Abstract
Methylation of specific histone residues is capable of both gene activation and silencing. Despite vast work on the function of methylation, most studies either present a static snapshot of methylation or fail to assign kinetic information to specific residues. Using liquid chromatography-tandem mass spectrometry on a high-resolution mass spectrometer and heavy methyl-SILAC labeling, we studied site-specific histone lysine and arginine methylation dynamics. The detection of labeled intermediates within a methylation state revealed that mono-, di-, and trimethylated residues generally have progressively slower rates of formation. Furthermore, methylations associated with active genes have faster rates than methylations associated with silent genes. Finally, the presence of both an active and silencing mark on the same peptide results in a slower rate of methylation than the presence of either mark alone. Here we show that quantitative proteomic approaches such as this can determine the dynamics of multiple methylated residues, an understudied portion of histone biology.
Keywords: Chromatin/Modification, Histones, Histones/Methylation, Histones/Modification, Methods/Mass Spectrometry, Proteomics
Introduction
Histones are decorated extensively with numerous post-translational modifications (PTMs)2 on several different residues (1). Located mostly in the unstructured N-terminal tails, these PTMs influence the expression of genes bound to the histones by the recruitment or displacement of non-histone transcriptional or regulatory proteins (2). Lysine methylation is notable among histone PTMs for its diversity of forms and for its binary-like influence over gene expression. Methyltransferases and demethylases catalyze specific conversions between unmodified (me0), mono- (me1), di- (me2), and trimethylation (me3) states. For instance, while G9a and Suv39h1 direct histone H3 lysine 9 (H3K9) me1/me2 and me3 production, respectively (3), JHDM2A (4), and JHDM3A (5) promote demethylation of H3K9me2 and H3K9me3, respectively. The binary influence on gene expression can be illustrated with chromatin immunoprecipitation (ChIP) studies that revealed H3K4 to be enriched in euchromatic regions and H3K9 in heterochromatic regions (6). Another residue that can be mono- and dimethylated on histones is arginine. Furthermore, whereas H4R3 methylation (7) and H3R2me1 (8) are associated with gene activation, H3R2me2 is associated with gene silencing (8).
These features suggest that histone lysine and arginine methylations are dynamically regulated processes. Nevertheless, most studies present histone methylation as a static condition at a particular time, with little regard to turnover or dynamics of the PTM. Indeed, the inability to resolve between transient and more long-lived methylations likely contributed to the traditional conception of histone methylation as irreversible (9). Past studies have used radiolabeling to track methylation turnover on a non-residue specific basis (10, 11) and other studies monitored the correlation between histone methylation and cell cycle progression (12–14), including a recent report (15) that tracked the modification profile of newly synthesized histones. For example, Pesavento et al. reported that newly synthesized H4 becomes progressively methylated at Lys-20 during the cell cycle (13). Additionally, McManus et al. (14) reported that levels of H3K9me1, me2, and me3 increase from interphase to metaphase, and decrease to initial levels at the start of the next mitotic cycle.
To our knowledge, there is no report of a quantitative model characterizing the steady-state kinetics of global methylation on a residue-specific basis. The development of methods to investigate methylation kinetics would elucidate the mechanisms by which this PTM can temporally and dynamically regulate gene expression and provide a sensitive metric to distinguish cells in different physiological states. The importance of understanding the dynamics of histone methylation is illustrated with past studies demonstrating that the shift from H3K4me2 and H3K36me2/3 to H3K4me3 and H3K79me2 defined the temporal transition from RNA polymerase II binding to release for transcript elongation (16).
To this end, we applied heavy methyl stable isotope labeling by amino acids in cell culture (SILAC) (17) to quantitatively probe the rates of histone methylation on a residue-specific basis in HeLa cells (Fig. 1A). The method relies on mass spectrometry (18) to detect the in vivo incorporation of labeled S-adenosyl methionine, which is metabolized from exogenous 13CD3-methionine, to methylation substrates. Using heavy methyl SILAC to quantify histone methylation dynamics, we report that different lysine and arginine residues, and different methylation states within the same residue, have different rates of formation. This and other findings in our studies advance the current understanding of histone methylation and provide a platform for further research into the dynamics of histone PTMs.
