Abstract
To prevent genetic code ambiguity due to misincorporation of amino acids into proteins, aminoacyl-tRNA synthetases have evolved editing activities to eliminate intermediate or final non-cognate products. In this work we studied the different editing pathways of class Ia leucyl-tRNA synthetase (LeuRS). Different mutations and experimental conditions were used to decipher the editing mechanism, including the recently developed compound AN2690 that targets the post-transfer editing site of LeuRS. The study emphasizes the crucial importance of tRNA for the pre- and post-transfer editing catalysis. Both reactions have comparable efficiencies in prokaryotic Aquifex aeolicus and Escherichia coli LeuRSs, although the E. coli enzyme favors post-transfer editing, whereas the A. aeolicus enzyme favors pre-transfer editing. Our results also indicate that the entry of the CCA-acceptor end of tRNA in the editing domain is strictly required for tRNA-dependent pre-transfer editing. Surprisingly, this editing reaction was resistant to AN2690, which inactivates the enzyme by forming a covalent adduct with tRNALeu in the post-transfer editing site. Taken together, these data suggest that the binding of tRNA in the post-transfer editing conformation confers to the enzyme the capacity for pre-transfer editing catalysis, regardless of its capacity to catalyze post-transfer editing.
Keywords: Aminoacyl tRNA Synthetase, Enzyme Catalysis, Enzyme Mechanisms, Mutant, Transfer RNA (tRNA), Aquifex aeolicus LeuRS, E. coli LeuRS, tRNA-dependent Pre-transfer Editing
Introduction
Aminoacyl-tRNA synthetases (aaRSs)3 are key enzymes involved in the translation of genetic information by catalyzing the formation of aminoacyl-tRNAs (1, 2). The aminoacylation reaction carried by aaRS is a two-step process. First, the amino acid is activated by ATP to yield an aminoacyl-adenylate intermediate. The second step consists in the transfer of the aminoacyl moiety to one of the two hydroxyl oxygens of the 3′-terminal nucleotide of tRNA to form the aminoacyl-tRNA and liberate AMP (3). The 20 aaRSs can be divided into 2 classes of 10 members each on the basis of conserved sequences and characteristic structural motifs (4). The accuracy of the tRNA aminoacylation reaction is essential to the fidelity of protein synthesis and, hence, cellular functions and viability (5–7). Each aaRS must select its cognate amino acid from the cellular pool of 20 different proteinaceous amino acids with an overall accuracy of approximately 1 error per 104–105 codons (8). Compared with tRNA selection, recognition of cognate amino acid is challenging for aaRSs because amino acids are small molecules with only a few interacting groups (9). Misactivation of amino acids isosteric to the cognate amino acid is observed by approximately half of all aaRSs because of the inherent physiochemical limitations on closely discrimination similar amino acid side chains (3). One typical example is leucyl-tRNA synthetase (LeuRS), which can misactivate isoleucine, methionine, homocysteine, α-amino butyrate, and norvaline (Nva) with different efficiencies (10).
To clear misactivated amino acid and misaminoacyl-tRNA, some synthetases have evolved an additional error-correcting mechanism known as “pre-transfer editing” to hydrolyze misaminoacyl-adenylate and “post-transfer editing” for removing mischarged tRNAs (3, 9). The post-transfer editing has been shown to occur in a distinct hydrolytic site that deacylates mischarged tRNAs while excluding the correct aminoacyl-tRNA (11). The editing site for class Ia subgroup aaRSs, including isoleucyl-tRNA synthetase, valyl-tRNA synthetase, and LeuRS, is located within the domain called connective peptide 1 (CP1), a discretely folded domain of about 200 residues inserted into the Rossmann-fold catalytic domain where aminoacylation occurs (11–14). A threonine-rich region with the CP1 domains marks the editing site (11, 15–17). The mechanism is believed to involve shuttling of the flexible CCA-3′ end of the tRNA from the synthetic site to the hydrolytic editing site located at a 30 Å distance (11, 18, 19). However, the mechanism, and even the actual active site of pre-transfer editing still remains controversial. The non-cognate aminoacyl-adenylates can be hydrolyzed via tRNA-dependent (20, 21) or tRNA-independent pathways by LeuRS and other aaRSs (22, 23). Fluorescence-based assays and mutational analysis together with x-ray crystallography studies showed that both pre- and post-transfer editing substrate analogs bind in overlapping sites in the CP1 domain of LeuRS and isoleucyl-tRNA synthetase and suggested that misactivated amino acid is translocated from the catalytic synthetic site to the editing domain in a tRNA-dependent manner (11, 15, 24–26). To accomplish this, a post-initiated pre-transfer editing model has been postulated for isoleucyl-tRNA synthetase. According to this hypothesis, after an initial post-transfer editing step the binding of tRNA triggers a conformational change in the editing site that makes it competent for pre-transfer editing (27). However, a clear structural basis for such a mechanism is unclear because no confined passageway between the synthetic and editing sites is apparent that might serve to prevent dissociation of misactivated aminoacyl-adenylates from the surface of the enzyme during translocation. On the other hand, several recent studies have shown that tRNA-dependent pre-transfer editing may occur in the synthetic active site. This has been first shown for class I glutaminyl-tRNA synthetase, which normally lacks a spatially separate editing domain (28). Similarly, class II prolyl-tRNA synthetase and seryl-tRNA synthetase also edit non-cognate aminoacyl-adenylates in their synthetic active site in a tRNA-independent way (23, 29). For LeuRS, the location of a tRNA-dependent pre-transfer editing activity within the synthetic site was also proposed (30). More recently, using a CP1-inactivated mutant of LeuRS, a tRNA-independent pre-transfer editing was suggested to occur in the synthetic site, whereas both tRNA-dependent pre-transfer and post-transfer editing occur in the CP1 domain (22).
Herein, we focus our study on tRNA-dependent pre-transfer editing of Aquifex aeolicus and Escherichia coli LeuRS (AaLeuRS and EcLeuRS) (22, 31). AaLeuRS is the only known heterodimeric LeuRS consisting of an α- and β-subunit of 634 and 289 amino acid residues, respectively (32). EcLeuRS is a single polypeptide enzyme of 860 amino acid residues. Here we show that both enzymes edit the non-cognate amino acid Nva, as demonstrated by the robust AMP formation in the thin-layer chromatography (TLC)-based editing assay. We examined the editing activity of CP1-inactivated mutants located in the Thr-rich region of the two enzymes (AaLeuRS-T273R and EcLeuRS-T252R). Despite a total loss of post-transfer editing, both mutated enzymes still retained high AMP formation rates, higher than in the absence of tRNA, suggesting that another editing pathway depending on the tRNA might exist in addition to the post-transfer editing pathway. The same effect was observed when AN2690 (5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole), an antifungal agent that binds into the CP1 editing site and traps tRNALeu, was used (33). We found that AN2690 inhibits aminoacylation and hydrolysis of mischarged tRNA but, unexpectedly, the compound only partially impaired AMP formation. On the contrary, when AaLeuRS-Y358D and EcLeuRS-Y330D mutants were used to block the tRNA entry into the CP1 editing pocket, both tRNA-dependent editing pathways were abolished. These data strongly suggested that prokaryotic AaLeuRS and EcLeuRS possess strong tRNA-dependent pre-transfer editing activity that crucially depends on the interaction of the CCA acceptor end within the CP1 domain. Our results also indicated that tRNA-dependent pre-transfer editing might cooperate with post-transfer editing to improve the fidelity of catalysis of LeuRS.
