Abstract
Maintenance of circadian alignment between an organism and its environment is essential to ensure metabolic homeostasis. Synchrony is achieved by cell autonomous circadian clocks. Despite a growing appreciation of the integral relation between clocks and metabolism, little is known regarding the direct influence of a peripheral clock on cellular responses to fatty acids. To address this important issue, we utilized a genetic model of disrupted clock function specifically in cardiomyocytes in vivo (termed cardiomyocyte clock mutant (CCM)). CCM mice exhibited altered myocardial response to chronic high fat feeding at the levels of the transcriptome and lipidome as well as metabolic fluxes, providing evidence that the cardiomyocyte clock regulates myocardial triglyceride metabolism. Time-of-day-dependent oscillations in myocardial triglyceride levels, net triglyceride synthesis, and lipolysis were markedly attenuated in CCM hearts. Analysis of key proteins influencing triglyceride turnover suggest that the cardiomyocyte clock inactivates hormone-sensitive lipase during the active/awake phase both at transcriptional and post-translational (via AMP-activated protein kinase) levels. Consistent with increased net triglyceride synthesis during the end of the active/awake phase, high fat feeding at this time resulted in marked cardiac steatosis. These data provide evidence for direct regulation of triglyceride turnover by a peripheral clock and reveal a potential mechanistic explanation for accelerated metabolic pathologies after prevalent circadian misalignment in Western society.
Keywords: Diseases/Metabolic, Gene/Regulation, Lipid/Triacylglycerol, Metabolism/Energy, Metabolism/Fatty Acid, Metabolism/Lipid, Tissue/Organ Systems/Heart, Chronobiology
Introduction
Striking time-of-day-dependent oscillations are observed in multiple cardiometabolic parameters in both animal models and humans. These parameters range from levels of circulating nutrients and endocrine factors, neural activity, glucose tolerance, insulin sensitivity, feeding behavior, and energy metabolism (both at the individual tissue and whole body levels) to cardiovascular function (1–5). Significant alterations in many of these oscillations are observed in metabolic disease states (e.g. obesity, diabetes mellitus, and cardiovascular disease), suggesting that circadian misalignment may play an important role in the etiology of multiple pathologies (5, 6). Recent molecular/genetic-based studies reinforce such a concept and suggest that intrinsic cellular circadian clocks play a pivotal role in mediating many, if not all, biological rhythms. Circadian clocks are transcriptionally based molecular mechanisms that generate self-sustained positive and negative feedback loops with a free running period of ∼24 h (7); this molecular mechanism has been identified within essentially all mammalian cells (both central and peripheral). Circadian clocks confer the selective advantage of anticipation. In doing so molecular clocks enable the cell to prepare for an external stimulus before its onset, thereby maintaining optimal synchrony with the environment. Given marked time-of-day-dependent rhythms in energy supply (e.g. dietary nutrient intake) and demand (e.g. physical activity), it is not surprising that the circadian clock mechanism is emerging as a major regulator of metabolism.
Evidence is accumulating in support of the hypothesis that mammalian circadian clocks influence fatty acid/lipid metabolism in a time-of-day-dependent manner. Genetic manipulation of either CLOCK or BMAL1, critical transcription factors at the heart of the clock mechanism, influences body weight gain and adiposity (8, 9). Additionally, CLOCK mutant mice show altered hepatic lipid accumulation (i.e. steatosis) after chronic high fat feeding and alcohol consumption (10, 11). However, these studies were performed in mouse models of ubiquitous clock dysfunction, raising questions of whether alterations in behavior (e.g. feeding/fasting cycles), digestion/absorption, and/or neurohumoral factors contribute toward observed lipid metabolism deviations as opposed to a cell autonomous clock directly playing an intrinsic role (e.g. the hepatic clock impacting hepatic steatosis). This concept was recently illustrated through elegant studies by Lamia et al. (12), who showed marked phenotypic differences in glucose tolerance depending on whether BMAL1 expression was genetically deleted in a ubiquitous or cell type-specific manner.
Our laboratory has recently generated a mouse model in which the cardiomyocyte circadian clock is selectively impaired through targeted expression of a dominant negative CLOCK protein using the myosin heavy chain α promoter; these mice are termed cardiomyocyte clock mutant (CCM)4 (13). Consistent with impairment of only the cardiomyocyte circadian clock, all behavioral and neurohumoral factors/parameters investigated to date are similar between wild-type (WT) and CCM littermates. Using this model, the cardiomyocyte circadian clock has been shown to play an important role in regulating multiple parameters of myocardial function (e.g. heart rate, cardiac output), metabolism (e.g. glycogenolysis), and transcription (1). In the latter case previous studies revealed an important role for the cardiomyocyte circadian clock in modulating the transcriptional responsiveness of the myocardium to acute elevations in fatty acid availability during fasting (13). Therefore, the purpose of the current studies was to test the hypothesis that the cardiomyocyte circadian clock plays a critical role in metabolic adaptation of the heart to chronic high fat feeding. The heart is a particularly advantageous organ for such studies, given appreciable similarity for mechanisms of metabolic adaptation (e.g. activation of peroxisome proliferator-activated receptors) and maladaptation (e.g. lipotoxicity) to fatty acids relative to tissues such as liver and skeletal muscle, the ability to simultaneously measure metabolic fluxes and organ (contractile) function, and the direct relevance to myocardial dysfunction associated with the cardiometabolic syndrome (14–16). Using this model, our studies provide evidence for direct regulation of triglyceride metabolism by this peripheral circadian clock.
EXPERIMENTAL PROCEDURES
Animals
Male WT and CCM mice (on the FVB/N background) were housed at the Children's Nutrition Research Facility of the Children's Nutrition Research Center at Baylor College of Medicine (Houston, TX) under temperature-, humidity-, and light-controlled conditions. A strict 12-h light/12-h dark cycle regime was enforced (lights on at 6 a.m.; zeitgeber time (ZT) 0). Mice received food and water ad libitum unless otherwise specified. Mice were housed in standard micro-isolator cages except during high fat feeding studies, during which mice were housed on wire-bottom cages to prevent consumption of bedding and feces. All animal experiments were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
Rodent Diets
A standard murine diet (Teklad Lab Animal Diets, Harlan Laboratories, Indianapolis, IN) a high fat diet (45% calories from fat, Research Diets, New Brunswick, NJ) and a control diet for the high fat feeding studies (10% calories from fat, Research Diets, New Brunswick, NJ) were utilized as specified for individual studies.
Non-invasive Mouse Monitoring/Measurements
Food intake was monitored manually during feeding protocols on a daily basis. Body weight was monitored in mice during feeding protocols at four-week intervals. Body composition was determined in mice using the Lunar PIXImus Densitometer (GE Medical Systems, Madison, WI). Echocardiography was performed to assess cardiac function; mice were anesthetized using 1.5% isoflurane with 95% O2 and placed on a heated platform, at which time M-mode measurements were recorded using a Visualsonic Vevo 770 Imaging System (VisualSonics, Toronto, Canada). Piximus and echocardiography measurements were performed 12 weeks and 16 weeks, respectively, after the initiation of feeding protocols.
Plasma Hormone and Substrate Measurements
Non-fasted plasma glucose, non-esterified fatty acids, insulin, and triglyceride concentrations were measured using commercially available kits (Thermo Scientific, Waltham, MA; Wako Diagnostics, Richmond, VA; Crystal Chem Inc., Downers Grove, IL; Thermo Scientific).
Gene Expression Microarray Analysis
Gene expression microarray analysis was performed using mouse Ref-6 BeadChips and the BeadStation System (Illumina, Inc., San Diego, CA). RNA was extracted from hearts using standard procedures (17). Microarrays were performed according to the manufacturer's guidelines and as described previously in detail (1, 18).
Quantitative RT-PCR
Candidate gene expression analysis was performed by quantitative RT-PCR using previously described methods (19, 20). Specific assays were designed for each gene from mouse sequences available in GenBank™. Primer and probe sequences are presented in Table 1. Standard RNA was made by the T7 polymerase method (Ambion, Austin, TX) using total RNA isolated from mouse hearts; the use of standard RNA allows absolute quantification of gene expression. Quantitative RT-PCR data are represented as mRNA molecules/ng of total RNA.
TABLE 1.