FIGURE 1.
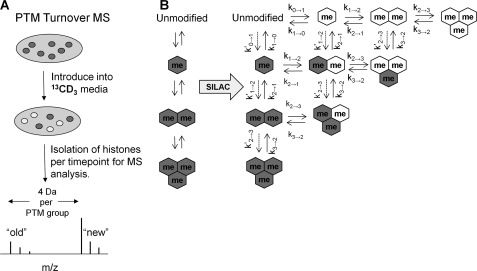
Schematic of experimental design for heavy methyl SILAC. A, suspension HeLa cultures were introduced into 13CD3-methionine supplemented media to allow for incorporation of the 13CD3 label (white) onto the methyl groups on histones. The labeled methyl can be distinguished from the unlabeled methyl (gray) using MS (4 Da shift per methyl incorporation). B, application of heavy methyl SILAC can be used to unravel methylation dynamics, as depicted in our idealized system. Gray hexagons indicate unlabeled “old” methyl, and white hexagons indicate labeled “new” methyl groups.
EXPERIMENTAL PROCEDURES
Cell Culture and SILAC
Throughout the experiment except during the transition into different media and sample collection, HeLa cells were maintained at 37 °C in 0.2 LPM CO2. Suspension cultures were initially maintained in minimum essential Joklik modified medium (SAFC Biosciences, KS) supplemented with 10% newborn calf serum (HyClone, UT), penicillin, streptomycin, and 1% GlutaMAX (Invitrogen). For the time course experiments, cultures were pelleted at 80 rcf in a refrigerated tabletop centrifuge. After decanting the media, cells were resuspended with Joklik media depleted of unlabeled methionine (HyClone) and supplemented with l-methionine-methyl-13CD3 (Sigma-Aldrich), which we term “labeled media.” We ran a parallel experiment with media supplemented with l-lysine-13C615N2 (Cambridge Isotope Laboratories Inc.) to determine the turnover for histone H1.4. The labeled media was supplemented with 5% dialyzed fetal bovine serum (Invitrogen), penicillin, streptomycin, and 1% GlutaMAX. Daily aliquots were taken for 7 days, and media was replenished everyday to maintain a cell density of 2–6 × 105 cells/ml throughout the experiment. Cells were pelleted at 600 rcf and washed in phosphate-buffered saline. The pellets were flash frozen and stored at −80 °C.
Histone Preparation for MS
Histones were isolated from cells as described in Ref. 19 and derivatized with propionic anhydride as described in Ref. 20. After derivatization, samples were diluted in 0.1% acetic acid for desalting before MS analysis using homemade C18 STAGE tips as previously described (21).
MS Analysis
Samples were loaded by an Eksigent AS2 autosampler onto 75 μm ID fused silica capillary columns packed with 15 cm of C18-reversed phase resin (Magic C18, 5 μm particles, 200 Å pore size, Michrom BioResources) and constructed with an electrospray tip for nanoflow reversed-phase high performance liquid chromatography tandem mass spectrometry on a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer (Thermo Electron). Peptides were resolved with a gradient from 5 to 35% Buffer B in a 110-min gradient (Buffer A: 0.1 m acetic acid, Buffer B: 70% acetonitrile in 0.1 m acetic acid) with a flow rate of ∼200 nl/min on an Agilent 1200 binary HPLC system. The Orbitrap was operated in data-dependent mode with a resolution of 30,000 for a full MS spectrum and 7 subsequent MS/MS spectra collected in the ion trap of fragments produced by collision-induced dissociation. To reduce duplicate MS/MS spectra, peptides selected for MS/MS interrogation were placed on an exclusion list for 30 s.
Peptide Quantification
All MS and MS/MS spectra were analyzed with Qual Browser (version 2.0.7, Thermo Scientific), and peptide abundances were obtained by chromatographic peak integration. Identities of all peptides were confirmed by manual inspection of MS/MS spectra (supplemental Fig. S1), and a list of all peptides quantified is provided (supplemental Table S1).