EXPERIMENTAL PROCEDURES
Preparation of Enzymes and RNA Substrates
AaLeuRS and EcLeuRS were overproduced in E. coli transformants containing their genes as His-tagged proteins and purified by nickel-nitrilotriacetic acid chromatography, as described previously (13, 33). The genes encoding the various mutations were constructed by the two-step PCR method, and the DNA sequences of all the mutants were confirmed by DNA sequencing. A. aeolicus tRNAGAGLeu (AatRNALeu) and E. coli tRNAGAGLeu (EctRNALeu), with an accepting activity of 1400 and 1300 pmol/A260, were prepared from an overproduction strain constructed in our laboratory, respectively (32, 34). Ile-EctRNALeu and Ile-AatRNALeu were obtained using EcLeuRS-Y330D mutant.
tRNA Charging and Deacylation
Aminoacylation activities of AaLeuRS or EcLeuRS were performed at 37 °C in a reaction mixture containing 100 mm Tris-HCl (pH 7.8), 30 mm KCl, 12 mm MgCl2, 0.5 mm dithiothreitol, 4 mm ATP, 20 μm AatRNALeu or EctRNALeu, 40 μm [3H]leucine (15 Ci/mmol), and 20 nm AaLeuRS or EcLeuRS with or without 100 μm AN2690 (Milestone Pharmtech USA Inc.) (13, 33). Misacylation assays were carried out in a similar system, except that 100 μm [3H]isoleucine (30 Ci/mmol) and 3 μm AaLeuRS or 0.5 μm EcLeuRS mutants were used. Hydrolytic editing assays of AaLeuRS or EcLeuRS and the mutants were performed at 37 °C in 100 mm Tris-HCl (pH 7.5), 30 mm KCl, 12 mm MgCl2, 0.5 mm dithiothreitol, and 1 μm [3H]Ile-tRNALeu (300 μCi/μmol), and the reactions were initiated with 20 nm enzyme. To measure deacylation of mischarged Ile-EctRNALeu (1 μm) in the presence of AN2690 (100 μm), uncharged EctRNALeu (5 μm) was added to the reaction mixture. Only uncharged tRNA can react with AN2690 and form the covalent adduct responsible for the enzyme inactivation.
AMP Formation
AMP formation by AaLeuRS was measured as previously described (22). The reaction mixture contained 100 mm Tris-HCl (pH 7.8), 30 mm KCl, 12 mm MgCl2, 5 mm dithiothreitol, 5 units/ml pyrophosphatase (Roche Applied Science), 3 mm ATP, 20 nm [α-32P]ATP (3000 Ci/mmol; Amersham Biosciences), and 15 mm Nva in the presence or absence of 5 μm tRNALeu. The reaction was initiated by the addition of 1 μm AaLeuRS or its mutants and incubated at 60 °C. For EcLeuRS, the assay was carried out at 37 °C and initiated by 0.2 μm enzyme. Aliquots (1.5 μl) were quenched in 6 μl of 200 mm sodium acetate (pH 5.0). Quenched aliquots (1.5 μl each) were spotted in duplicate on polyethyleneimine cellulose plates (Merck) pre-washed with water. Separation of aminoacyl-[32P]AMP, [32P]AMP, and [32P]ATP was performed by developing TLC plates in 0.1 m ammonium acetate and 5% acetic acid. The plates were visualized by phosphorimaging, and data were analyzed using Multi Gauge Version 3.0 software (FUJIFILM). The gray densities of [32P]AMP spots were compared with the gray density of known [32P]ATP concentrations. Rate constants were obtained from graphs of [32P]AMP formation plotted against time.
Determination of the Kd for tRNALeu by Tryptophan Fluorescence Quenching
Equilibrium titrations were performed at room temperature with 0.1 μm enzyme in 100 mm Tris-HCl (pH 7.8), 30 mm KCl, 12 mm MgCl2, and 0.5 mm dithiothreitol. Tryptophan fluorescence was excited at 280 nm. An emission wavelength of 340 nm was used to quantify binding after correction for dilution and for the inner filter effect. Control solutions of bovine serum albumin or tryptophan were performed to show that there was no fluorescence response to tRNA. The Kd values were determined by fitting fluorescence intensity change data versus tRNA concentration using Originpro 7.5 software.
RESULTS
Both E. coli and A. aeolicus LeuRSs Exhibit Strong tRNA-dependent Editing Pathways
In the presence of cognate amino acid, synthesis of aminoacyl-tRNA occurs after transient synthesis of an aminoacyl-adenylate molecule. It results in the release of a single AMP and pyrophosphate molecule. However, when a non-cognate amino acid is activated by an aaRS, it may be mischarged on the tRNA and then hydrolyzed in the editing domain or be directly hydrolyzed by the enzyme before the transfer step. Both hydrolytic pathways lead to the production of one AMP molecule per cycle. Therefore, the extra formation of AMP during the catalytic process is characteristic of editing events. AMP formation can be directly monitored on TLC (28).
The non-cognate amino acid Nva was used to assay the editing activity of AaLeuRS and EcLeuRS in the presence and absence of tRNA. The data in Table 1 showed that tRNA could significantly stimulate the editing activity measured by the increase of AMP formation due to the editing reaction. The observed rate constant of AMP formation (kobs) of AaLeuRS in the absence and presence of AatRNALeu was 0.086 and 1.43 s−1 (Table 1), respectively, consistent with our previous results (22). We then examined and measured the AMP formation rate of EcLeuRS in the presence of Nva. The kobs in the absence of tRNALeu was 0.33 s−1, whereas it reached 3.42 s−1 in the presence of tRNALeu (Table 2). These results showed that tRNALeu robustly stimulates editing activity of either AaLeuRS or EcLeuRS.
TABLE 1.
Observed rate constants of AaLeuRS and its mutants in AMP synthesis at 60 °C
Rates were determined using the AMP formation in the TLC assay described under “Experimental Procedures.” All rates represent the average of three trials with the S.D. indicated.
| LeuRS | tRNA | AMP formation kobs |
|---|---|---|
| s−1 | ||
| Wild type | − | (8.6 ± 1.4) × 10−2 |
| Wild type | + | 1.43 ± 0.25 |
| T273R | − | (9.1 ± 1.4) × 10−2 |
| T273R | + | 1.01 ± 0.17 |
| Y358D | − | (8.8 ± 1.4) × 10−2 |
| Y358D | + | (9.5 ± 1.6) × 10−2 |
| Y358E | − | (9.2 ± 1.4) × 10−2 |
| Y358E | + | (1.7 ± 1.6) × 10−1 |
TABLE 2.