Primer and probe sequences for quantitative RT-PCR measurements
TAMRA, 6-carboxytetramethylrhodamine; FAM, 6-carboxyfluorescein.
| Gene | Primer/probe | Sequence |
|---|---|---|
| agpat3 | Forward | 5′-TGGAAGACATCCCAGCAGAT-3′ |
| Reverse | 5′-CCCCTGGGAATACACCCTT-3′ | |
| Probe | 5′-FAM-AGGAGAAGGATGCCCTGCAAGAGATGTA-TAMRA-3′ | |
| atgl | Forward | 5′-GGTCCTCCGAGAGATGTGC-3′ |
| Reverse | 5′-TGGTTCAGTAGGCCATTCCTC-3′ | |
| Probe | 5′-FAM-CAGGGCTACAGAGATGGACTTCGATTCCTT-TAMRA-3′ | |
| cd36 | Forward | 5′-ATTGCGACATGATTAATGGCA-3′ |
| Reverse | 5′-GATGGACCTGCAAATGTCAGA-3′ | |
| Probe | 5′-FAM-AGATGCAGCCTCCTTTCCACCTTTTG-TAMRA-3′ | |
| dgat2 | Forward | 5′-GGCTGGCATTTGACTGGAA-3′ |
| Reverse | 5′-TGGTCAGCAGGTTGTGTGTCT-3′ | |
| Probe | 5′-FAM-AAGAAAGGTGGCAGGAGATCGCAGTG-TAMRA-3′ | |
| hsl | Forward | 5′-GCGCTGGAGGAGTGTTTTT-3′ |
| Reverse | 5′-TGTCCCCTGCAAGGCATAT-3′ | |
| Probe | 5′-FAM-TCTCCAGTTGAACCAAGCAGGTCACA-TAMRA-3′ | |
| pdk4 | Forward | 5′-TTCACACCTTCACCACATGC-3′ |
| Reverse | 5′-AAAGGGCGGTTTTCTTGATG-3′ | |
| Probe | 5′-FAM-CGTGGCCCTCATGGCATTCTTG-TAMRA-3′ | |
| s3-12 | Forward | 5′-AGGGCTGGGTGATATCTTTCA-3′ |
| Reverse | 5′-CCCCGGTCAGCAGAGAGTA-3′ | |
| Probe | 5′-FAM-CCATGACAACTGAGGAACAAGCTCAGC-TAMRA-3′ | |
| ucp3 | Forward | 5′-TGCTGAGATGGTGACCTACGA-3′ |
| Reverse | 5′-CCAAAGGCAGAGACAAAGTGA-3′ | |
| Probe | 5′-FAM-AAGTTGTCAGTAAACAGGTGAGACTCCAGCAA-TAMRA-3′ |
Myocardial Triglyceride Content Measurements
Myocardial triglyceride contents were measured from homogenate extracts using an enzymatic spectrophotometric kit according to the manufacturer's instructions (Wako Diagnostics).
Isolated Working Mouse Heart Perfusions
Myocardial contractile function and metabolism were determined ex vivo through isolated working mouse heart perfusions as previously described (1). All hearts were perfused in the working mode in a non-recirculating manner with a preload of 12.5 mm Hg and an after-load of 50 mm Hg. Standard Krebs-Henseleit buffer was supplemented with 8 mm glucose, 0.4 mm oleate conjugated to 3% bovine serum albumin (fraction V, fatty acid-free; dialyzed), 10 microunits/ml insulin, 1.5 mm lactate, 0.15 mm pyruvate, 0.05 mm l-carnitine, and 0.13 mm glycerol unless otherwise specified. Radiolabeled tracers ([U-14C]glucose (0.096 mCi/liter), [U-14C]lactic acid (0.009 mCi/liter), [9,10-3H]-oleate (0.067 mCi/liter), and [1-14C]-oleate (0.12 mCi/liter)) were utilized to monitor oxidative and non-oxidative substrate metabolism, as specified in individual experiments. Measurements of cardiac function (e.g. cardiac power), myocardial oxygen consumption, and coronary effluent were collected throughout the perfusion period at 5-min intervals as described previously (1). At the end of the perfusion period hearts were snap-frozen and stored at −80 °C before lipid extraction.
Lipid Extraction, Fractionation and Profiling
Myocardial lipids were extracted from both freshly isolated and ex vivo perfused hearts according to Bligh and Dyer (21). Lipids were separated into different fractions by thin-layer chromatography using silica gel plates. Specific standards were run in parallel for accurate identification of sample fractions. For lipid profile analysis of non-perfused hearts, total lipids were extracted from the homogenates after the addition of known amounts of internal standards. After fractionation, methyl esters were prepared using the boron trifluoride-methanol method described in Morrison and Smith (22) and quantified by gas-liquid chromatography (Hewlett-Packard 5890, Palo Alto, CA) on a DB-225 capillary column (Agilent Technologies/J&W Scientific, St. Louis, MO). Distinct fatty acids were identified by comparison to the retention times of fatty acid methyl ester standards.
Oil Red O Staining
Oil red O staining was performed on frozen heart sections as described in Carson (23). Digital images of the sections were captured using a Spot RT CCD (Diagnostic Instruments, Sterling Heights, MI).
Western Blotting
Protein expression was qualitatively measured through standard Western blotting techniques. Antibodies specific for adipose triglyceride lipase (ATGL; #2138, Cell Signaling Technology, Danvers, MA), hormone-sensitive lipase (HSL; #4107, Cell Signaling Technology), phospho-Ser-565-HSL (#4137, Cell Signaling Technology, AMPK-activated protein kinase α1 (AMPKα1; #07-350, Upstate Biotechnology Inc., Lake Placid, NY), AMPKα2 (#19131, Santa Cruz Biotechnology Inc., Santa Cruz, CA), and Ran-GTPase (#610342, BD Biosciences) were utilized for protein detection. Ran-GTPase was used as a loading control.
AMPK Activity Measurements
Frozen hearts were homogenized in lysis buffer and assayed for AMPK activity as described previously (24). To obtain isoform-specific measurements, protein (50 μg) was immunoprecipitated overnight with polyclonal antibodies specific to α1 or α2 AMPK bound to protein A/G-agarose before assay. Kinase activity was calculated as incorporated [32P]ATP (nmol) into SAMS peptide/g of protein/min.
Statistical Analysis
Statistical analysis of non-microarray data was performed using two-way or three-way ANOVA. Stata Version SE10.0 (Stata Corp., San Antonio, TX) was used to perform two-way ANOVA to investigate the main effects of genotype and diet, as well as diet and time. Three-way ANOVA was used to determine the main effects of genotype, diet, and perfusion conditions. A full model including second-order interactions was conducted for each experiment. Significant differences were determined using Type III sums of squares. The null hypothesis of no model effects was rejected at p < 0.05. Last, Bonferroni post hoc analyses were performed for pair-wise comparisons.
Statistical analysis of microarray data was performed with Stata version SE10.0. Two-way ANOVA was used to identify genes with statistically significant differences in mean expression (interpreted as main effect of diet, genotype, or diet-genotype interaction). p values were corrected for multiple testing using the Q-value system, with a false discovery rate level of 0.05. Gene lists were further analyzed using the Onto-Tools package of ontology and pathway analysis from Wayne State University.
RESULTS
CCM Hearts Exhibit Altered Responsiveness to Chronic High Fat Feeding
To test the hypothesis that the cardiomyocyte circadian clock is important for adaptation of the heart to chronic elevations in fatty acids, WT and CCM mice were fed a high fat diet for 16 weeks ad libitum. All parameters were subsequently measured at ZT18 (middle of the active phase), the time of day at which we have previously shown greatest transcriptional responsiveness of the rodent heart (and skeletal muscle) to fatty acids (13). Consistent with the cardiac-restricted nature of this genetic model, high fat feeding increased caloric intake, body weight, and percentage body fat by comparable levels in WT and CCM mice (Table 2). This feeding protocol also led to genotype-independent effects on multiple humoral factors (Table 2).
TABLE 2.