To describe unambiguously a specific methylated species, we denote each residue with X:Y, where X is the number of total methyls, and Y is the number of labeled methyls. For instance, H3K36me1:0 refers to unlabeled monomethylated H3K36, and H3K36me1:1 refers to labeled monomethylated H3K36. We define “methylation states” as the distinction between me1, me2, and me3, and “labeled intermediates” as the different degrees of labeling within states.
The relative abundances were calculated two ways, differing in what is considered the “total pool.” In the first method, the “total pool” represents all the methylation intermediates for a specific methylation state of a particular residue. Thus, the abundance of H3K36me1:0 would be determined with respect to H3K36me1:0 and me1:1. This reveals the distribution of differently labeled species within a methylation state and is more robust to variable ionization efficiencies of different methylation states within and between peptides. We justify the calculation with the logic that growth conditions should not change with 12CH3- or 13CD3-methionine, and that the total abundance of each methylated species does not change. For instance, the percentage of total H3 peptides monomethylated on Lys-36 should not change during the time course, but rather the distribution of labeled intermediates within the me1 state. We term values calculated from such a method as “relative distribution.” This was used to quantify half-maximal times of the formation of labeled peptides (t½), as described in supplemental Fig. S2. The half-maximal time provides a single measure of how quickly a particular species is formed, and thus is predictive of the overall rate of methylation for that species.
In the second method, the “total pool” represents all methylation states and intermediates on the tryptic digest fragment. Thus, the abundance of H3K79me1:0 would be determined with respect to H3K79me1:0, me1:1, me2:0, me2:1, and me2:2. This provides a more accurate comparison of abundances between different methylation states. We term values calculated from such a method as “relative levels,” and this was used for extrapolating kinetic parameters. Both “relative distribution” and “relative levels” are unit-less variables.
Kinetics Modeling
To quantify relative distributions, we applied another differential equation (supplemental Fig. S2), to determine the half-maximal times of the formation of the fully labeled species and used a Wilcoxon rank sum and Kruskal-Wallis test to compare half-maximal times between active and silencing marks. Relative distributions of the labeled methylation states and 13CD3-methionine incorporated protein from each replicate were fitted with MATLAB (7.8.0) (supplemental data and Fig. S1).
To quantify relative levels, we derived a set of first order differential equations (supplemental Fig. S2) to characterize the dynamics of methylated residues in an idealized system (Fig. 1B). The parameters are the rate of unmodified histones becoming bound to chromatin (α), sequential addition of a labeled methyl (k0→1, k1→2, k2→3), sequential demethylation (k3→2, k2→1, k1→0), and histone degradation for each methylation state (μ0, μ1, μ2, μ3 for unmodified, mono-, di-, and tri- methylation respectively). Additionally, not all peptides necessarily acquire a 13CD3 methyl group, as 12CH3 from unlabeled methionine resulting from macromolecule decomposition contributes to the sequential methylation. Thus, parameters (k′0→1, k′1→2, k′2→3), for the unlabeled methyl contribution are also considered.
Relative levels of all the species were fitted to the appropriate differential equations using MATLAB (7.8.0) (supplemental data and Fig. S2). Certain constraints were set to avoid non-biologically reasonable parameter values. In particular, none of the parameters could be less than 0. Additionally, all μ were set to be ≥ ln(2), which is the expected dilution rate resulting from cell division. After each of the 1000–10000 iterations of the program, a set of parameters is produced that minimizes the root mean squared difference (RMSD) between the observed data, and the expected solutions of the equations with the determined parameters. In the entire parameter space determined, we averaged only the sets of parameters that did not produce a RMSD greater than 125% of the minimum RMSD.