Observed rate constants of EcLeuRS and its mutants in AMP synthesis at 37 °C
Rates were determined using the AMP formation in the TLC assay described under “Experimental Procedures.” All rates represent the average of three trials with the S.D. indicated.
| LeuRS | tRNA | AN2690 | AMP formation kobs |
|---|---|---|---|
| s−1 | |||
| WT | − | − | (3.3 ± 0.4) × 10−1 |
| WT | + | − | 3.42 ± 0.51 |
| WT | − | + | (3.1 ± 0.4) × 10−1 |
| WT | + | + | 1.86 ± 0.28 |
| T252R | − | − | (3.5 ± 0.6) × 10−1 |
| T252R | + | − | 1.53 ± 0.27 |
| Y330D | − | − | (3.6 ± 0.53) × 10−1 |
| Y330D | + | − | (3.9 ± 0.63) × 10−1 |
| T252A/Y330D | − | − | (3.7 ± 0.56) × 10−1 |
| T252A/Y330D | + | − | (4.4 ± 0.72) × 10−1 |
A CP1 Mutation That Splits LeuRS Editing Pathways
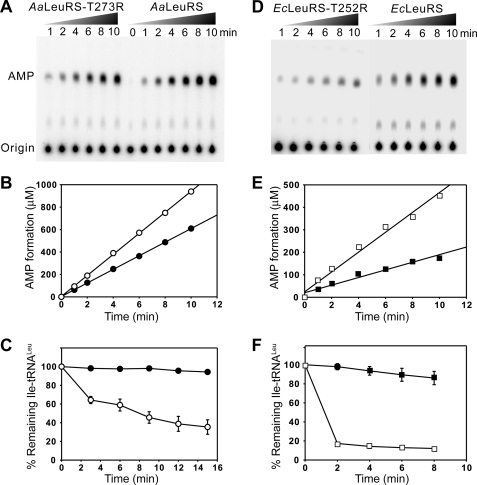
In theory, a robust editing capability may result from an efficient post-transfer editing activity that occurs after tRNA charging and/or from a rapid hydrolysis of misactivated amino acid in a pre-transfer editing step induced by tRNA. Both pathways lead to AMP accumulation. To distinguish between these possibilities, we specifically inactivated the post-transfer editing pathway by mutagenesis. We previously inactivated the CP1 domain from the AaLeuRS by mutating a conserved aspartic acid to alanine (D373A) (22). This residue was proposed to maintain the proper orientation of both pre- and post-transfer editing substrates into the CP1 domain (15). The mutation inactivated post-transfer editing but did not change tRNA-independent pre-transfer editing, suggesting that the later reaction was catalyzed within the aminoacylation active site (22). To confirm the result, one of the Thr residues from the threonine-rich region was selected for mutation (Fig. 1). Residue Thr-273 in AaLeuRS and the corresponding Thr-252 in EcLeuRS are key residues of the post-transfer editing activity of LeuRSs (17, 35, 36). In EcLeuRS, substitution of Thr-252 by smaller residues increased the size of the editing pocket, leading to a dramatic loss of editing specificity and deacylation of the cognate Leu-tRNALeu product (17, 35). Increasing the size of residue 252 had the opposite effect, preventing entry of non-cognate substrates into the editing pocket and, thus, reducing the enzyme fidelity (35, 36). Here we tested a new mutation combining both an increase of size and an addition of an extra positive charge. We mutated the crucial Thr residue to Arg and obtained the mutants AaLeuRS-T273R and EcLeuRS-T252R, respectively. Both mutants did not change the aminoacylation activity (supplemental Fig. S1), but the deacylation of Ile-tRNALeu was strongly impaired, consistent with a defect in post-transfer editing (Fig. 2, C and F). In parallel, we carried TLC-based assay in the presence of Nva and tRNALeu. Interestingly, AaLeuRS-T273R still exhibited 70% of AMP formation (kobs = 1.01 s−1) compared with the native enzyme (kobs = 1.43 s−1) (Table 1, Fig. 2, A and B). For EcLeuRS-T252R, the kobs of AMP formation was 1.53 s−1, about 45% that of native EcLeuRS (kobs = 3.42 s−1) (Table 2, Fig. 2, D and E). These values indicate that the Thr mutations have induced a decrease of AMP formation in the presence of tRNA, which should correspond to the loss of post-transfer editing activity. In the absence of tRNA, the kobs values of AaLeuRS-T273R and native AaLeuRS were comparable and reached only 0.091 and 0.086 s−1, respectively (Table 1). Those from EcLeuRS-T252R and native EcLeuRS were 0.35 and 0.33 s−1, respectively (Table 2). The latter results indicated that mutating the Thr residue crucial for post-transfer editing activity did not affect at all the tRNA-independent pre-transfer editing.
FIGURE 1.
Alignment of conserved regions within the editing (CP1) domain. Bold arrows indicate the residues involved in this study. Aa, A. aeolicus; Ec, E. coli; Hi, Haemophilus influenzae; Tt, T. thermophilus; Yp, Yersinia pestis; Xa, Xanthomonas axonopodis; Fn, Fusobacterium nucleatum; St, Salmonella typhimurium; Bs, Bacillus subtilis; Ba, Bacillus anthracis; Gs, Geobacillus stearothermophilus.
FIGURE 2.
The editing property of AaLeuRS, EcLeuRS, and mutated derivatives. A, shown is a TLC-based AMP formation assay of AaLeuRS or AaLeuRS-T273R in the presence of 5 μm AatRNALeu and 15 mm Nva. B, shown is a graphical representation of AMP formation. kobs of AMP formation was calculated from the slope and reported in Table 1. C, shown is hydrolysis of 1 μm Ile-AatRNALeu by 20 nm AaLeuRS or AaLeuRS-T273R. D, shown is AMP formation assay of EcLeuRS and EcLeuRS -T252R in the presence of 5 μm EctRNALeu and 15 mm Nva. E, shown is a graphical representation of the AMP formation. F, shown is hydrolysis of 1 μm Ile-EctRNALeu by 20 nm EcLeuRS or its EcLeuRS-T252R. ●, AaLeuRS-T273R; ○, AaLeuRS; ■, EcLeuRS-T252R; □, EcLeuRS.