Food intake, body weight, body fat composition, and plasma humoral factors in WT and CCM mice fed with either control or high fat diet
Values are expressed as the mean ± S.E. (n = 6). NEFA, non-esterified fatty acids.
| WT |
CCM |
|||
|---|---|---|---|---|
| Control | High fat | Control | High fat | |
| Caloric consumption (kcal/day) | 16.0 ± 0.67 | 19.85 ± 0.49a | 15.04 ± 1.03 | 18.29 ± 0.96a |
| Body weight (g) | 33.54 ± 0.58 | 38.53 ± 1.03a | 33.03 ± 0.40 | 40.35 ± 0.91a |
| Body fat composition (%) | 28.28 ± 2.87 | 36.39 ± 1.96a | 28.58 ± 0.99 | 35.69 ± 1.61a |
| Plasma NEFA (mm) | 0.065 ± 0.013 | 0.079 ± 0.013 | 0.093 ± 0.007 | 0.099 ± 0.023 |
| Plasma triglyceride (mg/dl) | 104.2 ± 20.57 | 28.71 ± 6.23a | 94.42 ± 13.74 | 28.67 ± 2.04a |
| Plasma glucose (mg/dl) | 280.3 ± 12.13 | 283.2 ± 7.31 | 265.5 ± 21.03 | 276.9 ± 16.41 |
| Plasma insulin (μg/l) | 2.57 ± 0.27 | 3.73 ± 0.68a | 2.78 ± 0.32 | 4.34 ± 0.64a |
| Plasma leptin (ng/ml) | 15.07 ± 3.35 | 30.31 ± 8.22a | 18.07 ± 2.91 | 45.95 ± 7.54a |
a Control versus high fat diet within a genotype.
In vivo cardiac function was next assessed by echocardiography in WT and CCM mice (Table 3). Consistent with previous reports, left ventricular posterior wall thickness in diastole (LVPWd), ejection fraction (EF), and fractional shortening (FS) were significantly higher in CCM versus WT hearts, independent of diet (Table 3) (1). Somewhat surprisingly, 16 weeks of high fat feeding did not significantly impact cardiac function in vivo in either WT or CCM mice (Table 3).
TABLE 3.
In vivo cardiac functional parameters in WT and CCM mice fed with either control or high fat diet
Values are expressed as the mean ± S.E. (n = 6). LVIDd, left ventricular internal diameter in diastole; LVIDs, left ventricular internal diameter in systole; IVSs, interventricular septum thickness in systole; LVPWd, left ventricular posterior wall thickness in diastole; EF, ejection fraction; FS, fractional shortening; HR, heart rate; bpm, beats per min.
| WT |
CCM |
|||
|---|---|---|---|---|
| Control | High fat | Control | High fat | |
| LVIDd (mm) | 3.99 ± 0.12 | 3.95 ± 0.09 | 3.80 ± 0.12 | 3.76 ± 0.12 |
| LVIDs (mm) | 2.63 ± 0.11 | 2.57 ± 0.10 | 2.14 ± 0.06a | 2.20 ± 0.11a |
| IVSs (mm) | 0.70 ± 0.07 | 0.76 ± 0.06 | 0.71 ± 0.06 | 0.75 ± 0.04 |
| LVPWd (mm) | 0.89 ± 0.11 | 1.09 ± 0.08 | 1.23 ± 0.11a | 1.14 ± 0.04 |
| EF (%) | 65.90 ± 0.95 | 67.86 ± 1.46 | 77.42 ± 1.21a | 74.48 ± 1.96a |
| FS (%) | 35.85 ± 0.72 | 37.41 ± 1.16 | 45.53 ± 1.16a | 42.88 ± 1.72a |
| HR (bpm) | 533 ± 20 | 499 ± 14 | 499 ± 16 | 478 ± 13 |
a WT versus CCM within a feeding group.
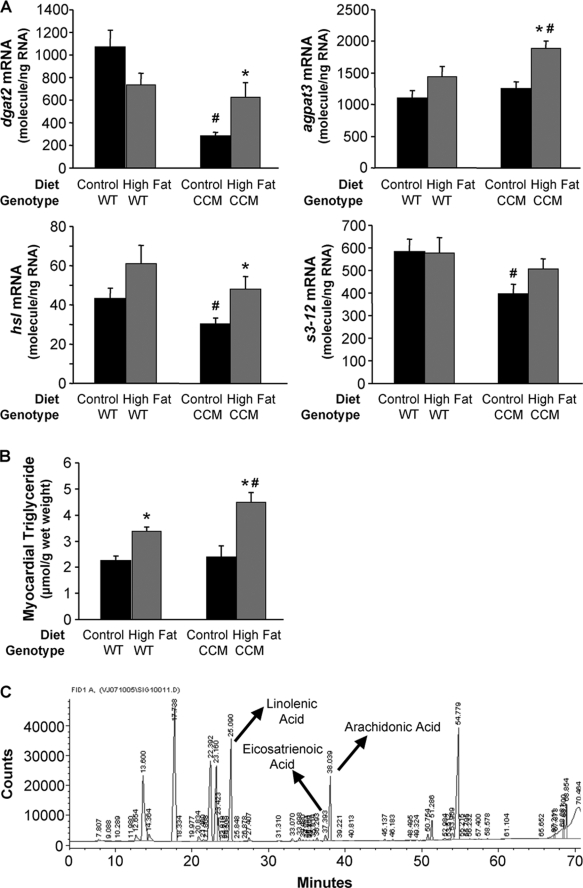
Given that the circadian clock mechanism is transcriptionally based, output initially manifests at the gene expression level (7, 25). Furthermore, we have previously reported that disruption of the cardiomyocyte circadian clock impairs acute transcriptional responsiveness of the heart to fatty acids during fasting (13). Therefore, gene expression microarray analysis was next performed to determine whether the cardiomyocyte circadian clock impacts the transcriptional response of the heart to chronic high fat feeding. Supplemental Fig. 1 illustrates those genes that differentially respond to chronic high fat feeding in CCM versus WT hearts (i.e. significant genotype-diet interaction); this gene list is included in supplemental Table 1. The list includes a number of genes known to influence triglyceride metabolism, consistent with previous observations that CCM hearts exhibit impaired fasting-induced triglyceride synthesis (13). As such, expression changes in multiple triglyceride metabolism genes were next measured by RT-PCR. On a control diet, CCM hearts exhibit reduced dgat2 (diacylglycerol acyltransferase 2; triglyceride synthesis), hsl (lipolysis), and s3-12 (lipid droplet-binding protein) mRNA levels relative to WT hearts (Fig. 1A). In addition, high fat feeding increased dgat2, hsl, and agpat3 (1-acylglycerol-3-phosphate O-acyltransferase 3; triglyceride synthesis) mRNA in CCM, but not WT, hearts (Fig. 1A). Consistent with differential transcriptional responsiveness of CCM hearts to high fat feeding, this dietary intervention led to a greater accumulation of myocardial triglyceride levels in CCM, versus WT, mice (Fig. 1B).
FIGURE 1.
High fat feeding-induced effects on metabolic gene expression (A), myocardial triglyceride (B), and distinct lipid species (C) were altered in CCM hearts. After 16 weeks of diet intervention, gene expression analyses of dgat2, agpat3, hsl, and s3-12 (A) were performed using quantitative RT-PCR in WT and CCM hearts isolated at ZT18. Myocardial triglyceride levels (B) were also measured. Lipid species were analyzed by gas chromatography after methyl esterification (C). Arachidonic acid, eicosatrienoic acid, and linolenic acid are indicated in the represented spectrum (C). Data are represented as the mean ± S.E. Statistical analysis was performed using a two-way ANOVA followed by a post hoc comparison with Bonferroni correction. * denotes control versus high fat diet within a genotype; # denotes p < 0.05 WT versus CCM within a feeding group (n = 13–19 for gene expression and n = 6 for myocardial triglyceride and lipididomic profiling).
Myocardial lipidomic profiling and metabolic flux assessment were performed to investigate further the role that the cardiomyocyte circadian clock plays in metabolic adaptation of the heart to high fat feeding. Lipidomic profiling revealed that stearic acid, linolenic acid, eicosatrienoic acid, and arachidonic acid levels were increased by a high fat diet in WT, but not CCM, hearts (Fig. 1C and Table 4). Differential responsiveness to high fat feeding based on genotype (i.e. diet-genotype interaction) was specific for linolenic acid, eicosatrienoic acid, and arachidonic acid (Fig. 1C and Table 4).
TABLE 4.