RESULTS
Effect of 13CD3-Methionine Isotope on Cell Physiology and General Histone Protein Turnover Rates
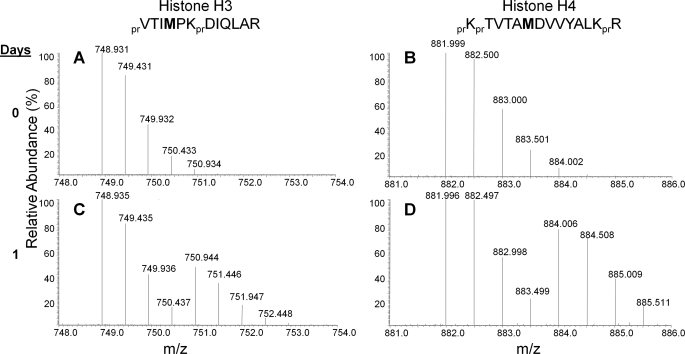
To track and quantify newly methylated histones, we cultured HeLa cells in media with 13CD3-methionine. HeLa cell morphology appeared normal throughout the time course, indicating that the presence of the heavy isotope itself does not affect the biology of our system. Because most newly synthesized core histones H3 and H4 during S phase (23) incorporate the labeled methionine (13CD3, +4 Da shift per methyl group incorporation), half-maximal labeling for all histones should occur after one mitotic cycle. The rate of label incorporation was determined by examining the peptides from H1.4 (residues 33–53, KASGPPVSELITKAVAASKER) H3 (residues 117–128, VTIMPKDIQLAR) and H4 (residues 79–92, KTVTAMDVVYALKR) (Fig. 2). The peptides mentioned above for H3 and H4 will incorporate a single labeled methionine during protein synthesis after incubation in the labeled 13CD3-methionine media and thus can be monitored for protein turnover. In Fig. 2A, we observed a peak at 748.932 m/z corresponding to the “old” unlabeled H3 117–128 peptide signal, and after 1 day, observed that this peptide signal is decreasing concurrently with the appearance of the “new” synthesized and isotopically labeled peptide at 750.945 m/z (Fig. 2C). Similarly, signals corresponding to the “old” and “new” H4 79–92 peptide can be detected at 881.998 and 884.010 m/z, respectively, as well (Fig. 2, B and D).
FIGURE 2.
Monitoring general protein methylation rates on histones by MS. Full mass spectra of the [M+2H]2+ ions of the old unlabeled (charge = +2, with the molecular ion carrying two additional protons) containing H3 and H4 peptides (A and B) and newly synthesized peptides (C and D) that incorporate heavy 13CD3-methionine after 1 day of introduction into labeled media. Note that after 1 day, nearly equal abundances for the isotopic distributions corresponding to the “old” and “new” synthesized peptide are observed. pr, propionyl amide group from chemical derivatization of histone proteins.
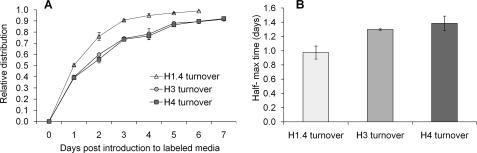
As histone H1 variants do not contain methionine, we used a different SILAC experiment to monitor turnover of histone H1.4. For H1.4, we labeled cells with isotopically heavy lysine (15N213C6-lysine), and thus could monitor protein turnover of the H1 variant by analysis of many peptides. We chose the fully lysine labeled peptide 33–53 for estimation of H1.4 turnover (data not shown) as it gave robust signals in the MS. It should be noted that all the Lys and Arg residues on these three “general protein turnover” peptides from H3, H4, and H1.4 were not detected to be post-translationally modified in our HeLa cells and thus serve as ideal peptides to monitor overall protein turnover independently of PTM dynamics. Using this approach, we found that t½ was approximately 1 day for H3, H4, and H1.4, and this is consistent with the nearly 24 h doubling time of HeLa cells under physiological conditions, with H1.4 exhibiting slightly faster t½ than H3 and H4 (Table 1 and Fig. 3B). Previous studies of Drosophila histones revealed that H1 is deposited on newly replicated chromatin after H3 and H4, which are deposited at nearly the same time (24), Because our HeLa cultures are asynchronous, we expect that such differences in mitotic timing average over any differences in histone deposition during DNA replication.
TABLE 1.
