proteomics Server). The table presents the locations of the glycosylated sites in TF as identified in our study. It also serves as a comparison between the hypothetical mass of a non-glycosylated peptide versus the carbohydrate composition and mass of the corresponding glycopeptide (Tables 4 and 5). All four potential sites for N-linked glycosylation containing the Asn-X-Ser/Thr/Cys motif (boldface letters), where X could be any amino acid except Pro, are found on distinct peptides. Only one site is in the cytosolic domain, Asn-261, whereas the remaining three sites are in the extracellular domain. The TF peptides recovered by MS and identified by Sequest search results are marked with numbers 1, 2, and 3, which correspond to peptides recovered in rTF1–243, rTF1–263, and pTF, respectively (supplemental Table 1). These peptides accounted for 77.4% of the amino acid sequence coverage for TF1–243, 73.7% for TF1–263, and 70.7% for pTF. Small peptides and single amino acids were identified as miscleaved products attached to the larger, neighboring peptide. For example, peptides SK and DVK were identified as miscleaved products on the peptide CFYTTDTECDLTDEIVK. Fragment 10 in supplemental Table 1 was too large for the mass range used in our method. However, digestion of TF with GluC (marked with an asterisk) produced Sequest-identified fragments within the sequence of fragment 10. The addition of these GluC peptides increased our amino acid coverage. Our inability to obtain 100% coverage is not surprising. We also hypothesize that the other fragments not identified by MS were too hydrophilic (fragment 17) and not retained by the C18 column, resulting in these peptides eluting from the column with the other non-retained species in the sample, such as the salt in the digestion buffer. Our experience has shown that non-retained peptides also experience poor ionization due to the coelution with the digestion buffer and a decrease in sensitivity due to an inability to be concentrated on the head of the column (data not shown).
TABLE 4.
Carbohydrate composition of glycopeptides for rTF1–263
| Site | Composition | m/z | z | Glycopeptide |
|---|---|---|---|---|
| Da | ||||
| Asn-11 | GlcNAc1 | 915 | 2 | 1,829 |
| Fuc1GlcNAc1 | 988 | 2 | 1,975 | |
| (Core)Fuc1 | 1,333 | 2 | 2,665 | |
| (Core)Man1 | 1,504 | 2 | 3,007 | |
| (Core)Man2 | 1,585 | 2 | 3,169 | |
| (Core)Man3 | 1,666 | 2 | 3,331 | |
| Asn-124 | GlcNAc1 | 633 | 2 | 1,265 |
| (Core)HexNAc1 | 1,078 | 2 | 2,155 | |
| (Core)Hex1HexNAc1 | 1,160 | 2 | 2,319 | |
| (Core)Hex2HexNAc1 | 1,241 | 2 | 2,481 | |
| (Core)Hex2HexNAc2 | 1,342 | 2 | 2,683 | |
| (Core)Man2 | 1,139 | 2 | 2,277 | |
| (Core)Man3 | 1,220 | 2 | 2,439 | |
| (Core)Man4 | 1,301 | 2 | 2,601 | |
| (Core)Man5 | 1,382 | 2 | 2,763 | |
| (Core)Man6 | 1,463 | 2 | 2,925 | |
| (Core)Man7 | 1,544 | 2 | 3,087 | |
| (Core)Man8 | 1,625 | 2 | 3,249 | |
| Asn-137 | GlcNAc1 | 561 | 2 | 1,121RAsn |
| 584 | 2 | 1,167Asn | ||
| (Core)Fuc1 | 663 | 2 | 1,325RAsn | |
| 1,002 | 2 | 2,003Asn | ||
| (Core)Fuc1HexNAc1 | 720 | 3 | 2,158RAsn | |
| 1,104 | 2 | 2,207Asn | ||
| (Core)Fuc1HexNAc2 | 788 | 3 | 2,362RAsn | |
| (Core)Fuc1Hex1HexNAc2 | 856 | 3 | 2,566RAsn | |
| 1,287 | 2 | 2,573Asn | ||
| (Core)Man2 | 911 | 3 | 2,731RAsn | |
| 1,091 | 2 | 2,181Asn | ||
| (Core)Man3 | 780 | 3 | 2,338RAsn | |
| 1172 | 2 | 2,343Asn | ||
| (Core)Man4 | 834 | 3 | 2,501RAsn | |
| 1,253 | 2 | 2,505Asn | ||
| (Core)Man5 | 888 | 3 | 2,662RAsn | |
| 1,334 | 2 | 2,667Asn | ||
| (Core)Man6 | 942 | 3 | 2,770RAsn | |
| 1,415 | 2 | 2,829Asn | ||
| 996 | 3 | 2,986RAsn |
TABLE 5.