Myocardial fatty acid composition in WT and CCM mice fed with either control or high fat diet
Values are expressed as the mean ± S.E. (nmol/g tissue; n = 6).
| WT |
CCM |
|||
|---|---|---|---|---|
| Control | High fat | Control | High fat | |
| (14:0) Myristic acid | 333 ± 18 | 329 ± 23 | 402 ± 14a | 336 ± 21b |
| (16:0) Palmitic acid | 12113 ± 348 | 11123 ± 526 | 12613 ± 545 | 11671 ± 190 |
| (18:0) Stearic acid | 12865 ± 183 | 15065 ± 321b | 13446 ± 455 | 14507 ± 299 |
| (20:0) Arachidic acid | 245 ± 22 | 181 ± 18b | 195 ± 17 | 153 ± 18 |
| (22:0) Behenic acid | 131 ± 18 | 113 ± 17 | 101 ± 12 | 106 ± 20 |
| (16:1) Palmitoleic acid | 817 ± 146 | 323 ± 61b | 968 ± 121 | 305 ± 33b |
| (18:1) Oleic acid | 10004 ± 637 | 10909 ± 965 | 9703 ± 456 | 9394 ± 251 |
| (20:1) Gadoleic acid | 405 ± 34 | 308 ± 26b | 289 ± 23a | 230 ± 7a,b |
| (22:1) Erucic acid | 92.9 ± 7.8 | 85.1 ± 3.7 | 82.8 ± 6.8 | 64.8 ± 4.5a,b |
| (18:2) n-6 Linolenic acid | 12142 ± 422 | 13248 ± 273b | 11747 ± 618 | 12008 ± 273a |
| (20:2) n-6 | 326 ± 20 | 803 ± 34b | 294 ± 30 | 754 ± 16b |
| (20:3) n-6 Eicosatrienoic acid | 673 ± 23 | 806 ± 49b | 581 ± 51 | 562 ± 27a |
| (20:4) n-6 Arachidonic acid | 6196 ± 169 | 8172 ± 157b | 6525 ± 186 | 6969 ± 168a |
| (22:4) n-6 | 532 ± 42 | 856 ± 44b | 565 ± 25 | 792 ± 18b |
| (22:5) n-6 Docosapentaenoic acid | 1559 ± 125 | 1505 ± 143 | 1403 ± 129 | 1206 ± 70 |
| (18:3) n-3 α-Linolenic acid | 461 ± 15 | 338 ± 27b | 483 ± 28 | 375 ± 15b |
| (20:5) n-3 Eicosapentaenoic acid | 65.9 ± 15.7 | 131 ± 14b | 70.0 ± 4.3 | 123 ± 20b |
| (22:5) n-3 Docosapentaenoic acid | 1009 ± 112 | 1269 ± 283b | 1136 ± 80 | 1214 ± 421b |
| (22:6) n-3 Docosahexaenoic acid | 17844 ± 325 | 17525 ± 819 | 17518 ± 824 | 17967 ± 875 |
a WT versus CCM within a feeding group.
b Control versus high fat diet within a genotype.
Hearts were next perfused in the working mode ex vivo for assessment of metabolic fluxes. Table 5 shows steady state contractile function parameters for hearts isolated from WT and CCM mice. All hearts were initially perfused for 30 min in the presence of 0.4 mm oleate followed by 30 min in the presence of 1.2 mm oleate. In contrast to the in vivo measurements, high fat feeding decreased cardiac power and efficiency in WT hearts, and this effect was not seen in the CCM hearts (although these parameters were already reduced in CCM hearts at ZT18, as reported previously at ZT18) (Table 5) (1). The differences observed between in vivo and ex vivo measurements of contractile function are likely due to neurohumoral compensation in vivo; ex vivo measurements are a better assessment of intrinsic myocardial function. Heart rate is also significantly lower in CCM hearts, as reported previously (Table 5) (1)).
TABLE 5.
Ex vivo cardiac functional parameters from hearts isolated from WT and CCM mice fed with either control or high fat diet and perfused initially at 0.4 mm oleate followed by an acute fatty acid challenge of 1.2 mm oleate
CP, cardiac power; OC, oxygen consumption; CE, cardiac efficiency; HR, heart rate; DP, developed pressure; RPP, rate pressure product; MVO2, myocardial oxygen consumption rate; bpm, beats per min.
| WT |
CCM |
|||||||
|---|---|---|---|---|---|---|---|---|
| Control |
High fat |
Control |
High fat |
|||||
| 0.4 mm Oleate | 1.2 mm Oleate | 0.4 mm Oleate | 1.2 mm | 0.4 mm Oleate | 1.2 mm Oleate | 0.4 mm Oleate | 1.2 mm Oleate | |
| CP (mW) | 1.42 ± 0.08 | 1.40 ± 0.09 | 0.95 ± 0.12a | 0.96 ± 0.13a | 0.96 ± 0.08b | 1.03 ± 0.10b | 1.08 ± 0.10 | 1.08 ± 0.12 |
| MVO2 (μmol/min/g dry wt) | 49.8 ± 1.1 | 52.3 ± 3.6 | 48.2 ± 5.9 | 49.5 ± 6.1 | 46.0 ± 3.3 | 54.9 ± 5.3 | 48.8 ± 2.3 | 53.0 ± 2.7 |
| CE (CP/MVO2) | 0.03 ± 0.001 | 0.03 ± 0.002 | 0.02 ± 0.002a | 0.02 ± 0.001a | 0.02 ± 0.002 | 0.02 ± 0.002b | 0.02 ± 0.003 | 0.02 ± 0.003 |
| HR (bpm) | 336 ± 9 | 339 ± 6 | 320 ± 13 | 337 ± 15 | 222 ± 20b | 242 ± 23b | 263 ± 17b | 263 ± 28b |
| DP (mm Hg) | 15.2 ± 1.8 | 15.3 ± 2.1 | 14.1 ± 0.9 | 13.8 ± 1.1 | 18.6 ± 1.3 | 18.6 ± 1.6 | 16.0 ± 0.9 | 16.5 ± 1.0 |
| RPP (bpm·mm Hg) | 5143 ± 720 | 5216 ± 802 | 4520 ± 421 | 4692 ± 510 | 4078 ± 395 | 4364 ± 433 | 3842 ± 477 | 3970 ± 633 |
Values are expressed as mean ± S.E. (n = 5–8).
a Control versus high fat diet.
b WT versus CCM.
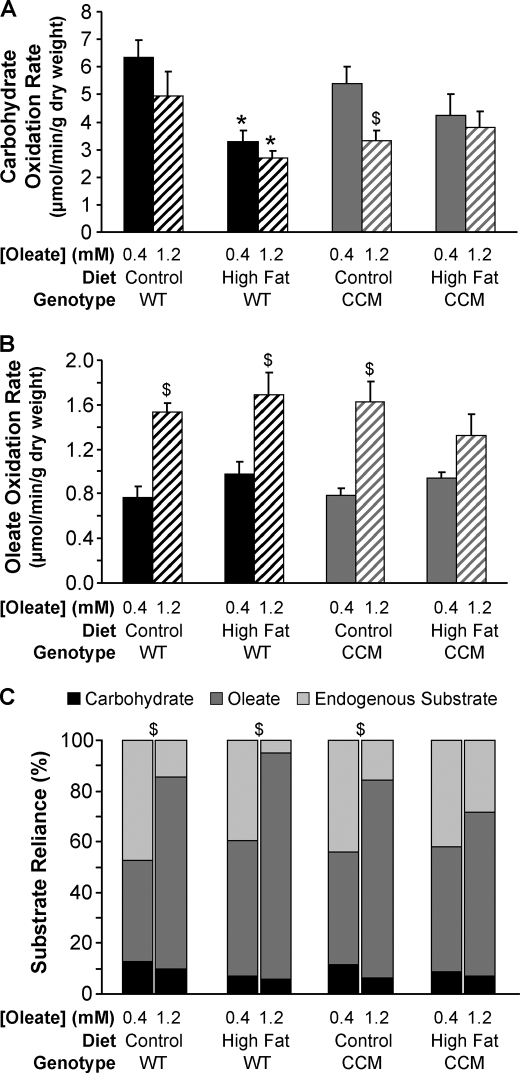
Consistent with previously published studies, high fat feeding decreased carbohydrate oxidation rates in WT hearts (Fig. 2A) (26). However, this decrease did not reach statistical significance in the CCM hearts (Fig. 2A). No statistically significant effects were observed for oleate oxidation rates with regard to diet or genotype (Fig. 2B). When challenged acutely with elevated fatty acids (i.e. 0.4 mm oleate to 1.2 mm oleate) ex vivo, hearts isolated from WT mice fed a control or high fat diet exhibit increased reliance on oleate oxidation and decreased reliance on endogenous substrate utilization (Fig. 2C). In contrast, hearts from CCM mice fed the high fat diet did not show this acute response, suggesting that the CCM hearts are not able to switch off endogenous substrate utilization under these conditions (Fig. 2C). The endogenous substrate being utilized is most likely triglyceride, consistent with elevated myocardial triglyceride levels observed in this group (Fig. 1B), suggesting that CCM hearts are unable to reduce endogenous triglyceride utilization during acute increases in fatty acid availability.