The average half-maximal times (t½, days) of fully labeled histones
Although the H3K9 data combine the relative abundances of the peptide regardless of Lys-14 acetylation, the Lys-27 and Lys-36 data do not combine the relative abundance from peptides that are also modified at Lys-36 or Lys-27, respectively. A, active, S, silent, and n, not definitively determined for specific methylation state of the residue.
| PTM | t½ | S.E. | Type | PTM | t½ | S.E. | Type |
|---|---|---|---|---|---|---|---|
| Overall H3 | 1.298 | 0.007 | – | H3K36me2:2 | 0.571 | 0.000 | A(48) |
| H3K4me1:1 | 0.959 | 0.129 | A(33) | H3K79me1:1 | 1.105 | 0.070 | A(49) |
| H3K9me1:1 | 0.342 | 0.001 | A(33) | H3K79me2:2 | 3.609 | 0.283 | A(49) |
| H3K9me2:2 | 1.031 | 0.060 | S(33) | Overall H4 | 1.385 | 0.072 | – |
| H3K9me3:3 | 1.344 | 0.339 | S(33) | H4K20me1:1 | 0.297 | 0.005 | A(33) |
| H3K18me1:1 | 1.207 | 0.116 | N | H4K20me2:2 | 1.467 | 0.001 | S(18) |
| H3K27me1:1 | 0.470 | 0.005 | A(33) | H4K20me3:3 | 4.809 | 1.483 | S(34) |
| H3K27me2:2 | 1.145 | 0.001 | S(33) | H4R3me1:1 | 2.788 | 1.806 | N |
| H3K27me3:3 | 3.128 | 0.032 | S(33) | Overall H1.4 | 0.976 | 0.065 | – |
| H3K36me1:1 | 0.751 | 0.085 | A(33) | H1.4K25me1:1 | 1.264 | 0.166 | N |
FIGURE 3.
Overall protein turnover rates of histones H3, H4, and H1. 4. A, relative distributions of the formation of the newly synthesized H1.4 (triangle), H3 (circle), H4 (square) peptides that contain either a heavy 13CD3-methionine (H3 and H4) or l-lysine-13C615N2 (H1.4) isotope during the time course (days). The calculation of relative distributions is described in the text (e.g. H3 turnover = (H3M120met1:1+H3M120met1:1ox)/(H3M120met1:1+H3M120met1:1ox+H3M120met1:0+H3M120met1:0ox, notation as detailed in supplemental Table S1)). H1.4, H3, and H4 values are averages from two technical replicates, with vertical bars indicating the S.E. B, average half-maximal time of the H1.4, H3, H4 peptides, with vertical bars indicating the S.E.
Interestingly, from past studies that produced viable single knockouts of individual H1 variants in mice, researchers reasoned that the variants act redundantly in chromatin packing (25). A possible mechanism is suggested by fluorescence recovery after photobleaching studies that find H1 variants rapidly bind and unbind to the linker DNA (26). We hypothesize that the slightly greater rate of H1.4 protein turnover, relative to the core histones, hint at this rapid exchange of linker histones between nucleosomes. Other radiolabeling studies on mouse kidney cells also found H1 histones to have half-lives as much as three times shorter than H3 or H4, consistent with our observation of a very slight difference in half-lives for H1.4 versus H3 or H4 (27). Yet, it is uncertain whether such results in terminally differentiated cells can be accurately recapitulated in the immortalized HeLa cell line.