Taken together, the data showed that the substitution of the conserved Thr by Arg induced inactivation of the post-transfer editing and decrease of AMP formation corresponding to this pathway. In theory, the remaining AMP formation measured with these mutants should result from the sum of the enzyme-catalyzed pre-transfer editing pathways and the spontaneous hydrolysis of Nva-AMP. However, the latter reaction is generally low and negligible compared with the total AMP accumulation rate (22). Thus, in addition to the post-transfer editing pathway, both AaLeuRS and EcLeuRS possess tRNA-dependent pre-transfer editing pathways that contribute to 70 and 45% of the total AMP formation. Such a pathway would hydrolyze non-cognate adenylates at the pre-transfer level. Compared with the previous results obtained with the AaLeuRS-D373A mutant, the level of the tRNA-dependent pre-transfer editing here measured with the Thr mutant was significantly higher (71% here versus 13% in Ref. 22). This might indicate that the T273R substitution did not inactivate the editing activity as did the D373A mutant. Although both types of mutants totally abolished the Ile-tRNALeu deacylation in vitro, the remaining AMP formation rate was not equivalent in the presence of tRNALeu. This strongly suggested that the enzymes carrying the D373A and T273R mutations did not exhibit the same CP1 editing site conformation in the presence of tRNA, one having greater tRNA-dependent pre-transfer editing activity. In conclusion, the T273R and T252R mutations have separated the post-transfer and pre-transfer editing pathways, leading to enzymes possessing exclusively pre-transfer editing activities.
AN2690 Specifically Targets Post-transfer Editing from EcLeuRS but Not AaLeuRS
It was recently shown that the benzoxaborole antifungal compound AN2690 inhibits yeast cytoplasmic LeuRS by forming a covalent adduct with the 3′-adenosine of tRNALeu at the editing site, thus locking the enzyme in an inactive conformation (33). AN2690 is also active on human cytoplasmic LeuRS (37) and Candida albicans LeuRS (38). In the present work AN2690 was tested against AaLeuRS and EcLeuRS. AaLeuRS was rather resistant to the compound, and the aminoacylation activity was only slightly reduced by 100 μm AN2690 (data not shown). This resistance can hardly be interpreted according to the fact that all of the residues that interact with AN2690 in the Thermus thermophilus enzyme (33) also exist in the Aquifex enzyme. Additionally, the AaLeuRS did not contain any of the mutations that led to AN2690 resistance in the Saccharomyces cerevisiae enzyme (33), suggesting that the resistance may result from subtle differences between the structures of the two CP1 domains. On the other hand, AN2690 critically reduced the aminoacylation efficiency of EcLeuRS in a dose-dependent and tRNA-dependent manner as previously observed (33). With 100 μm AN2690, the aminoacylation activity of EcLeuRS was 5-times reduced (Fig. 3A and data not shown), and the hydrolytic rate of mischarged Ile-tRNALeu was drastically reduced (Fig. 3B). By the TLC assay, in the presence of 100 μm AN2690, 15 mm Nva, and 5 μm tRNALeu, the kobs of AMP formation was 1.86 s−1, about 54% that of the editing activity without AN2690 (3.42 s−1). This AMP formation rate in the presence of AN2690 and tRNA was close to the value measured with EcLeuRS-T252R mutant (kobs = 1.53 s−1), which was deprived of post-transfer editing activity. Thus, both the T252R mutation and AN2690 compound induced comparable effects at the editing level and confirm the existence of a significant tRNA-dependent pre-transfer editing in EcLeuRS. In the absence of tRNALeu, AN2690 had no effect on the kobs of AMP formation (kobs = 3.1 × 10−1 s−1 versus 3.3 × 10−1 for the native), indicating that AN2690 does not affect the tRNA-independent editing pathway of EcLeuRS (Table 2, Fig. 3, C and D).
FIGURE 3.
Effect of AN2690 on the aminoacylation and editing properties of EcLeuRS. A, shown is aminoacylation of 20 μm EctRNALeu by 20 nm EcLeuRS in the presence (○) or absence (●) of AN2690 (100 μm). B, shown is hydrolysis of 1 μm Ile-EctRNALeu by 20 nm EcLeuRS (▼) in the presence of 5 μm EctRNALeu (○) or in the presence of 5 μm EctRNALeu and 100 μm AN2690 (●). C, shown is AMP formation in the presence or absence of tRNA (5 μm EctRNALeu) with 100 μm AN2690 and 15 mm Nva. D, shown is a graphical representation of the TLC data shown in panel C in the presence (○) or absence (●) of EctRNALeu.
Binding of tRNA to the Editing Site May Be Required for tRNA-dependent Pre-transfer Editing
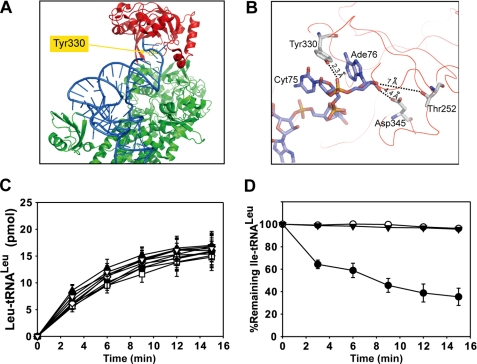
Our results showed that the pre-transfer editing of the EcLeuRS was maintained active when the CP1-editing site was inactivated by mutation T252R and when the 3′-CCA end of tRNALeu was trapped into the CP1 domain of EcLeuRS by AN2690. Next, we wanted to test if the presence of the tRNA in the CP1 domain was required for pre-transfer editing. We designed mutants intended to prevent binding of the CCA end of the tRNA in the CP1 domain. Inspection of the crystal structure of T. thermophilus LeuRS in complex with tRNA in the post-transfer editing conformation revealed an obvious candidate in the aromatic residue Tyr-332, which is located at 2.3 Å distance from the phosphate group of Ade76 (Fig. 4, A and B) (19). Tyr-332 corresponds to Tyr-358 of AaLeuRS and Tyr-330 of EcLeuRS (Fig. 1). Tyr-358 of AaLeuRS was mutated to Ala, Leu, Arg, Lys, Asp, Glu, Phe, Trp, and Thr. All the mutants exhibited full amino acylation activity (Fig. 4C). Among them, only mutants Y358D and Y358E were unable to deacylate Ile-tRNALeu (Fig. 4D) and formed Ile-tRNALeu as a result of the loss of the post-transfer editing (see later). These two mutants were then tested for their total editing activity by measuring the AMP formation. In the presence of tRNA, the kobs of AMP formation of AaLeuRS-Y358D and -Y358E was only 0.095 and 0.17 s−1, respectively (Table 1), which was much lower than native AaLeuRS (1.43 s−1) and very close to the tRNA-independent pre-transfer editing rate (0.086 s−1, Table 1). In the absence of tRNA the AMP formation rates were very similar to the rate of the native enzyme (0.088 and 0.092 s−1, Table 1), indicating that both mutants only displayed tRNA-independent pre-transfer editing and lost tRNA-dependent pathways including pre- and post-transfer editing.
FIGURE 4.