FIGURE 2.
High fat feeding-induced responses in myocardial carbohydrate oxidation rate (A), oleate oxidation rate (B), and substrate reliance (C). After 16 weeks of diet intervention, carbohydrate oxidation rate (A), oleate oxidation rate (B), and substrate reliance (C) were measured in WT and CCM hearts, isolated at ZT18, and perfused ex vivo in the working mode initially with 0.4 mm for 30 min and subsequently with 1.2 mm oleate for 30 min. [U-14C]Glucose (0.096 mCi/liter) and [U-14C]lactic acid (0.009 mCi/liter) were included in the perfusion buffer to determine carbohydrate oxidation rate and reliance. [9,10-3H]Oleate (0.067 mCi/liter) was included in the perfusion buffer to determine oleate oxidation rate and reliance. Data are represented as the mean ± S.E. Statistical analysis was performed using a three-way ANOVA followed by a post hoc comparison with Bonferroni correction. * denotes control versus high fat diet within a genotype; # denotes p < 0.05 WT versus CCM within a feeding group; $ denotes p < 0.05 0.4 mm oleate versus 1.2 mm oleate within a genotype and feeding group (n = 5–8).
CCM Hearts Exhibit Altered Diurnal Rhythms in Triglyceride Metabolism
Transcriptional, metabolite, and metabolic flux parameters measured thus far for the ad libitum high fat feeding studies in WT and CCM mice are consistent with cardiomyocyte circadian clock regulation of myocardial triglyceride metabolism. Multiple indices of triglyceride turnover were, therefore, investigated in WT and CCM hearts at distinct times of the day (ZT0, ZT6, ZT12, and ZT18) to directly test this hypothesis. These included measures of metabolite levels, metabolic fluxes, gene expression, protein expression, and kinase activities. For these studies, all mice were fed a normal murine chow diet in an ad libitum fashion.
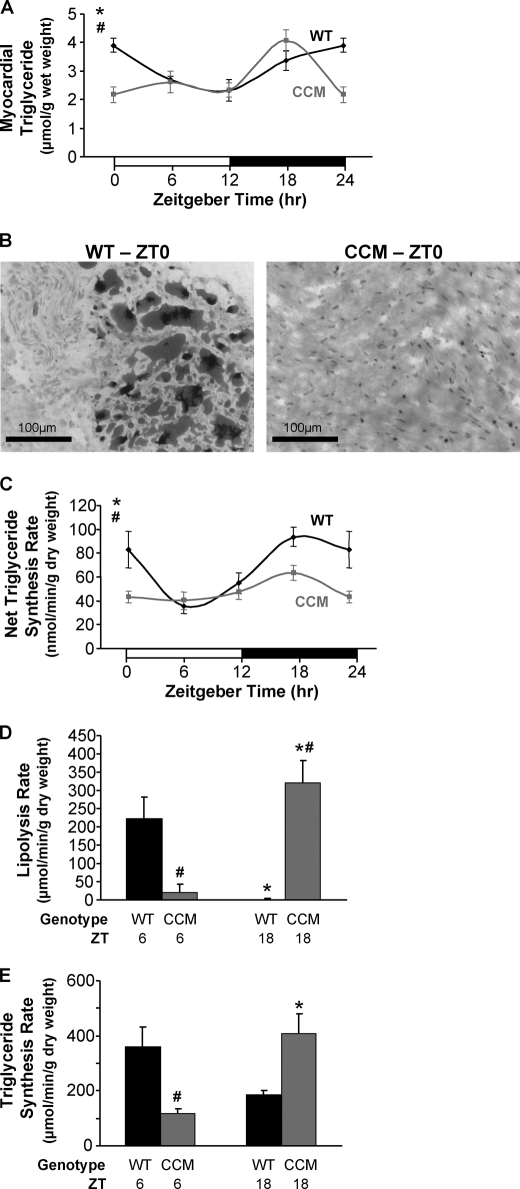
Myocardial triglyceride levels were investigated in WT and CCM hearts over the course of the day. WT hearts exhibited a significant diurnal variation in triglyceride levels, peaking at the wake-to-sleep transition (i.e. ZT0); this peak was attenuated in CCM hearts (Fig. 3, A and B). Interestingly, myocardial triglyceride levels are essentially identical in WT and CCM hearts at ZT18, suggesting that initial active phase-associated triglyceride synthesis in vivo is driven in large part by extracardiac factors (e.g. post-prandial rise in circulating insulin levels). To control for putative acute neurohumoral influences, ex vivo perfused hearts were next utilized. Here, net triglyceride synthesis rates were assessed by measuring net incorporation of [3H]oleate into triglyceride in WT and CCM hearts. WT hearts exhibited diurnal variations in net triglyceride synthesis, peaking at ZT18, in the middle of active/awake phase, with an ∼2.5-fold increase from trough to peak (Fig. 3C). This diurnal variation was severely attenuated in CCM hearts (Fig. 3C).
FIGURE 3.
Diurnal variations in myocardial triglyceride levels (A and B), net triglyceride synthesis rate (C), lipolysis rate (D), and triglyceride synthesis rate (E) in WT hearts were altered in CCM hearts. Myocardial triglyceride levels (A) were measured in WT and CCM hearts isolated at ZT 0, 6, 12, and 18. Oil red O staining (B) was performed on frozen sections from WT and CCM hearts isolated at ZT0. The black bar denotes 100 μm. Net incorporation of [3H]oleate into triglyceride (C) was measured in ex vivo perfused WT and CCM hearts isolated at ZT0, 6, 12, and 18; after the 40-min perfusion with buffer containing [9,10-3H]oleate (0.067 mCi/liter), the hearts were freeze-clamped and stored until analysis. Myocardial lipolysis (D) and triglyceride synthesis (E) were determined in ex vivo perfused WT and CCM hearts isolated at ZT6 and ZT18 using a pulse-chase method. The hearts were initially pulsed with 1.2 mm oleate and tracer amounts of [1-14C]oleate (0.12 mCi/liter) for 1 h followed by a 15-min wash with nonradioactive buffer and subsequently chased with buffer containing 0.4 mm oleate and tracer amounts of [9,10-3H]oleate (0.067 mCi/liter) for an additional 45 min. Data are represented as the mean ± S.E. Statistical analysis was performed using a two-way ANOVA followed by a post hoc comparison with Bonferroni correction. * denotes p < 0.05 time effect, and # denotes p < 0.05 genotype effect (n = 6 for myocardial triglyceride, n = 3–4 for Oil red O staining, n = 6–7 for net triglyceride synthesis, and n = 2–4 for lipolysis and triglyceride synthesis measurements).
An important question is whether diurnal variations in net incorporation of oleate into triglyceride were due to changes in synthesis and/or degradation rates. Therefore, triglyceride turnover rates were studied in WT and CCM hearts using a pulse-chase analysis ex vivo, as previously described for rat hearts (27). In this study WT and CCM hearts were isolated at ZT6 and ZT18, the trough and peak of net triglyceride synthesis observed earlier. Hearts were initially pulsed with buffer containing 1.2 mm oleate labeled with 14C tracer followed by a wash with nonradioactive buffer and, last, chased with buffer containing 0.4 mm oleate with 3H tracer. Consistent with low rates of net triglyceride synthesis at ZT6, lipolysis rates were highest at this time in WT hearts (Fig. 3D). In contrast, lipolysis rates were undetectable for WT hearts at ZT18, consistent with high rates of net triglyceride synthesis observed at this time (Fig. 3D). Triglyceride synthesis rates followed the same pattern as lipolysis rates in WT hearts and, therefore, do not readily explain net triglyceride synthesis rhythms (Figs. 3, D and E). Importantly, CCM hearts exhibit markedly different patterns of triglyceride turnover compared with WT hearts (Figs. 3, D and E). Collectively, these data suggest that cardiomyocyte circadian clock-mediated diurnal variations in lipolysis significantly contribute toward time-of-day-dependent myocardial triglyceride turnover.