Detection of Progressing Methylation State Intermediates
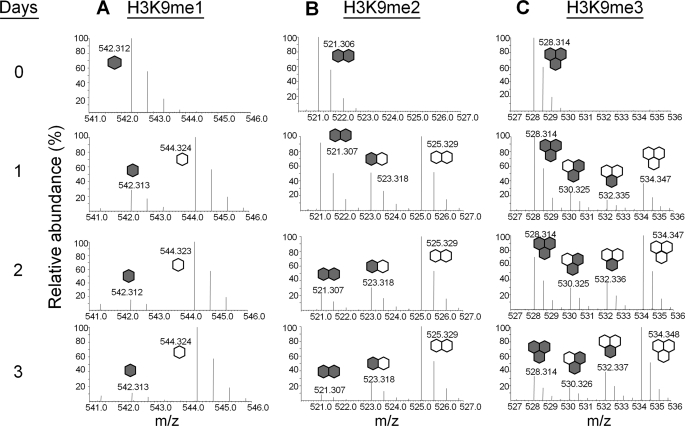
As diagramed in Fig. 1A, our experiments were performed by introducing HeLa cells grown in standard Joklik media into Joklik media that had 13CD3-methionine replacing natural methionine. Incorporation of a methyl group on the “new” labeled histones induces a +4 Da shift per methyl group. Thus, we expect to see +4, +8, and +12 Da shifts on newly labeled mono-, di-, and trimethylated peptides, respectively. Fig. 4 shows the incorporation of labeled methyl groups (me1, me2, and me3) onto histone H3K9 on the [M+2H]2+ ions detected from the peptide KSTGGKAPR. In Fig. 4A, we see at Day 0 the peptide signal coming only from the unlabeled “old” H3K9me1:0 peptide at 542.312 m/z, and as we progress through subsequent days in labeled media, we detect an increase in the “new” labeled H3K9me1:1 peptide at 544.324 m/z as expected (+4 Da shift on a 2+ charged peptide equals net +2 Da shift observed), and a decrease in the “old” H3K9me1:0 peptide.
FIGURE 4.
Rate of histone H3 Lys-9 methylation. Full mass spectra of the [M+2H]2+ ions from the H3K9 peptide, KSTGGKAPR, showing Lys-9 (A) mono- (B) di- and (C) trimethylation from H3 protein extracted from the first 3 days after pulse 13CD3-methionine labeling. Note the presence of peaks in the H3K9me2 and H3K9me3 fragment with an intermediate mass between the masses of the unlabeled and fully labeled peptides. Gray hexagons, unlabeled methyl, and white hexagons, labeled methyl.
However, we made an interesting observation when analyzing MS data of H3K9me2 and H3K9me3 peptides. As shown in Fig. 4B, we detected the unlabeled “old” H3K9me2:0 peptide at 521.306 m/z at Day 0, and upon incubation of cells in the labeled media we detected an increase in the fully labeled “new” H3K9me2:2 peptide at 525.329 (+8 Da shift on a +2 charged peptide equals net +4 Da m/z shift) over several days. We also detected another peak at 523.318 m/z (Day 1) that co-eluted with the two other H3K9me2 peptides. Similarly, we also detected the “old” H3K9me3:0 peptide at 528.314 m/z and the newly fully labeled H3K9me3:3 peptide at 534.347 m/z (Fig. 4C). However, we detected two peaks at 530.325 and 532.335 m/z, respectively, that also co-elute with the unlabeled and fully labeled H3K9me3 peptides. We performed MS/MS experiments on these peptides, as shown in Fig. 5 for the peptide at 523.318 m/z co-eluting with the H3K9me2 peptides, and found these peptides possessed both unlabeled and labeled methyl groups on the Lys-9 residue. Fig. 5 displays the MS/MS spectrum for the 523.318 m/z species, and the fragmentation pattern is consistent with Lys-9 containing both one unlabeled and one labeled methyl group (b1 ion at 217 Da). We reasoned that this peptide originated from an H3 protein that was previously monomethylated on Lys-9, and was methylated to the dimethyl state after transfer into the 13CD3-labeled Joklik media. This methylated peptide represents an “intermediate” species that would have been missed by any other approach. In our quantitative analysis below, we also consider pathways where demethylation could lead to labeled intermediates.
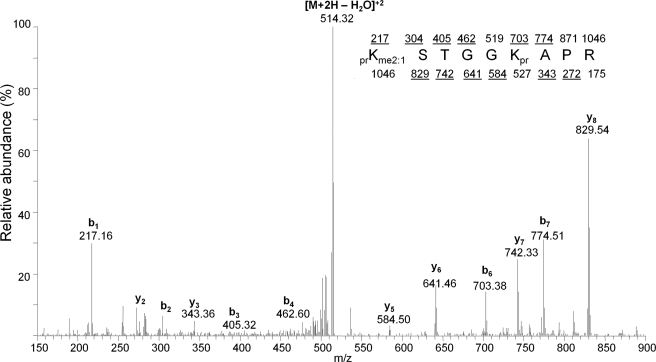
FIGURE 5.