Carbohydrate composition of glycopeptides for pTF
| Site | Composition | m/z | z | Glycopeptide |
|---|---|---|---|---|
| Da | ||||
| Asn-11 | (Core)Fuc1Hex1HexNac2 | 1,619 | 2 | 3,237 |
| (Core)Fuc1Hex2HexNac3 | 1,801; 1,201 | 2; 3 | 3,601 | |
| (Core)Fuc1Hex2HexNac1SA1 | 1,744 | 2 | 3,487 | |
| (Core)Fuc1Hex1HexNac2SA1 | 1,764 | 2 | 3,527 | |
| (Core)Fuc1Hex2HexNac2SA1 | 1,845; 1,230 | 2; 3 | 3,689 | |
| (Core)Fuc1Hex2HexNac3SA1 | 1,946; 1,297 | 2; 3 | 3,890 | |
| (Core)Fuc1Hex2HexNac2SA2 | 1,990; 1,327 | 2; 3 | 3,979 | |
| (Core)Fuc1Hex2HexNac3SA2 | 1,395 | 3 | 4,183 | |
| (Core)Fuc1Hex3HexNac4SA1 | 1,419 | 3 | 4,255 | |
| (Core)Fuc1Hex3HexNac3SA2 | 1,449 | 3 | 4,345 | |
| (Core)Fuc1Hex3HexNac3SA3 | 1,546 | 3 | 4,636 | |
| (Core)Fuc1Hex3HexNac3SA4 | 1,643 | 3 | 4,927 | |
| (Core)Fuc1Hex3HexNac4SA4 | 1,711 | 3 | 5,131 | |
| (Core)Fuc1Hex4HexNac4SA4 | 1,764 | 3 | 5,290 | |
| Asn-124 | (Core)Hex1 | 1,058 | 2 | 2,115 |
| (Core)Hex2 | 1,139 | 2 | 2,277 | |
| (Core)Hex1HexNac1 | 1,160; 774 | 2; 3 | 2,320 | |
| (Core)Hex2HexNac1 | 1,240; 828 | 2; 3 | 2,480 | |
| (Core)Hex1HexNac2 | 1,263 | 2 | 2,525 | |
| (Core)Hex3HexNac1 | 1,321; 882 | 2; 3 | 2,642 | |
| (Core)Hex2HexNac2 | 1,342; 895 | 2; 3 | 2,683 | |
| (Core)Hex3HexNac2 | 1,423; 949 | 2; 3 | 2,845 | |
| (Core)Hex4HexNac2 | 1,504; 1,003 | 2; 3 | 3,007 | |
| (Core)Hex3HexNac3 | 1,524; 1,017 | 2; 3 | 3,048 | |
| (Core)Hex4HexNac3 | 1,071 | 3 | 3,211 | |
| (Core)Hex1HexNac1SA1 | 1,305; 871 | 2; 3 | 2,610 | |
| (Core)Hex2HexNac1SA1 | 1,366; 924 | 2; 3 | 2,771 | |
| (Core)Hex3HexNac1SA1 | 1,467; 978 | 2;3 | 2,933 | |
| (Core)Hex2HexNac2SA1 | 1,489; 993 | 2; 3 | 2,976 | |
| (Core)Hex3HexNac2SA1 | 1,569; 1,046 | 2; 3 | 3,135 | |
| (Core)Hex3HexNac3SA1 | 1,671; 1,114 | 2; 3 | 3,340 | |
| (Core)Hex3HexNac2SA2 | 1,143 | 3 | 3,427 | |
| (Core)Hex3HexNac3SA2 | 1,211 | 3 | 3,631 | |
| Asn-137 | (Core)Fuc1HexNac1 | 1,183 | 2 | 2,365Asn |
| (Core)Fuc1Hex1HexNac1 | 1,263 | 2 | 2,525Asn | |
| (Core)Fuc1Hex1HexNac2 | 1,286 | 2 | 2,571RAsn | |
| 1,364 | 2 | 2,727RAsn | ||
| (Core)Fuc1Hex2HexNac2 | 1,367 | 2 | 2,733Asn | |
| 1,445 | 2 | 2,889RAsn | ||
| (Core)Fuc1Hex2HexNac3 | 1,469 | 2 | 2,937Asn | |
| 1,546 | 2 | 3,091RAsn | ||
| (Core)Fuc1Hex2HexNac3 | 1,628 | 2 | 3,255RAsn | |
| (Core)Fuc1Hex1HexNac1SA1 | 1,408 | 2 | 2,815RAsn | |
| (Core)Fuc1Hex2HexNac2SA1 | 1,513; 1,009 | 2; 3 | 3,025Asn | |
| 1,590; 1,061 | 2; 3 | 3,180RAsn | ||
| (Core)Fuc1Hex2HexNac3SA1 | 1,614; 1,077 | 2; 3 | 3,229Asn | |
| 1,129 | 3 | 3,383RAsn | ||
| (Core)Fuc1Hex2HexNac2SA2 | 1,658; 1,106 | 2; 3 | 3,315Asn | |
| 1,158 | 3 | 3,472RAsn | ||
| (Core)Fuc1Hex3HexNac3SA2 | 1,131 | 3 | 3,391Asn | |
| 1,773 | 2 | 3,545RAsn | ||
| (Core)Fuc1Hex1HexNac3SA2 | 1,279 | 3 | 3,835RAsn | |
| (Core)Fuc1Hex2HexNac3SA2 | 1,226 | 3 | 3,676RAsn | |
| (Core)Fuc1Hex3HexNac4SA1 | 1,199 | 3 | 3,596Asn | |
| 1,251 | 3 | 3,750RAsn | ||
| (Core)Fuc1Hex2HexNac2SA3 | 1,803 | 2 | 3,605Asn | |
| (Core)Fuc1Hex3HexNac4SA2 | 1,296 | 3 | 3,886Asn | |
| 1,347 | 3 | 4,039RAsn | ||
| (Core)Fuc1Hex3HexNac3SA3 | 1,378 | 3 | 4,132RAsn | |
| (Core)Fuc1Hex4HexNac4SA3 | 1,401 | 3 | 4,201RAsn | |
| (Core)Fuc1Hex4HexNac5SA2 | 1,469 | 3 | 4,405RAsn |
Carbohydrate Analysis
To determine the presence of carbohydrates at potential glycosylation sites of TF proteins, reduced and alkylated peptides from both the G and D TF proteins were manually inspected using density maps. Supplemental Fig. 1 shows the density maps of rTF1–263G in A and rTFD in B. A number of doubly or triply charged peptide ions observed in the density map of the G form disappeared from the spectrum of the D form, suggesting that these peptides were glycosylated.