Spatial location of mutated residues and effects of various mutants on aminoacylation and deacylation. A, structure of TtLeuRS in complex with tRNALeu (in blue) (2BYT). Residue numbering is from EcLeuRS. Tyr-330 is shown in yellow (spatial equivalent of AaY358). The editing domain (CP1) is colored in red, and the synthetic and C-terminal domain is in green. B, a view of the CP1 domain shows the last nucleotides from the tRNA and three crucial residues from the enzyme. Numbering is from EcLeuRS. Distances between nucleotides and residues are indicated. C, tRNALeu aminoacylation performed in the reaction mix containing 20 μm tRNALeu and 20 nm AaLeuRS (●) or mutated derivatives: ♦, AaLeuRS-Y358A; □, AaLeuRS-Y358L; △, AaLeuRS-Y358R; ■, AaLeuRS-Y358K; ○, AaLeuRS-Y358D; ▼, AaLeuRS-Y358E; ◇, AaLeuRS-Y358F; ▲, AaLeuRS-Y358W; ▽, AaLeuRS-Y358T. D, hydrolysis of 1 μm Ile-AatRNALeu by 20 nm AaLeuRS (●), AaLeuRS-Y358D (○), or AaLeuRS-Y358E (▼) is shown.
Likewise, we constructed the corresponding Y330D mutant of EcLeuRS. The mutation abolished both tRNA-dependent editing activities as AaLeuRS-Y358D, in agreement with the effect observed with AaLeuRS mutants (Table 2).
Although the crystallographic structure suggested the existence of a short-distance interaction between Tyr-330 and the phosphate group of Ade76, the effect of the Y330D mutation on tRNA binding was verified by fluorescence titration (supplemental Fig. S2). Whereas the wild-type EcLeuRS exhibited a dissociation constant of 0.4 μm for tRNALeu, the mutated Y330D protein showed a 2-fold higher value (Kd = 0.81 μm) corresponding to a variation of binding energy of −0.42 kcal/mole (39), consistent with a loss of binding energy of a hydrogen bond (39). As an additional control, the Kd for tRNALeu of mutant T252R was measured under the same conditions. This residue is located far from and is not supposed to interact with the acceptor end of the tRNA (Fig. 4B). A Kd value of 0.41 μm was found that is very similar to the Kd of the native enzyme. The steady-state parameters of Y330D mutant during the aminoacylation reaction were also measured (supplemental Table S1). The wild-type EcLeuRS and Y330D mutants exhibited nearly the same kcat (4.25 and 4.04 s−1, respectively), whereas the Km value of the mutant was slightly increased (3.3 μm for the mutant versus 2.6 μm for the wild-type LeuRS), which is consistent with the increase of the Kd value (see above). The results confirmed the initial hypothesis that Tyr-330 residue might interact with the last residue of the tRNA as observed in the crystallographic structure (15, 19).
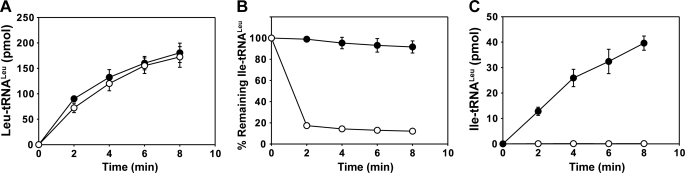
In the end we constructed the double mutant of EcLeuRS-T252A/Y330D to accumulate the negative effects of two individual mutations involved in editing and verify that the Y330D mutation prevents CCA binding in the editing site. Mutation T252A enlarges the part of the editing site that recognizes the amino acid moiety, resulting in the hydrolysis of all charged tRNAs, including Leu-tRNALeu (17, 35). As a consequence, the aminoacylation activity of T252A was considerably reduced (17, 35). Interestingly, for the T252A/Y330D double mutant, the inactivating effect of mutation T252A on tRNA charging was suppressed by mutation Y330D, which is consistent with the expected effect of Y330D on the tRNA entry in the CP1 domain (Fig. 5A). The T252A/Y330D double mutant could not hydrolyze Ile-tRNALeu (Fig. 5B) but catalyzed formation of a certain amount of Ile-tRNALeu (Fig. 5C). The AMP formation rate catalyzed by T252A/Y330D in the absence and presence of tRNA was similar to that of Y330D alone (Table 2), suggesting that the double mutant also loses both tRNA-dependent editing pathways. These data can be compared with other results recently reported (40). The aminoacylation activity of the T252A mutant was rescued by one Gly → Pro mutant located within the β-strands of the hinge domain that connects the CP1 domain to the Rossmann-fold domain. This Pro mutation induced defects in post-transfer editing, and by combining this mutation with the T252A mutation that is defective in Leu-tRNALeu formation (see above), the authors were able to rescue the Leu-tRNALeu formation, similar to what we observed here with the T252A/Y330D double mutant. The authors hypothesized that the charged tRNA could not be translocated from the aminoacylation to the editing active site, and consequently it could escape to the hyperhydrolytic activity of T252A mutant (40). Here we suggest that preventing the CP1-tRNALeu interaction by mutating Tyr-330 to Asp is another way to prevent translocation of the CCA end, to bypass the hydrolytic activity of T252A mutant, and to rescue the aminoacylation activity of the enzyme.
FIGURE 5.
Aminoacylation and deacylation properties of EcLeuRS T252A/Y330D double mutant. A, aminoacylation of 20 μm EctRNALeu by 20 nm EcLeuRS (○) or EcLeuRS-T252A/Y330D (●) is shown. B, deacylation of 1 μm Ile-EctRNALeu by 20 nm EcLeuRS (○) or EcLeuRS-T252A/Y330D (●) is shown. C, shown is a mischarging experiment of 20 μm EctRNALeu with isoleucine by 0.5 μm EcLeuRS (○) or EcLeuRS-T252A/Y330D (●).
Altogether, these data suggest that the Y330D mutation prevents adequate binding of the tRNA CCA end in the editing site. This effect might result from the electrostatic repulsion between the phosphate of Ade76 and the mutated residue Y330D. Consequently, the aminoacyl-tRNA would not be checked in the editing site but directly released from the enzyme, which explains the loss of tRNA-deacylation activity. Concerning the loss of tRNA-dependent editing pathways of mutants carrying the Y330D substitution, one can see a correlation between the disruption of the interaction with the acceptor end of the tRNA and the loss of tRNA-dependent pre-transfer editing activity. This suggests that the CP1 orientation changes that are observed when comparing the apoenzyme and the enzyme in complex with tRNA (15, 19) may be crucial for tRNA-dependent pre-transfer editing activity.