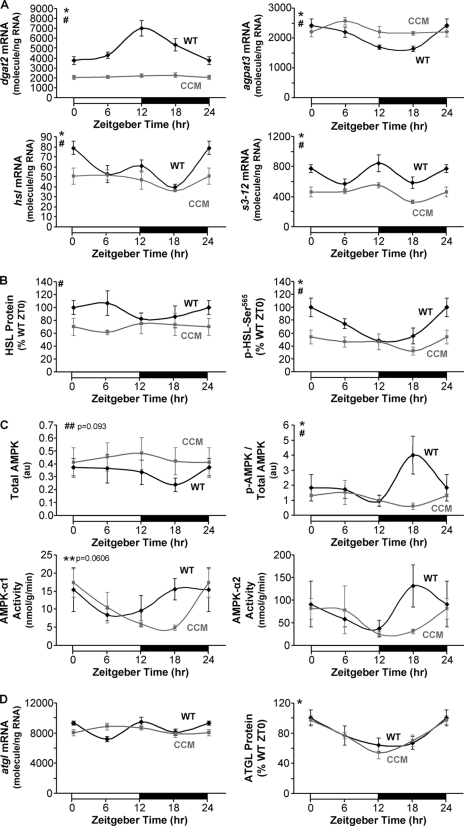
The possible mechanism(s) by which the cardiomyocyte circadian clock regulates myocardial triglyceride turnover was next investigated. Quantitative RT-PCR analyses revealed that dgat2, agpat3, hsl, and s3-12 mRNA levels exhibit diurnal variations in WT, but not CCM, hearts (Fig. 4A). Consistent with hsl mRNA data, we observed a slight, but significant repression of HSL total protein levels in CCM hearts (Fig. 4B). AMPK-mediated phosphorylation of HSL at Ser-565 results in inhibition of this lipase (28). WT hearts exhibited a significant diurnal variation in phospho-Ser-565-HSL that was absent in CCM hearts (Fig. 4B). Consistent with these observations, phospho-AMPK exhibited a diurnal variation in WT hearts that was significantly attenuated in CCM hearts (Fig. 4C). In addition, activity of the α1 subunit of AMPK showed a trend for diurnal variation peaking at ZT18 (p = 0.0606), consistent with the increase in phospho-Ser-565-HSL level during and after this time point. Collectively, these data suggest that the cardiomyocyte circadian clock influences HSL both transcriptionally and post-translationally via AMPK. Recent studies have exposed ATGL as an important enzyme in triglyceride metabolism both within adipose tissue and the myocardium (29). We, therefore, decided to investigate atgl mRNA and ATGL total protein levels in WT and CCM hearts. Gene expression of atgl neither oscillated in WT hearts nor was significantly different in CCM hearts (Fig. 4D). Consistent with the latter, ATGL total protein levels were not different between WT and CCM hearts (Fig. 4D). Interestingly, ATGL total protein levels exhibit an approximate 2-fold oscillation in both WT and CCM hearts (Fig. 4D), suggesting time-of-day-dependent post-transcriptional regulation that is likely mediated by systemic factors (i.e. independent of the cardiomyocyte circadian clock).
FIGURE 4.
Diurnal variations in myocardial enzymes involved in triglyceride metabolism were attenuated in CCM hearts at both the mRNA (A) and protein (B and C) levels. Gene expression analyses of dgat2 (A), agpat3 (A), hsl (A), s3-12 (A), and atgl (D) were performed using quantitative RT-PCR in WT and CCM hearts isolated at ZT 0, 6, 12, and 18. Protein and phosphoprotein expression levels (B, C, and D) of triglyceride metabolism enzymes were determined by Western blot analysis. AMPK-α1 and AMPK-α2 activities (C) were also measured. Data are represented as the mean ± S.E. Statistical analysis was performed using a two-way ANOVA followed by a post hoc comparison with Bonferroni correction. * denotes p < 0.05 time effect, and # denotes p < 0.05 genotype effect (n = 6 for gene expression and n = 4–6 for protein, phosphoprotein, and activity measurements).
Time of Day at Which Dietary Fat Is Consumed Profoundly Influences Myocardial Steatosis
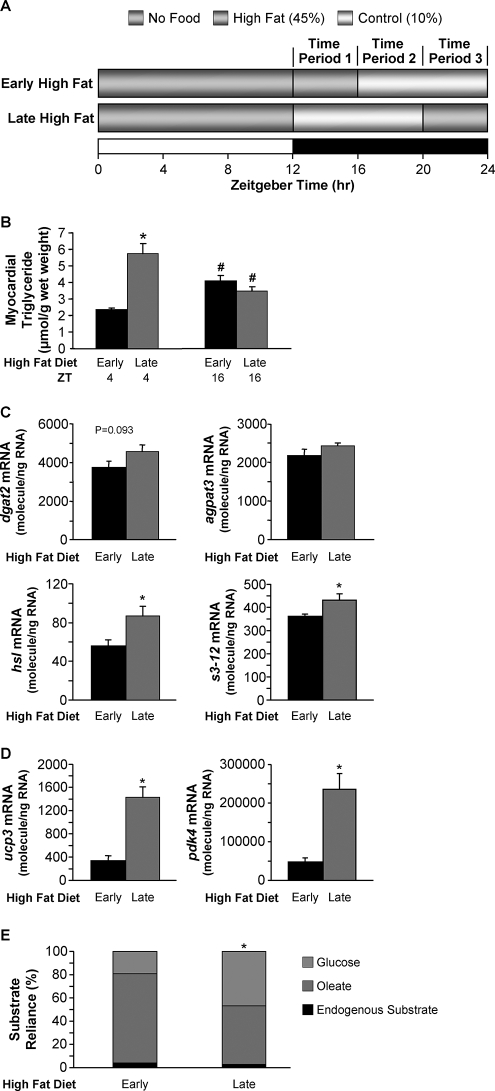
Data presented thus far show diurnal variations in myocardial triglyceride metabolism, which are mediated by the cardiomyocyte circadian clock. In an attempt to reveal the potential significance of these findings, WT mice were fed dietary fat in a time-of-day-dependent manner, and the effects on myocardial steatosis were investigated. As illustrated in Fig. 5A, mice were randomly assigned to one of two feeding regimes for 16 weeks. For both groups, mice were allowed access to food only during the normal awake (dark) phase. The 12-h dark phase was divided into three distinct 4-h time periods (TPs; which could be considered comparable with periods of breakfast, lunch, and dinner for TP1, TP2, and TP3, respectively). For the early high fat group mice were provided a high fat diet only during TP1 (and control diet during TP2 and TP3). For the late fat group mice were provided a high fat diet only during TP3 (and control diet during TP1 and TP2). Consistent with a higher propensity for net triglyceride synthesis at the end of the dark phase (Fig. 3C), hearts isolated from the late high fat group (at ZT4, during the light phase) possess higher triglyceride levels (i.e. greater steatosis) compared with hearts isolated from early high fat-fed mice (∼2.5-fold; Fig. 5B). In contrast, myocardial triglyceride levels were not significantly different between early and late high fat-fed groups during the dark phase (ZT16; Fig. 5B), suggesting continuous turnover of this lipid pool in late high fat-fed mice. Indeed, expression of genes promoting both lipolysis (e.g. hsl) and triglyceride synthesis (e.g. s3-12) were increased in hearts from the late high fat group versus the early high fat group (Fig. 5C). Hearts from the late high fat group also exhibit increased expression of known fatty acid-responsive genes (Fig. 5D) and rates of endogenous substrate utilization (compared with the early high fat group; Fig. 5E) during the light phase (i.e. ZT4), consistent with increased myocardial triglyceride stores acting as a significant source of endogenous fatty acids at this time (when lipolysis rates are higher in the mouse heart). Associated with increased triglyceride turnover (an energetically wasteful process) in late fat-fed mouse hearts, cardiac efficiency tended to be lower in this group (Table 6).
FIGURE 5.