To determine the composition of carbohydrates at potential glycosylation sites, the MS/MS spectra of ions seen in supplemental Fig. 1A but not in supplemental Fig. 1B were manually investigated. For example, supplemental Fig. 2 shows the MS/MS scan for an ion with m/z of 1,382 that appeared in rTF1–263G but not in rTF1–263D. Two sets of fragmentation ladders can be distinguished. One ladder represents the oxonium ions that fragment off of the glycopeptides during collision-induced dissociation and show up as a singly charged species (49, 58). Starting at mass 528, which has been shown to be an oxonium ion for N-glycans (58), we observed increasing mass steps of 162 up to a mass of 1500. The mass steps of 162 represent a loss in the number of Man on the glycan that start out as Man8GlcNAc1 (m/z = 1500) and end as Man2GlcNAc1 (m/z 528). The second observed ladder represents the doubly charged glycopeptide that shows losses of Man starting at m/z 1301, which represents a loss of 81 from the selected ion 1,382. Mass steps of 81 continue down to m/z 734, representing a loss of eight Man residues. A mass step of 101.7 from m/z 734 to 632.3 represents the loss of a GlcNAc, which we initially hypothesize as coming from the chitobiose core. The loss of this GlcNAc suggests that m/z 632 is the doubly charged glycopeptide with only one GlcNAc, commonly referred to as the Y1 ion (59). The most intense product ion in the spectrum is twice the mass of m/z 632 and potentially represents the singly charged Y1 ion.
The MS/MS spectra of the D forms allowed for the identification of ions that were observed in the MS/MS spectra of the G forms. For example, Sequest data analysis of the rTF1–263D sample identified m/z 632 as the peptide VNVTVEDER with a GlcNAc. The mass change by GlcNAc molecule, which remains attached to an Asn residue after treatment with Endo F enzymes, acts as a marker designating specific glycosylation sites in the D sample. This mass is seen in the MS/MS spectrum for ion 1,382 in supplemental Fig. 2 and thereby identifies ion 1,382 as being a glycopeptide with amino acid sequence VNVTVEDER. Supplemental Fig. 3 shows an example of a hybrid glycopeptide with sialic acid (SA) found in the pTF sample. As in supplemental Fig. 2, the most abundant ion is 1264, and the doubly charged ladder ends at m/z 632, which indicates that the peptide is VNVTVEDER.
N-Glycan Composition of rTF1–263
Table 4 presents the carbohydrate composition of rTF1–263. Three of the four potential sites for N-linked carbohydrates were found to be glycosylated including Asn-11, -124, and -137. Because all N-linked carbohydrates contain a specific trimannosyl diacetylchitobiose core structure composed of -Asn-GlcNAc2Man3, we will refer to that structure as the “core” and will discuss the carbohydrate composition and heterogeneity of sugar molecules that extend from and do not include the core. Proteolytic fragment 1 generated by the tryptic digest of TF (supplemental Table 1), which includes Asn-11, shows that the carbohydrate composition is dominated by high Man sugars, with the greatest number of 3 Man residues attached to the core (m/z = 1,666, z = 2). The smallest modification is one GlcNAc molecule on the Asn (m/z = 915, z = 2). Two fucosylated species are found, one a fucosylated core (m/z = 1333, z = 2) and the other a fucosylated GlcNAc on the Asn (m/z = 988, z = 2). The range of glycan masses on Asn-11 in rTF1–263 is from 202 to 1,866 Da.