tRNA-dependent Pre-transfer Editing and Post-transfer Editing Are Both Required to Maintain Aminoacylation Specificity
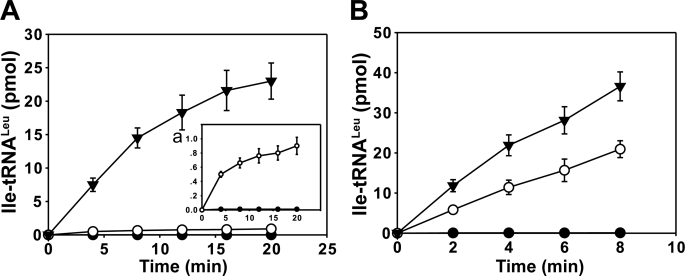
The mutagenesis study revealed some crucial residues for post-transfer and pre-transfer editing. The mutants could also be used to compare the contribution of the different pathways to aminoacylation fidelity. Toward that end, we compared the misacylation of tRNALeu by Ile by the different mutants. The misacylation can be regarded as the net result of the various proofreading activities of the enzyme. Among the constructed mutants, the Tyr mutants (AaLeuRS-Y358D and EcLeuRS-Y330D) were the most severely damaged in both tRNA-dependent editing pathways including tRNA-dependent pre- and post-transfer editing (Tables 1 and 2), whereas the Thr mutants (AaLeuRS-T273R and EcLeuRS-T252R) only lose their post-transfer editing (Fig. 2). As expected, the mischarging curves showed that the Tyr mutants (AaLeuRS-Y358D and EcLeuRS-Y330D) were the less specific and exhibited the highest misacylation rates (Fig. 6). The Thr mutants (AaLeuRS-T273R and EcLeuRS-T252R), which still carried tRNA-dependent pre-transfer editing, were more specific than the Tyr mutants and exhibited lower misacylation rates. For instance, the misacylation rates were 2-times lower for EcLeuRS-T252R mutant compared with EcLeuRS-Y330D (Fig. 6B) or 15-times lower for AaLeuRS-T273R compared with AaLeuRS-Y358D (Fig. 6A). These data showed that the LeuRS aminoacylation specificity could not be achieved by the tRNA-dependent pre-transfer editing. The pathway hydrolyzed a certain amount of incorrect aminoacyl-adenylate intermediates, but at the end the fidelity of aminoacylation is guaranteed by the sum of tRNA-independent and tRNA-dependent pre- and post-transfer editing.
FIGURE 6.
Isoleucylation of tRNALeu by AaLeuRS, EcLeuRS, and their editing-defective mutants. A, shown is isoleucylation of 20 μm AatRNALeu by 3 μm AaLeuRS (●), AaLeuRS-T273R (○), or AaLeuRS-Y358D (▼) and isoleucylation of 20 μm AatRNALeu by 3 μm AaLeuRS or AaLeuRS-T273R, which is also shown in inset a. B, shown is isoleucylation of 20 μm EctRNALeu with isoleucine by 0.5 μm EcLeuRS (●), EcLeuRS-T252R (○), or EcLeuRS-Y330D (▼).
DISCUSSION
Relative Contributions of the Different Editing Pathways to Aminoacylation Fidelity
It was previously reported that tRNA could robustly stimulate the editing activity of LeuRSs in the presence of non-cognate amino acids (22, 31). In theory, stimulation of editing activity by tRNA can occur at the post-transfer level (post-transfer editing) or pre-transfer level (tRNA-dependent pre-transfer editing). However, the total contribution of tRNA-dependent pre-transfer editing to the total editing of LeuRS was not well documented. In the present study different editing conditions, several mutants, and an antifungal compound with antibiotic properties were used to decipher the contribution of the editing pathways of A. aeolicus and E. coli LeuRSs.
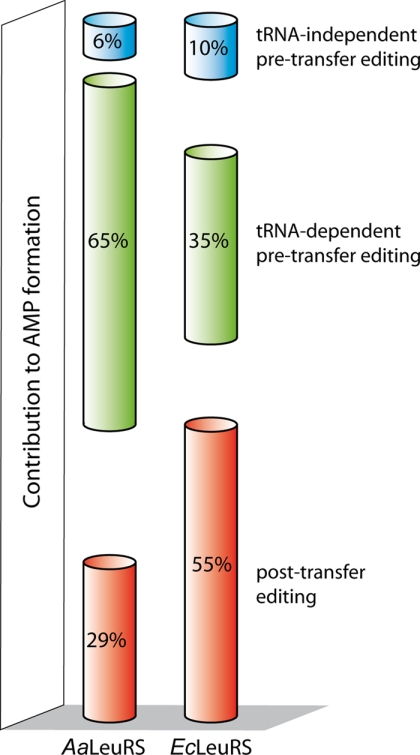
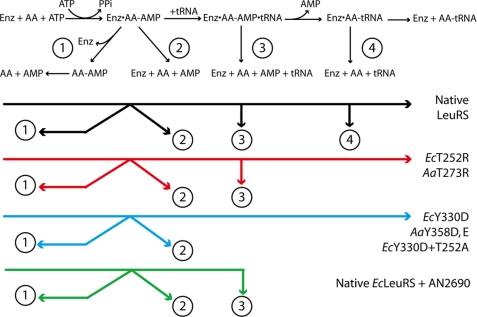
In the presence of the non-cognate amino acid Nva, LeuRSs generated high amounts of AMP due to the repetitive cycles of synthesis-editing. To separate the contribution of tRNA-dependent editing from the tRNA-independent editing, the AMP formation rates in the presence or absence of tRNA were compared. It appeared that tRNA-independent editing is a minor pathway that only contributes to 6 and 10% of the AMP synthesis of A. aeolicus and E. coli LeuRSs, respectively (Fig. 7, pathway 2 in Fig. 8). Included in these AMP formation rates is the spontaneous hydrolysis rate of Nva-AMP that results from its release of the enzyme (pathway 1 in Fig. 8), but the rate is so low that its contribution to AMP accumulation is negligible (22). Thus, the remaining 94 and 90% of AMP synthesis correspond to the tRNA-dependent editing pathways, which can basically be separated in the post-transfer editing and pre-transfer editing (tRNA-dependent) reactions (pathways 3 and 4 in Fig. 8).
FIGURE 7.
Histogram summarizing the relative contributions of each editing pathway in AMP formation. Percentages were calculated from kobs values of AMP formation reported in Tables 1 and 2. tRNA-independent pre-transfer editing was measured in the absence of tRNA. tRNA-dependent pre-transfer editing was obtained with AaLeuRS-T273R and EcLeuRS-T252R mutants in the presence of tRNA deduced from the tRNA-independent pre-transfer editing contribution. Post-transfer editing was the difference between the native enzymes and the post-transfer defective mutants (or for EcLeuRS in the presence of AN2690), both in the presence of tRNA.
FIGURE 8.
Schematic presentation of editing pathways and effects of the different mutations or AN2690 addition. Numbers refer to the editing pathways. Pathway 1 is the spontaneous chemical hydrolysis after release from the enzyme. Pathway 2 is the tRNA-independent pre-transfer editing. Pathway 3 is the tRNA-dependent pre-transfer editing. Pathway 4 is the post-transfer editing (tRNA-dependent). The remaining synthetic and editing pathways of each mutant and AN2690 are shown in the lower part of the figure. AA, aminoacyl.