Sequencing of H3K9me2-labeled intermediate. MS/MS spectrum of the [M+2H]2+ ion at 523.318 m/z, which was determined to be generated from the peptide prKme2STGGKprAPR. Expected mono-isotopic b- (top row) and y-type (bottom row) ion fragment masses are displayed, with fragments that could be annotated as underlined. We found that the Lys-9 residue contained both a single unlabeled and 13CD3-labeled methyl group. [M+2H-H2O]2+ refers to the +2 peptide with loss of water. pr, propionyl amide group from chemical derivatization of histone proteins.
Additionally, the two “intermediate” H3K9me3 species at 530.325 and 532.335 m/z, respectively, were determined after MS/MS experiments (supplemental Table S1 and Fig. S2) to be H3K9me3 peptides containing 2 “old” and 1 “new” methyl labels, and 1 “old” and 2 “new” methyl labels, respectively. In other words, the species at 530.325 m/z was an H3K9me2 peptide that was progressively methylated to the trimethyl state, while the species at 532.335 m/z was an H3K9me1 peptide that was progressively methylated to the trimethyl state. In addition to H3K9, we monitored the methylation status of histone H3K4, H3K18, H3K27, H3K36, H3K79, H4R3, H4K20, and H1.4K25 (supplemental Fig. S3). Thus, our method is useful both for monitoring the disappearance of the old methylated histones and appearance of the newly methylated histones, and for tracking any “intermediate” species that indicate progressive methylation in histone dynamics.
Methylation Rates within and between Residues
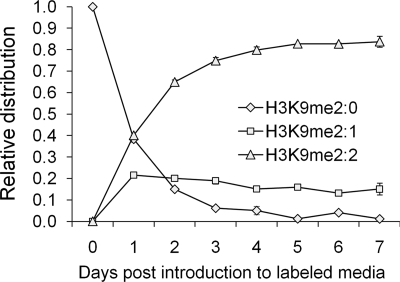
Using the methodology outlined above, we plotted the abundance of the peptides corresponding to the disappearance of “old” methylation, synthesis of “new” methylation and any “intermediate” methylation species for the H3, H4, and H1 residues. Fig. 6 shows the relative distribution of the three H3K9me2-labeled species. The H3K9me2:0 peptide is nearly completely gone by Day 7. H3K9me2:2 increases in abundance and reaches an asymptote of over 80%, while the “intermediate” H3K9me2:1 reaches a maximum at Day 1 and levels off in abundance over several days. The H3K9me2:2 peptide does not reach 100% abundance because of methylation from the unlabeled intracellular methionine pool; if the cells continued to grow in the labeled media, we expect that eventually this pool will become labeled and the H3K9me2:2 peptide should reach 100% abundance. The intermediate species often have different rates than “old” or “new” methyl peptides. As previously mentioned for H3K9me2 (Fig. 6), H3K9me2:2 becomes the primary labeled species with the me2 state by Day 2 and levels off in abundance after Day 3. The trend that the fully labeled species within a methylation state has the greatest relative abundance after the time course is observed for all residues analyzed (Fig. 6 and supplemental Fig. S4).
FIGURE 6.
Kinetics of dimethylated H3K9-labeled intermediates. Relative distribution of the three labeled intermediates of H3K9me2 peptide after pulse labeling. H3K9me2:0, old, H3K9me2:1, intermediate, and H3K9me2:2, new. Vertical bars denote S.E. Lines do not reflect differential equation fitting and are simply visual guides.