Similarly to Asn-11, Asn-124 is also modified with high Man glycopeptides with the highest number of 8 Man residues (m/z = 1,625, z = 2). The smallest glycopeptide is composed of one GlcNAc molecule attached to the Asn (m/z = 633, z = 2). Unlike Asn-11, this site lacks fucosylated glycans. This site is also modified by hybrid carbohydrates ranging from one HexNAc or Hex2HexNAc2 residues attached to the core (m/z = 1078, z = 2 and m/z = 1342, z = 2, respectively). We define HexNAc as either GlcNAc or GalNAc, both with a mass of 203.20 Da. We define hexose as either mannose, glucose, or galactose, all three with a mass of 162 Da. The range of glycan masses on Asn-124 in rTF1–263 is from 203 to 2188 Da.
Asn-137 is found in both the proteolytic fragment resulting from trypsin digest as listed in supplemental Table 1 and a miscleaved fragment where Arg-135 from fragment 12 is attached to fragment 13 (RAsn). Asn-137 is modified by both high Man and fucosylated structures. The biggest glycan is composed of 6 Man residues attached to the core (m/z = 1,415, z = 2 and m/z = 996, z = 3). The fucosylated glycopeptides range from a fucosylated core (m/z = 1,002, z = 2 and m/z = 720, z = 3) to a fucosylated core plus Hex1HexNAc2 (m/z = 1,287, z = 2 and m/z = 911, z = 30). As observed with Asn-11 and Asn-124, Asn-137 is also found to be modified by one GlcNAc attached to the Asn. The range of glycan masses on Asn-137 in rTF1–263 is from 203 Da to 1,864 Da.
N-Glycan Composition of pTF
Table 5 presents the carbohydrate composition of pTF. Similar to rTF1–263, the natural protein is glycosylated on Asn-11, -124, and -137 in the extracellular domain, whereas Asn-261 in the cytosolic region is not modified. High Man glycans, a predominant feature of rTF1–263, are absent on all three sites in pTF. Asn-11 is modified predominantly by complex carbohydrates. All carbohydrates on this site have a fucosylated core. The majority of the structures are sialylated with a minimum of 1 and a maximum of 2 SA residues. The largest glycan on Asn-11 is composed of Hex3HexNAc3SA2 (m/z = 1,211, z = 3). The smallest glycan consists of a fucosylated core plus Hex1HexNAc2 (m/z = 1,619, z = 2). The remaining carbohydrates have a wide heterogeneous composition with varying numbers of Hex, HexNAc, and SA molecules. The range of glycan masses on Asn-11 in pTF is from 1,611 to 3,666 Da.
Asn-124 is modified by hybrid and sialylated complex carbohydrates. This site is not fucosylated, as was also observed for Asn-124 in rTF1–263. The largest glycan is a sialylated structure consisting of Hex3HexNAc3SA2 (m/z = 1,211, z = 3). The smallest is one Hex attached to the core (m/z = 1,058, z = 2). The remaining carbohydrate compositions are heterogeneous and vary in the numbers of Hex, HexNAc, and SA molecules attached to the core. The range of glycan masses on Asn-124 in pTF is from 1,055 to 2,571 Da.
The glycan composition of Asn-137 of pTF is similar to that of Asn-11. As observed in rTF1–263, the glycopeptides exist as the correctly cleaved peptide and a miscleaved peptide with one Arg residue attached to Asn-137. The miscleavage could be a result of the inaccessibility of this site to trypsin due to the presence of carbohydrates on Asn-137, which is next to the assigned cleavage site. All of the carbohydrates on this site contain a fucosylated core. The largest glycan is composed of a fucosylated core plus Hex4HexNAc5SA2 (m/z = 1,469, z = 3). The smallest is a fucosylated core plus one HexNAc (m/z = 1,183, z = 2). The diversity of the other carbohydrate structures is wide and found on both the correctly cleaved and miscleaved peptides either doubly or triply charged. On Asn-137 in pTF, the range of glycan masses is from 1,241 to 3,285 Da.