Consumption of a high fat diet at the end of the active/awake phase results in marked elevations in myocardial triglyceride (B), metabolic gene expression (C), myocardial fatty acid gene expression (D), and alterations in myocardial substrate reliance (E). After 16 weeks of diet intervention, myocardial triglyceride levels (B) were measured in hearts isolated at ZT4 and ZT16. Gene expression analyses of dgat2 (C), agpat3 (C), hsl (C), s3-12 (C), ucp3 (D), and pdk4 (D) were performed using quantitative RT-PCR in WT hearts isolated at ZT4. Substrate reliance (E) was measured in hearts perfused ex vivo in the working mode under basal conditions at ZT4. In this study hearts were perfused only with Krebs-Henseleit buffer containing 8 mm glucose, 0.4 mm oleate conjugated to 3% bovine serum albumin, and tracer amounts of [U-14C]glucose (0.12 mCi/liter) and [9,10-3H]oleate (0.067 mCi/liter) for glucose and oleate oxidation measurements, respectively. Data are represented as the mean ± S.E. Statistical analysis was performed using a two-way ANOVA. * denotes p < 0.05 diet effect; # denotes p < 0.05 time effect within a feeding group (n = 5–6 for gene expression, n = 6 for myocardial triglyceride, and n = 6–7 for ex vivo measurements).
TABLE 6.
Ex vivo cardiac functional parameters from hearts isolated from WT mice fed with either an early or a late high fat diet
Values are expressed as the mean ± S.E. (n = 6–7). CP, cardiac power; OC, oxygen consumption; CE, cardiac efficiency; HR, heart rate; DP, developed pressure; RPP, rate pressure product; MVO2, myocardial oxygen consumption rate; bpm, beats per min.
| Early high fat | Late high fat | |
|---|---|---|
| CP (milliwatts) | 1.14 ± 0.10 | 1.07 ± 0.15 |
| MVO2 (μmol/min/g dry wt) | 45.4 ± 4.9 | 53.8 ± 6.6 |
| CE (CP/MVO2) | 0.026 ± 0.003 | 0.020 ± 0.002 |
| HR (bpm) | 307 ± 11 | 316 ± 29 |
| DP (mm Hg) | 15.6 ± 1.8 | 14.8 ± 1.0 |
| RPP (bpm·mm Hg) | 4741 ± 496 | 4679 ± 550 |
DISCUSSION
The purpose of this study was to determine whether a cell autonomous circadian clock plays a significant role in the chronic metabolic adaptation of a peripheral tissue to fatty acids. Accordingly, we investigated whether an in vivo model of cardiomyocyte-specific circadian clock disruption (i.e. CCM mice) was associated with differential responsiveness of the heart to high fat feeding. We found that CCM mice exhibit altered myocardial responsiveness to high fat feeding at the levels of the transcriptome and lipidome as well as at metabolic flux and contractile function levels. Subsequent studies confirmed that the cardiomyocyte circadian clock directly regulates triglyceride turnover, primarily at the level of lipolysis. Mechanistic studies suggest that the cardiomyocyte circadian clock regulates HSL both transcriptionally and post-translationally. Consistent with circadian clock-mediated augmentation of net triglyceride synthesis at the end of the active period, high fat feeding at this time results in marked cardiac steatosis. Taken together, these data reveal that a peripheral cell autonomous circadian clock directly regulates triglyceride metabolism. Dysregulation of peripheral circadian clocks during metabolic disease states may, therefore, contribute toward multiple features of the cardiometabolic syndrome through disparities in triglyceride turnover.
Circadian clocks and metabolism are closely interlinked (30). Indeed, several metabolism-based loops have now been described for the mammalian circadian clock. It is, therefore, not surprising that genetic models of clock component mutation/ablation often present profound metabolic phenotypes. For instance, ubiquitous genetic mouse models of CLOCK mutation or BMAL1 ablation (central components of the mammalian circadian clock system) markedly influence whole-body metabolism (9, 31). The CLOCKΔ19 mutant mouse is both hyperphagic and obese and exhibits abnormalities in circulating glucose, lipids, and hormones/adipokines, consistent with a cardiometabolic syndrome phenotype (9). Additionally, abnormalities in intestinal nutrient uptake have been reported in these mice, consistent with circadian clock regulation of digestion and absorption (32). Interestingly, CLOCKΔ19 mutant mice appear to have increased insulin sensitivity, underscoring a complex metabolic phenotype (9). This complexity is illustrated further by differential responsiveness of hepatic triglyceride accumulation after distinct nutritional challenges (10). CLOCKΔ19 mutant mice exhibit attenuated high fat feeding-induced hepatic steatosis despite reports that the same model shows increased sensitivity to alcohol-induced hepatic steatosis (10). The complex metabolic phenotype exhibited in these mice likely reflects the impact that this ubiquitous mutation has on multiple tissues/systems, both peripheral and central. A striking difference in the metabolic phenotype of BMAL1 null mice, compared with CLOCKΔ19 mutant mice, is regarding adiposity; BMAL1 null mice are lean and exhibit marked lipoatrophy with age (8). Consistent with this observation, in vitro studies have shown that BMAL1 plays an important role in adipogenesis (33).
To better define the roles of central versus peripheral circadian clocks on metabolic regulation in vivo, several laboratories have begun to develop cell type-specific models of clock disruption. For example, through overexpression of REV-ERBα (a negative regulator of BMAL1) in hepatocytes, Kornmann et al. (34) revealed the contribution of the hepatocyte circadian clock on metabolic gene expression oscillations in the liver over the course of the day. More recently, Lamia et al. (12) have reported that the hepatocyte circadian clock plays a central role in whole body glucose homeostasis through generation of a hepatocyte-specific BMAL1 null mouse. Along the same lines, we have utilized a mouse model with cardiomyocyte-specific circadian clock disruption (i.e. CCM mice), exposing critical roles for this peripheral circadian clock in modulating the acute transcriptional responsiveness of the myocardium to fatty acids as well as regulating glycogenolysis (1, 13). In addition, the present study revealed a critical role for this peripheral clock mechanism in metabolic adaptation of the heart to high fat feeding.
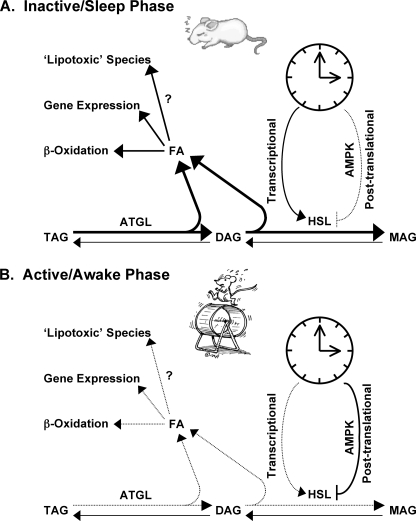
Transcriptome and lipidome analysis of hearts isolated from WT versus CCM mice fed a high fat diet for 16 weeks suggested that the cardiomyocyte circadian clock distinctly influences both fatty acid and triglyceride metabolism (supplemental Fig. 1 and Table 4). Subsequent metabolite measurements (Fig. 1B) and metabolic flux analyses (Fig. 2) provided data consistent with increased triglyceride turnover in hearts isolated from high fat-fed CCM mice. To better define the influence that the cardiomyocyte circadian clock plays on myocardial triglyceride turnover, WT and CCM hearts were next investigated at multiple time points over the course of the day. As reviewed previously, for a process to be defined as cardiomyocyte circadian clock-regulated, the process should oscillate in a diurnal/circadian manner in WT hearts, and these oscillations should be attenuated/abolished in CCM hearts. Consistent with these criteria, diurnal variations in myocardial triglyceride levels (Fig. 3A) as well as net triglyceride synthesis (Fig. 3C) are markedly attenuated in CCM hearts. This appears to be due to circadian clock-mediated regulation of lipolysis as opposed to synthesis (Fig. 3D). Lipolysis rates are increased during the sleep phase in WT but not CCM hearts. During the active phase, lipolysis is not detectable in WT hearts, which would promote post-prandial storage of excess dietary nutrients. Circadian clock regulation of lipolysis is likely mediated at least in part through transcriptional and post-translational control of HSL. Our data are consistent with the model presented in Fig. 6 wherein the cardiomyocyte circadian clock regulates HSL activity through transcriptional and post-translational (e.g. AMPK-dependent) mechanisms. Although ATGL does not appear to be under transcriptional control by the circadian clock, we cannot discount post-translational regulation of this important triglyceride lipase (through alterations in phosphorylation status or levels of activating protein) via the clock mechanism.