Separation of the post-transfer editing and pre-transfer editing (tRNA-dependent) reactions was obtained using CP1 mutants that selectively shut down post-transfer editing. A conserved Thr residue from the Thr-rich region was mutated to Arg. The resulting AaLeuRS-T273R and EcLeuRS-T252R exhibited full aminoacylation activity, but no post-transfer editing activity, as shown by the deacylation assay. This could be explained by the fact that the larger residue Arg created steric hindrance at the entrance of the post-transfer editing site for misacylated tRNA, leading to loss of post-transfer editing as already observed with other large residues (17, 35). Nevertheless, despite the loss of post-transfer editing, both mutated A. aeolicus and E. coli LeuRSs produced 71 and 45% of the initial AMP level, respectively (Fig. 7), indicating that tRNA-dependent pre-transfer editing is a robust editing pathway that efficiently hydrolyzes misactivated amino acid (pathway 3 in Fig. 8). Previous work on AaLeuRS also revealed the existence of a tRNA-dependent pre-transfer editing; however, the latter was estimated to account for only 13% of the total AMP formation rate (22). This substantial difference was likely due to the D373A mutation used to inactivate the CP1 domain (22) instead of the T273R (or EcLeuRS-T252R) used in the present work. This difference suggests that each mutation has a significantly different impact on the CP1 domain. In TtLeuRS, the equivalent residue of Asp-373 has been shown to be crucial for the interaction with the amino group of Nva in the adenylate molecule (15), whereas the equivalent residue of Thr-273 forms part of the pocket intended to exclude from deacylation the cognate Leu-adenylate. For unexplained reasons, the mutation of Asp-373 to Ala more strongly impacted tRNA-dependent pre-transfer editing than the T273R mutation. One hypothesis may be that tRNA-dependent pre-transfer editing occurs within the CP1 domain, and Asp-373 may play a more consequential role in catalysis than Thr-273. Alternatively, D373A mutation may bring a subtle conformation change in the active site of CP1, leading to inappropriate interaction with tRNA, which subsequently reduced tRNA-dependent pre-transfer editing activity. One may suggest inactivating the CP1 domain by deleting the entire CP1 domain, as was recently reported (30). Nevertheless, this kind of large deletion may destabilize the protein and induced long-distance effects on the aminoacylation and amino acid activation activities as observed (30). The data suggest that the best way to inactivate a CP1 domain may be to combine several mutations of the crucial residues (Thr-252 and Asp-345 for instance) that should not disturb the enzyme conformation and synthetic activity.
A Mutation in the CP1-editing Domain That Affects tRNA Binding Abolishes Both the Post- and the Pre-transfer tRNA-dependent Editing Pathway
When a mutation was introduced in the CP1 domain (AaLeuRS-Y358D or EcLeuRS-Y330D) to prevent proper binding of tRNA, both tRNA-dependent pre-transfer editing and post-transfer editing were abolished (Fig. 4, pathways 3 and 4 in Fig. 8). Such an effect could be interpreted in two ways. One interpretation would be that both post- and pre-transfer tRNA-dependent editing pathways reside into the CP1-editing domain, but this would be contradicted by the Thr mutants described above that specifically abolished post-transfer editing and did not affect AMP formation resulting from tRNA-dependent pre-transfer editing. One might argue that pre- and post-transfer substrates bind different binding sites in the CP1-editing domain; however, it was shown that both substrates use essentially the same binding site (15). Another explanation would be that tRNA binding triggers a conformation change that may confer to the enzyme the ability to hydrolyze non-cognate adenylates. In that case the question of the location of the pre-transfer editing reaction would remain inconclusive, and we cannot exclude the possibility that the reaction occurs within the synthetic site as recently proposed (30).
AN2690 Covalently Binds CCA in the CP1-editing Domain and Specifically Inhibits Post-transfer Editing in Addition to tRNA Charging
AN2690 was used as an additional probe to explore the LeuRS editing mechanism. AN2690 is an antibiotic that binds the non-cognate amino acid binding pocket in the editing site. This compound inhibits LeuRS by forming a covalent adduct with the 3′-adenosine of tRNALeu at the editing site, thus locking the enzyme in an inactive conformation (33). Theoretically, when the adduct of AN2690 is formed in the CP1 domain, the binding of Nva-AMP as described earlier is no longer possible because of their overlapping binding sites (15, 33). When tRNALeu was covalently bound in the editing domain with AN2690, we observed the post-transfer editing was inhibited as shown by the decrease of AMP formation and of Ile-tRNALeu deacylation (Fig. 3). However, a robust AMP formation resistant to AN2690 and tRNA-dependent still existed (kobs = 1.86 s−1, Table 2). The AMP formation rate was close to the rate measured with CP1-inactivating mutation T252R, also deprived of deacylation activity (kobs = 1.53 s−1, Table 2), suggesting that a similar pathway was impaired in both cases. In other words, AN2690 mimics the effect of the T252R mutation in inactivating the post-transfer editing while preserving intact tRNA-dependent pre-transfer editing (Fig. 8). However, the aminoacylation activity was lost in the presence of AN2690 but not T252R mutation.
CCA Binding in the CP1 Domain Activates the Pre-transfer Competent State
In summary, our results showed that the tRNA-dependent pre-transfer editing of LeuRS was kept intact when the CP1-editing site was inactivated either by the Thr mutations (EcT252R and AaT273R) or by AN2690 that entrapped the CCA end of tRNALeu into the CP1 domain. An active CP1 domain was not required for tRNA-dependent pre-transfer editing, but a proper binding of the tRNA-CCA end in the CP1 domain was essential for non-cognate adenylate hydrolysis. Indeed, changing one essential interaction between the tRNA-terminal adenine and EcLeuRS residue Tyr-330 completely abolished tRNA-dependent pre-transfer editing to a basal level corresponding to the tRNA-independent pre-transfer editing (kobs = 0.36 s−1, Table 2). Fluorescence titration confirmed that the mutation Y330D impacted tRNA binding. The result revealed a 2-fold increase of the Kd for tRNALeu (supplemental Fig. S2) corresponding to a binding energy variation of −0.46 kcal/mol, a value that might correspond to the loss of one hydrogen bond with the CCA end.
In summary, the data indicate the entry of the 3′ end of tRNA into the editing site confers to the enzyme a conformation for the pre-transfer editing catalysis. Significant conformation changes were observed when comparing the complexed and uncomplexed LeuRS structures (19), but their involvement in activating the pre-transfer editing site can hardly be proved. Concerning the location of the tRNA-dependent pre-transfer editing site, one cannot eliminate a possible location into the synthetic site as observed with synthetases deprived of separated editing domains (23, 28, 29). This would explain to some extent that the CP1-inactived LeuRSs here tested were still active in tRNA-dependent pre-transfer editing. In several aspects, the tRNA-dependent pre-transfer editing mystery appears like the tRNA-dependent amino acid activation catalyzed by Arg-, Glu-, and glutaminyl-tRNA synthetase. This phenomenon has been studied for decades because the intermediates could not be isolated and studied separately. Both activation and editing reactions require tRNA to form a kind of transient ribonucleoprotein providing the three-dimensional conformation required for the synthetic or hydrolytic catalysis. To further study in the mechanism of tRNA-dependent pre-transfer editing, it would be necessary to identify crucial residues directly and specifically involved in the hydrolysis of misactivated amino acid. Molecular dynamics simulations may also assist in understanding the molecular trigger of the editing pathways, the adenylate trajectories, and the editing process. Investigations of the editing process may require fast kinetics approaches to understand the different steps and transient conformational changes (41). Detailed analysis of the editing process may require isolation of as yet unstudied labile and transient intermediates.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Lee and Benjamin Beauchamp for carefully reading the manuscript.