DISCUSSION
In this study, we compare the function and carbohydrate composition of rTF1–243, rTF1–263, and natural pTF. The data presented show that pTF is ∼5-fold more active than any rTF, and a significant cause can be attributed to glycosylation of the protein. After deglycosylation, natural human pTF is virtually equivalent in activity to deglycosylated rTF1–263. It has been demonstrated that TF is posttranslationally modified by N-linked glycosylation (12); however, the contribution of the carbohydrates to natural TF activity is controversial. It has been suggested that the activity of purified TF from bovine brain is inhibited by concanavalin A (37). It has also been observed that treatment of murine macrophages with tunicamycin inhibits TF activity in FXa generation (36). It has also been suggested that carbohydrates are not important for TF activity (30–34). These suggestions possibly arise from observations that the non-glycosylated recombinant forms are functional, as observed in this study for rTF1–243. One study compared the activity of truncated, soluble, recombinant forms of TF that do not interact with membrane and suggested that the activity is not carbohydrate-dependent (35). That observation is in line with our data where carbohydrates do not affect the activity of TF protein in a membrane-independent interaction (fluorogenic assay). A previously described mutagenesis study involving C186S and C209S demonstrated a decrease of activity in the mutant protein (34). The mutation also abolished the ability of the cell to fully glycosylate the protein, which could be a contributing factor in the decreased activity.
Despite the general consensus that TF proteins from different sources show heterogeneity in carbohydrate composition, based on their ability to bind concanavalin A (60), the extent and the type of glycans have not yet been studied. In the present study, we determined that N-linked carbohydrates modify both rTF and natural pTF. Three of the four potential sites for N-linked glycosylation, Ans-11, Asn-124, and Asn-137, contain glycans. An example of the analysis was shown in supplemental Figs. 2 and 3, where one can see the difference in MS/MS spectra between a high Man glycan attached to Asn-124 on rTF1–263 (Fig. 2) and a complex glycan attached to Asn-124 on pTF (Fig. 3). This type of MS analysis is limited to the glycan composition and cannot be used to define the structure of the glycans. Glycan structure analysis is best carried out by releasing the glycan from the protein, followed by permethylation and MSn analysis (61–63). Our goal in this study was to detect and define any differences in the type of glycans attached to each site that is glycosylated. The MS analysis we performed is consistent with previously published work that focused on similar goals (49, 64).
Our analysis could only identify N-linked glycans and not O-linked carbohydrates. The enzymes we used to remove carbohydrates from the protein do not remove O-linked carbohydrates. Hence, O-linked glycopeptides would remain unchanged in the D samples and would not be highlighted by their apparent disappearance, as was the case for N-linked carbohydrates (see supplemental Fig. 1). However, earlier work has found no evidence of O-linked carbohydrates (30). All oligosaccharides, thus studied, have a typical trimannosyl diacetylchitobiose core composed of Hex3GlcNAc2 found in N-linked carbohydrates. Some carbohydrates contain a fucosylated core on the Asn-linked GlcNAc, and in the case of pTF, SA may be present.
We have chosen to convey the heterogeneity of glycosylation at each site for each protein by describing the composition of the glycan and the glycopeptide mass for rTF1–263G and natural pTFG (Tables 4 and 5, respectively). An earlier report by Jiang et al. (65) suggests that there will be a bias for detecting glycopeptides without SA over glycopeptides with SA when using electrospray ionization in positive ion mode. This bias would probably extend to the non-glycosylated form of the glycopeptide as well. The possible bias in detecting the non-glycosylated peptides and glycopeptides without SA does not detract from our ability to define the differences in glycan composition between the two sources of TF. Further work is required to define how much of a bias exists in ionization efficiency between non-glycosylated, neutral, and SA-containing glycopeptides when using electrospray ionization in positive ion mode.
Three main differences exist between the carbohydrate composition of rTF1–263 and pTF. First, the carbohydrate composition of rTF1–263 predominates in high Man glycans on all three sites. In contrast, carbohydrates of pTF are highly processed and are completely devoid of high Man sugars. Hybrid structures composed of only Man and HexNAc molecules are found mainly on Asn-124 in pTF and to some extent on Asn-124 in rTF1–263. Interestingly, neither the recombinant nor the natural form of TF is fucosylated at this site, which is the only prominent feature that the two TFs share. The second main difference is the presence of SA on all three glycosylated sites of pTF including Asn-11, Asn-124, and Asn-137, a unique feature of the natural pTF. This sugar is present on both fucosylated (Asn-11 and Asn-137) and non-fucosylated (Asn-124) cores and is absent from rTF1–263. The inability of insect cells to sialylate carbohydrates was previously reported (66). Third, fucosylation is much more prevalent in pTF than in rTF1–263, where the majority of sugars contain a fucosylated core on Asn-11 and Asn-137. The overabundance of high Man glycans in rTF1–263 dominates over the few fucosylated glycopeptides in the recombinant protein. The range of glycan masses in TF is quite diverse in both the recombinant and natural form, with an extensively wider and higher numerical range in pTF.