FIGURE 6.
Proposed regulation of triglyceride metabolism by peripheral circadian clocks during the mouse sleep (A) and active/awake (B) phase. The cardiomyocyte circadian clock regulates HSL activity through transcriptional and post-translational mechanisms, likely an AMPK-dependent process. AMPK, AMP-activated protein kinase; DAG, diacylglycerol; MAG, monoacylglycerol; TAG, triacylglycerol.
Mammals consume dietary nutrients with often predictable temporal patterns. One common feature between the feeding behaviors of different mammals is that nutrient intake occurs normally when the animal is awake. Humans generally tend to consume the majority of dietary nutrients during discrete mealtimes. In contrast, laboratory rodents (e.g. mice) will eat throughout the 24-h period when fed ab libitum, although the greatest food intake (approximately two-thirds) is during the active (dark) phase. In an attempt to enforce a human meal-feeding regime on mice without causing nutrient deprivation (and, therefore, activation of a starvation-like metabolic response), mice were allowed access to food only during the dark phase. During this time half the mice were allowed access to a high fat diet during the beginning of the dark phase, whereas the other half consumed the high fat diet only during the end of the dark phase. This feeding regime (as depicted in Fig. 5A) was intended to simulate a high fat breakfast (i.e. early high fat-fed group) or a high fat dinner (i.e. late high fat-fed group). The results were remarkable. Consumption of the high fat diet at the end of the dark phase resulted in marked cardiac steatosis relative to high fat diet consumption during the beginning of the dark phase. This myocardial triglyceride pool appears to be relatively labile, apparently generating fatty acids species for induction of fatty acid-responsive genes (Fig. 5D) and β-oxidation (Fig. 5E). Whether the triglyceride-derived fatty acyl groups are also channeled into so-called lipotoxic pathways remains to be investigated. Clearly these data have broad implications toward human health and disease. Dietary lipid consumption in the evening likely promotes triglyceride accumulation in extra-cardiac tissues also, such as adipose, liver, and skeletal muscle. If so, this could potentially contribute toward cardiometabolic syndrome development.
In summary, the present study has exposed an important role for a peripheral circadian clock in the metabolic adaptation of an organ to high fat feeding. Furthermore, these studies are consistent with direct regulation of triglyceride turnover by a peripheral circadian clock. The findings support the hypothesis that circadian clocks play critical roles in metabolic regulation, and disruption of these cell autonomous mechanisms likely contributes toward multiple metabolic disease states, including obesity, diabetes mellitus, and cardiovascular disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL-074259 (NHLBI).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Table 1.
- CCM
- cardiomyocyte clock mutant
- ZT
- zeitgeber time
- ATGL
- adipose triglyceride lipase
- HSL
- sensitive lipase
- TP
- time period
- WT
- wild type
- RT
- reverse transcription
- ANOVA
- analysis of variance.
REFERENCES
- 1.Bray M. S., Shaw C. A., Moore M. W., Garcia R. A., Zanquetta M. M., Durgan D. J., Jeong W. J., Tsai J. Y., Bugger H., Zhang D., Rohrwasser A., Rennison J. H., Dyck J. R., Litwin S. E., Hardin P. E., Chow C. W., Chandler M. P., Abel E. D., Young M. E. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1036–H1047 [DOI] [PubMed] [Google Scholar]
- 2.Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. (2000) Genes Dev. 14, 2950–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durgan D. J., Moore M. W., Ha N. P., Egbejimi O., Fields A., Mbawuike U., Egbejimi A., Shaw C. A., Bray M. S., Nannegari V., Hickson-Bick D. L., Heird W. C., Dyck J. R., Chandler M. P., Young M. E. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H2385–H2393 [DOI] [PubMed] [Google Scholar]
- 4.Martino T., Arab S., Straume M., Belsham D. D., Tata N., Cai F., Liu P., Trivieri M., Ralph M., Sole M. J. (2004) J. Mol. Med. 82, 256–264 [DOI] [PubMed] [Google Scholar]
- 5.van den Buuse M. (1999) Physiol. Behav. 68, 9–15 [DOI] [PubMed] [Google Scholar]
- 6.Young M. E., Bray M. S. (2007) Sleep Med. 8, 656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edery I. (2000) Physiol. Genomics 3, 59–74 [DOI] [PubMed] [Google Scholar]
- 8.Bunger M. K., Walisser J. A., Sullivan R., Manley P. A., Moran S. M., Kalscheur V. L., Colman R. J., Bradfield C. A. (2005) Genesis. 41, 122–132 [DOI] [PubMed] [Google Scholar]
- 9.Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudo T., Tamagawa T., Kawashima M., Mito N., Shibata S. (2007) J. Biol. Rhythms 22, 312–323 [DOI] [PubMed] [Google Scholar]
- 11.Kudo T., Tamagawa T., Shibata S. (2009) J. Circadian. Rhythms 7, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamia K. A., Storch K. F., Weitz C. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durgan D. J., Trexler N. A., Egbejimi O., McElfresh T. A., Suk H. Y., Petterson L. E., Shaw C. A., Hardin P. E., Bray M. S., Chandler M. P., Chow C. W., Young M. E. (2006) J. Biol. Chem. 281, 24254–24269 [DOI] [PubMed] [Google Scholar]
- 14.Young M. E., Razeghi P., Cedars A. M., Guthrie P. H., Taegtmeyer H. (2001) Circ. Res. 89, 1199–1208 [DOI] [PubMed] [Google Scholar]
- 15.Szczepaniak L. S., Victor R. G., Orci L., Unger R. H. (2007) Circ. Res. 101, 759–767 [DOI] [PubMed] [Google Scholar]
- 16.Barger P. M., Kelly D. P. (2000) Trends Cardiovasc. Med. 10, 238–245 [DOI] [PubMed] [Google Scholar]
- 17.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 18.Lockridge J. B., Sailors M. L., Durgan D. J., Egbejimi O., Jeong W. J., Bray M. S., Stanley W. C., Young M. E. (2008) J. Lipid Res. 49, 1395–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson U. E., Heid C. A., Williams P. M. (1996) Genome Res. 6, 995–1001 [DOI] [PubMed] [Google Scholar]
- 20.Heid C. A., Stevens J., Livak K. J., Williams P. M. (1996) Genome Res. 6, 986–994 [DOI] [PubMed] [Google Scholar]
- 21.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 22.Morrison W. R., Smith L. M. (1964) J. Lipid Res. 5, 600–608 [PubMed] [Google Scholar]
- 23.Carson F. L. (1997) Histotechnology: A Self-instructional Text, 2nd Ed., pp. 159–161, ASCP Press, Chicago [Google Scholar]
- 24.Minokoshi Y., Kim Y. B., Peroni O. D., Fryer L. G., Müller C., Carling D., Kahn B. B. (2002) Nature 415, 339–343 [DOI] [PubMed] [Google Scholar]
- 25.Hardin P. E. (2004) J. Biol. Rhythms 19, 348–360 [DOI] [PubMed] [Google Scholar]
- 26.Wilson C. R., Tran M. K., Salazar K. L., Young M. E., Taegtmeyer H. (2007) Biochem.J. 406, 457–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saddik M., Lopaschuk G. D. (1991) J. Biol. Chem. 266, 8162–8170 [PubMed] [Google Scholar]
- 28.Donsmark M., Langfort J., Holm C., Ploug T., Galbo H. (2004) Biochem. Biophys. Res. Commun. 316, 867–871 [DOI] [PubMed] [Google Scholar]
- 29.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 30.Taegtmeyer H. (2000) J. Am. Coll. Cardiol. 36, 1386–1388 [DOI] [PubMed] [Google Scholar]
- 31.Rudic R. D., McNamara P., Curtis A. M., Boston R. C., Panda S., Hogenesch J. B., Fitzgerald G. A. (2004) PLoS. Biol. 2, e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan X., Hussain M. M. (2009) J. Lipid Res. 50, 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornmann B., Schaad O., Bujard H., Takahashi J. S., Schibler U. (2007) PLoS. Biol. 5, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.