This work was supported by Natural Science Foundation of China Grants 30670463 and 30930022, National Key Basic Research Foundation of China Grant 2006CB910301, the 973 Project of China Grant 2005CB724600, Committee of Science and Technology in Shanghai Grant 09JC1415900, and the Exchange Program and Programme International de Coopération Scientifique from CNRS Grant 3606.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1 and S2.
- aaRS
- aminoacyl-tRNA synthetase
- LeuRS
- leucyl-tRNA synthetase
- EcLeuRS
- E. coli LeuRS
- AaLeuRS
- A. aeolicus LeuRS
- Nva
- norvaline
- CP1
- connective peptide 1.
REFERENCES
- 1.Schimmel P. (1987) Annu. Rev. Biochem. 56, 125–158 [DOI] [PubMed] [Google Scholar]
- 2.Woese C. R., Olsen G. J., Ibba M., Söll D. (2000) Microbiol. Mol. Biol. Rev. 64, 202–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ling J., Reynolds N., Ibba M. (2009) Annu. Rev. Microbiol. 63, 61–78 [DOI] [PubMed] [Google Scholar]
- 4.Eriani G., Delarue M., Poch O., Gangloff J., Moras D. (1990) Nature 347, 203–206 [DOI] [PubMed] [Google Scholar]
- 5.Hendrickson T. L., de Crécy-Lagard V., Schimmel P. (2004) Annu. Rev. Biochem. 73, 147–176 [DOI] [PubMed] [Google Scholar]
- 6.Lee J. W., Beebe K., Nangle L. A., Jang J., Longo-Guess C. M., Cook S. A., Davisson M. T., Sundberg J. P., Schimmel P., Ackerman S. L. (2006) Nature 443, 50–55 [DOI] [PubMed] [Google Scholar]
- 7.Nangle L. A., Motta C. M., Schimmel P. (2006) Chem. Biol. 13, 1091–1100 [DOI] [PubMed] [Google Scholar]
- 8.Loftfield R. B., Vanderjagt D. (1972) Biochem. J. 128, 1353–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubowski H., Goldman E. (1992) Microbiol. Rev. 56, 412–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinis S. A., Fox G. E. (1997) Nucleic Acids Symp. Ser. 36, 125–128 [PubMed] [Google Scholar]
- 11.Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. (2000) Cell 103, 793–803 [DOI] [PubMed] [Google Scholar]
- 12.Lin L., Hale S. P., Schimmel P. (1996) Nature 384, 33–34 [DOI] [PubMed] [Google Scholar]
- 13.Chen J. F., Guo N. N., Li T., Wang E. D., Wang Y. L. (2000) Biochemistry 39, 6726–6731 [DOI] [PubMed] [Google Scholar]
- 14.Zhao M. W., Zhu B., Hao R., Xu M. G., Eriani G., Wang E. D. (2005) EMBO J. 24, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 16.Hendrickson T. L., Nomanbhoy T. K., de Crécy-Lagard V., Fukai S., Nureki O., Yokoyama S., Schimmel P. (2002) Mol. Cell 9, 353–362 [DOI] [PubMed] [Google Scholar]
- 17.Xu M. G., Li J., Du X., Wang E. D. (2004) Biochem. Biophys. Res. Commun. 318, 11–16 [DOI] [PubMed] [Google Scholar]
- 18.Silvian L. F., Wang J., Steitz T. A. (1999) Science 285, 1074–1077 [PubMed] [Google Scholar]
- 19.Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 20.Baldwin A. N., Berg P. (1966) J. Biol. Chem. 241, 831–838 [PubMed] [Google Scholar]
- 21.Schimmel P., Schmidt E. (1995) Trends Biochem. Sci. 20, 1–2 [DOI] [PubMed] [Google Scholar]
- 22.Zhu B., Yao P., Tan M., Eriani G., Wang E. D. (2009) J. Biol. Chem. 284, 3418–3424 [DOI] [PubMed] [Google Scholar]
- 23.Gruic-Sovulj I., Rokov-Plavec J., Weygand-Durasevic I. (2007) FEBS Lett. 581, 5110–5114 [DOI] [PubMed] [Google Scholar]
- 24.Nomanbhoy T. K., Hendrickson T. L., Schimmel P. (1999) Mol. Cell 4, 519–528 [DOI] [PubMed] [Google Scholar]
- 25.Nomanbhoy T. K., Schimmel P. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5119–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop A. C., Beebe K., Schimmel P. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordin B. E., Schimmel P. (2003) Biochemistry 42, 12989–12997 [DOI] [PubMed] [Google Scholar]
- 28.Gruic-Sovulj I., Uter N., Bullock T., Perona J. J. (2005) J. Biol. Chem. 280, 23978–23986 [DOI] [PubMed] [Google Scholar]
- 29.Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., Musier-Forsyth K. (2006) J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 30.Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Englisch S., Englisch U., von der Haar F., Cramer F. (1986) Nucleic Acids Res. 14, 7529–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu M. G., Chen J. F., Martin F., Zhao M. W., Eriani G., Wang E. D. (2002) J. Biol. Chem. 277, 41590–41596 [DOI] [PubMed] [Google Scholar]
- 33.Rock F. L., Mao W., Yaremchuk A., Tukalo M., Crépin T., Zhou H., Zhang Y. K., Hernandez V., Akama T., Baker S. J., Plattner J. J., Shapiro L., Martinis S. A., Benkovic S. J., Cusack S., Alley M. R. (2007) Science 316, 1759–1761 [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Wang E. D., Wang Y. L. (1998) Science in China (series C) 41, 225–23118425626 [Google Scholar]
- 35.Mursinna R. S., Lincecum T. L., Jr., Martinis S. A. (2001) Biochemistry 40, 5376–5381 [DOI] [PubMed] [Google Scholar]
- 36.Mursinna R. S., Martinis S. A. (2002) J. Am. Chem. Soc. 124, 7286–7287 [DOI] [PubMed] [Google Scholar]
- 37.Yao P., Zhou X. L., He R., Xue M. Q., Zheng Y. G., Wang Y. F., Wang E. D. (2008) J. Biol. Chem. 283, 22591–22600 [DOI] [PubMed] [Google Scholar]
- 38.Seiradake E., Mao W., Hernandez V., Baker S. J., Plattner J. J., Alley M. R., Cusack S. (2009) J. Mol. Biol. 390, 196–207 [DOI] [PubMed] [Google Scholar]
- 39.Carter P. J., Winter G., Wilkinson A. J., Fersht A. R. (1984) Cell 38, 835–840 [DOI] [PubMed] [Google Scholar]
- 40.Mascarenhas A. P., Martinis S. A. (2009) FEBS Lett. (2009)583, 3443–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Francklyn C. S., First E. A., Perona J. J., Hou Y. M. (2008) Methods. 44, 100–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.