As determined from the crystal structure of TF, generated with the extracellular domain of TF, two sites, including Asn-11 and Asn-137, are solvent-exposed, whereas Asn-124 is buried on the C-module close to the membrane (32, 67). Asn-137 is located within a region containing residues that are part of the extensive region of FVIIa ligand binding and include Arg-135, Leu-133, and Phe-140. Mutational analysis of Arg-135 and Phe-140 revealed that mutation of these residues results in a decreased ligand affinity but does not alter its catalytic function toward a small substrate (68). These data may support our observation where removal of the sugar moiety does not alter the catalytic efficiency of the complex, as demonstrated by the unaltered ability of FVIIa to cleave the fluorogenic substrate when complexed with either the G or D form of TF. The location of Asn-124 in the structure of TF renders it potentially important in the recognition and binding of FX. Because the residue is located close to the membrane and is in close proximity to the Cys-186–Cys-209 disulfide, where the interaction of TF and FX takes place, glycosylation on that residue of TF may influence its function in macromolecular substrate recognition (67).
Although contradictory reports exist as to the affect of glycosylation on the activity of TF protein, we have shown that carbohydrates significantly influence natural TF function. The unique carbohydrate compositions render the natural human protein different from the recombinant counterpart with respect to glycosylation. A common feature of pTF with other coagulation proteins, such as FV, anti-thrombin III, tissue factor pathway inhibitor, thrombin-activatable fibrinolysis inhibitor, and FVII proteins, is the presence of SA (20–23, 69, 70). This sugar may be important for protein-protein and protein-membrane interactions. Whether the presence of the carbohydrate is important for FX binding by the TF·FVIIa complex or for the interaction with the membrane cannot be deduced from our study. Nevertheless, our data support the observation that the presence and possibly the composition of carbohydrates play a role in TF function.
Supplementary Material
Acknowledgments
We thank Dr. Dwight E. Matthews for helpful analysis and discussion. The LC/MS instrumentation and the materials used for LC/MS analysis were provided by support made possible by Vermont Genetics Network Grant P20RR16462 from the IDeA Networks of Biomedical Research Excellence Program of the National Center for Research Resources, a component of the National Institutes of Health.
This work was supported, in whole or in part, by National Institutes of Health Grant P01HL46703 (Project 2). This work was presented in part at the XXII Congress of the International Society on Thrombosis and Haemostasis, July 11–16, 2009, Boston, MA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
R. Jenny, personal communication.
- TF
- tissue factor
- rTF
- recombinant TF
- pTF
- placental TF
- FV, FVa, FVII, FVIIa, FVIIIa, FIX, FIXa, and FX
- factor V, Va, VII, VIIa, VIIIa, IX, IXa, and X, respectively
- FXase
- factor Xase
- rFVIIa
- recombinant FVIIa
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- HBS
- Hepes-buffered saline
- mAb
- monoclonal antibody
- PNGase F
- peptide-N-glycosidase F
- FLI
- fluorescence-linked immunoassay
- Endo
- endoglycosidase
- G
- glycosylated
- D
- deglycosylated
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- LC
- liquid chromatography
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- Fuc
- fucose
- SA
- sialic acid
- RAsn
- miscleaved fragment where Arg-135 from fragment 12 is attached to fragment 13
- HexNAc
- N-acetylhexosamine
- Hex
- hexose.
REFERENCES
- 1.Mackman N. (2006) Blood Cells Mol. Dis. 36, 104–107 [DOI] [PubMed] [Google Scholar]
- 2.van 't Veer C., Mann K. G. (1997) J. Biol. Chem. 272, 4367–4377 [DOI] [PubMed] [Google Scholar]
- 3.Mann K. G. (2003) Chest 124, 4S–10S [DOI] [PubMed] [Google Scholar]
- 4.Spicer E. K., Horton R., Bloem L., Bach R., Williams K. R., Guha A., Kraus J., Lin T. C., Nemerson Y., Konigsberg W. H. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5148–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toomey J. R., Smith K. J., Stafford D. W. (1991) J. Biol. Chem. 266, 19198–19202 [PubMed] [Google Scholar]
- 6.Broze G. J., Jr. (1982) J. Clin. Invest. 70, 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komiyama Y., Pederson A. H., Kisiel W. (1990) Biochemistry 29, 9418–9425 [DOI] [PubMed] [Google Scholar]
- 8.Paborsky L. R., Caras I. W., Fisher K. L., Gorman C. M. (1991) J. Biol. Chem. 266, 21911–21916 [PubMed] [Google Scholar]
- 9.Belting M., Dorrell M. I., Sandgren S., Aguilar E., Ahamed J., Dorfleutner A., Carmeliet P., Mueller B. M., Friedlander M., Ruf W. (2004) Nat. Med. 10, 502–509 [DOI] [PubMed] [Google Scholar]
- 10.Astrup T. (1965) Thromb. Diath. Haemorrh. 14, 401–416 [PubMed] [Google Scholar]
- 11.Drake T. A., Morrissey J. H., Edgington T. S. (1989) Am. J. Pathol. 134, 1087–1097 [PMC free article] [PubMed] [Google Scholar]
- 12.Egorina E. M., Sovershaev M. A., Osterud B. (2008) Thromb. Res. 122, 831–837 [DOI] [PubMed] [Google Scholar]
- 13.Dorfleutner A., Ruf W. (2003) Blood 102, 3998–4005 [DOI] [PubMed] [Google Scholar]
- 14.Mody R. S., Carson S. D. (1997) Biochemistry 36, 7869–7875 [DOI] [PubMed] [Google Scholar]
- 15.Rudd P. M., Woods R. J., Wormald M. R., Opdenakker G., Downing A. K., Campbell I. D., Dwek R. A. (1995) Biochim. Biophys. Acta 1248, 1–10 [DOI] [PubMed] [Google Scholar]
- 16.Hansen L., Blue Y., Barone K., Collen D., Larsen G. R. (1988) J. Biol. Chem. 263, 15713–15719 [PubMed] [Google Scholar]
- 17.Peterson C. B., Blackburn M. N. (1985) J. Biol .Chem. 260, 610–615 [PubMed] [Google Scholar]
- 18.Kornfeld R., Kornfeld S. (1985) Annu. Rev. Biochem. 54, 631–664 [DOI] [PubMed] [Google Scholar]
- 19.Peter-Katalinić J. P. (2005) Methods Enzymol. 405, 139–171 [DOI] [PubMed] [Google Scholar]
- 20.Silveira J. R., Kalafatis M., Tracy P. B. (2002) Biochemistry 41, 1672–1680 [DOI] [PubMed] [Google Scholar]
- 21.Munzert E., Heidemann R., Büntemeyer H., Lehmann J., Müthing J. (1997) Biotechnol. Bioeng. 56, 441–448 [DOI] [PubMed] [Google Scholar]
- 22.Fenaille F., Groseil C., Ramon C., Riandé S., Siret L., Chtourou S., Bihoreau N. (2008) Glycoconj. J. 25, 827–842 [DOI] [PubMed] [Google Scholar]
- 23.Agarwala K. L., Kawabata S., Takao T., Murata H., Shimonishi Y., Nishimura H., Iwanaga S. (1994) Biochemistry 33, 5167–5171 [DOI] [PubMed] [Google Scholar]
- 24.Yang L., Manithody C., Rezaie A. R. (2009) J. Thromb. Haemost. 7, 1696–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bach R., Rifkin D. B. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6995–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dietzen D. J., Page K. L., Tetzloff T. A. (2004) Blood 103, 3038–3044 [DOI] [PubMed] [Google Scholar]
- 27.Osterud B., Rao L. V., Olsen J. O. (2000) Thromb. Haemost. 83, 861–867 [PubMed] [Google Scholar]
- 28.Reinhardt C., von Brühl M. L., Manukyan D., Grahl L., Lorenz M., Altmann B., Dlugai S., Hess S., Konrad I., Orschiedt L., Mackman N., Ruddock L., Massberg S., Engelmann B. (2008) J. Clin. Invest. 118, 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pendurthi U. R., Ghosh S., Mandal S. K., Rao L. V. (2007) Blood 110, 3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paborsky L. R., Harris R. J. (1990) Thromb. Res. 60, 367–376 [DOI] [PubMed] [Google Scholar]
- 31.Paborsky L. R., Tate K. M., Harris R. J., Yansura D. G., Band L., McCray G., Gorman C. M., O'Brien D. P., Chang J. Y., Swartz J. R. (1989) Biochemistry 28, 8072–8077 [DOI] [PubMed] [Google Scholar]
- 32.Harlos K., Martin D. M., O'Brien D. P., Jones E. Y., Stuart D. I., Polikarpov I., Miller A., Tuddenham E. G., Boys C. W. (1994) Nature 370, 662–666 [DOI] [PubMed] [Google Scholar]
- 33.Waxman E., Ross J. B., Laue T. M., Guha A., Thiruvikraman S. V., Lin T. C., Konigsberg W. H., Nemerson Y. (1992) Biochemistry 31, 3998–4003 [DOI] [PubMed] [Google Scholar]
- 34.Rehemtulla A., Ruf W., Edgington T. S. (1991) J. Biol. Chem. 266, 10294–10299 [PubMed] [Google Scholar]
- 35.Stone M. J., Ruf W., Miles D. J., Edgington T. S., Wright P. E. (1995) Biochem. J. 310, 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shands J. W., Jr. (1985) Blood 65, 169–175 [PubMed] [Google Scholar]
- 37.Pitlick F. A. (1975) J. Clin. Invest. 55, 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bona R., Lee E., Rickles F. (1987) Thromb. Res. 48, 487–500 [DOI] [PubMed] [Google Scholar]
- 39.Parhami-Seren B., Butenas S., Krudysz-Amblo J., Mann K. G. (2006) J. Thromb. Haemost. 4, 1747–1755 [DOI] [PubMed] [Google Scholar]
- 40.Bajaj S. P., Rapaport S. I., Brown S. F. (1981) J. Biol. Chem. 256, 253–259 [PubMed] [Google Scholar]
- 41.Butenas S., Ribarik N., Mann K. G. (1993) Biochemistry 32, 6531–6538 [DOI] [PubMed] [Google Scholar]
- 42.Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 43.Lawson J. H., Butenas S., Mann K. G. (1992) J. Biol. Chem. 267, 4834–4843 [PubMed] [Google Scholar]
- 44.Butenas S., Lawson J. H., Kalafatis M., Mann K. G. (1994) Biochemistry 33, 3449–3456 [DOI] [PubMed] [Google Scholar]
- 45.Krishnaswamy S., Field K. A., Edgington T. S., Morrissey J. H., Mann K. G. (1992) J. Biol. Chem. 267, 26110–26120 [PubMed] [Google Scholar]
- 46.Klammer A. A., MacCoss M. J. (2006) J. Proteome Res. 5, 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guzzetta A. W., Basa L. J., Hancock W. S., Keyt B. A., Bennett W. F. (1993) Anal. Chem. 65, 2953–2962 [DOI] [PubMed] [Google Scholar]
- 48.Valmu L., Alfthan H., Hotakainen K., Birken S., Stenman U. H. (2006) Glycobiology 16, 1207–1218 [DOI] [PubMed] [Google Scholar]
- 49.Iacob R. E., Perdivara I., Przybylski M., Tomer K. B. (2008) J. Am. Soc. Mass. Spectrom. 19, 428–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wuhrer M., Koeleman C. A., Hokke C. H., Deelder A. M. (2005) Anal. Chem. 77, 886–894 [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., Wu S. L., Hancock W. S. (2006) Biotechnol. Prog. 22, 873–880 [DOI] [PubMed] [Google Scholar]
- 52.Irungu J., Go E. P., Zhang Y., Dalpathado D. S., Liao H. X., Haynes B. F., Desaire H. (2008) J. Am. Soc. Mass. Spectrom. 19, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yates J. R., 3rd, Eng J. K., McCormack A. L., Schieltz D. (1995) Anal. Chem. 67, 1426–1436 [DOI] [PubMed] [Google Scholar]
- 54.Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 55.Trimble R. B., Tarentino A. L. (1991) J. Biol. Chem. 266, 1646–1651 [PubMed] [Google Scholar]
- 56.Plummer T. H., Jr., Elder J. H., Alexander S., Phelan A. W., Tarentino A. L. (1984) J. Biol. Chem. 259, 10700–10704 [PubMed] [Google Scholar]
- 57.Demain A. L., Vaishnav P. (2009) Biotechnol. Adv. 27, 297–306 [DOI] [PubMed] [Google Scholar]
- 58.Huddleston M. J., Bean M. F., Carr S. A. (1993) Anal. Chem. 65, 877–884 [DOI] [PubMed] [Google Scholar]
- 59.Harvey D. J., Bateman R. H., Green M. R. (1997) J. Mass Spectrom. 32, 167–187 [DOI] [PubMed] [Google Scholar]
- 60.van den Besselaar A. M., Bertina R. M. (1984) Thromb. Haemost. 52, 192–195 [PubMed] [Google Scholar]
- 61.Reinhold V. N., Reinhold B. B., Costello C. E. (1995) Anal. Chem. 67, 1772–1784 [DOI] [PubMed] [Google Scholar]
- 62.Weiskopf A. S., Vouros P., Harvey D. J. (1998) Anal. Chem. 70, 4441–4447 [DOI] [PubMed] [Google Scholar]
- 63.Ashline D., Singh S., Hanneman A., Reinhold V. (2005) Anal. Chem. 77, 6250–6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Go E. P., Irungu J., Zhang Y., Dalpathado D. S., Liao H. X., Sutherland L. L., Alam S. M., Haynes B. F., Desaire H. (2008) J. Proteome Res. 7, 1660–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang H., Desaire H., Butnev V. Y., Bousfield G. R. (2004) J. Am. Soc. Mass. Spectrom. 15, 750–758 [DOI] [PubMed] [Google Scholar]
- 66.Marchal I., Jarvis D. L., Cacan R., Verbert A. (2001) Biol. Chem. 382, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin D. M., Boys C. W., Ruf W. (1995) FASEB J. 9, 852–859 [DOI] [PubMed] [Google Scholar]
- 68.Schullek J. R., Ruf W., Edgington T. S. (1994) J. Biol. Chem. 269, 19399–19403 [PubMed] [Google Scholar]
- 69.Valnickova Z., Christensen T., Skottrup P., Thøgersen I. B., Højrup P., Enghild J. J. (2006) Biochemistry 45, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 70.Mori Y., Hamuro T., Nakashima T., Hamamoto T., Natsuka S., Hase S., Iwanaga S. (2009) J. Thromb. Haemost. 7, 111–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.