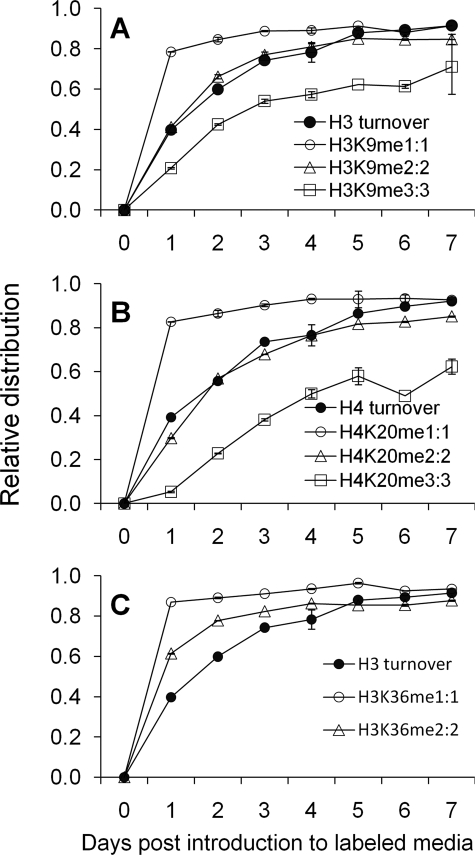
We next compared the methylation rates of labeled species on different histone methylation residues to general overall histone turnover as described in Fig. 2. For example, comparisons made with the fully labeled peptides for H3K9, H4K20, and H3K36 peptides are shown in Fig. 7. Among our findings is the difference in the rate of label incorporation between the three methylation states of H3K9 (Table 1). From quantifying half-maximal times, we find that H3K9me1 reaches its final fully labeled state faster than histone H3. H3K9me2 and H3K9me3 have similar half-maximal times, and both states have t½ similar to histone H3 (Table 1). A similar pattern for each of the H4K20 methylation states was also observed, where H4K20me2 has a similar t½ as overall histone H4 protein as shown in Fig. 7B, consistent with previous evidence that H4K20 methylation does not appreciably turnover in vivo (14). In contrast, H3K36 methylation is much different, as both me1 and me2 have faster half-maximal rates than the histone H3 protein turnover (Fig. 7C and Table 1). Additionally, H3K27 methylation followed the overall trend of me1 > me2 > me3 (Table 1). The changes in t½ across different methylation states are not additive. For instance, H3K9me2 has a t½ nearly three times as long as H3K9me1, while H3K9me3 has a half-maximal time four times as long as H3K9me1. Thus, it is unlikely that differences in t½ between the methylation states are due to the additional time required to sequentially add methyl groups to a single residue. Half-maximal times for all methylation sites detected on histones H3, H4, and H1.4 are listed in Table 1.
FIGURE 7.
Comparison between methylation state dynamics and total histone turnover. The relative distribution of fully labeled (A) H3K9me, (B) H4K20me, and (C) H3K36me compared with the overall turnover of histone H3 or H4. Vertical bars denote S.E. Lines do not reflect differential equation fitting and are simply visual guides.
After quantifying t½ and categorizing the residues by their known epigenetic function, we observed that the silencing marks H3K27me3:3 and H4K20me3:3 have the slowest rates of all residues examined (Table 1). Our study also revealed the active mark H3K9me1 as having among the fastest rates. Indeed, active marks had significantly faster half-maximal times than repressive marks (p = 0.0200). The relative distributions of all labeled intermediates and fully labeled species for each histone peptide compared with overall turnover are shown in supplemental Fig. S4.
Influence of Multiple PTMs on Methylation Rates
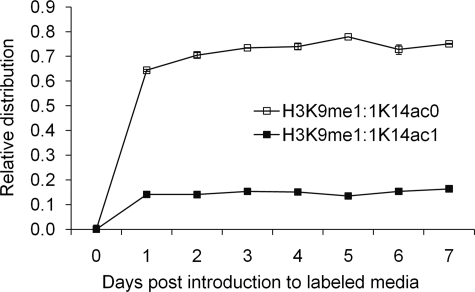
When calculating the relative distributions for each labeled species, we combined the relative abundances of all the peptides that had the particular modification of interest without regard to modifications on other residues on the same peptide. This pertains to the H3 peptide KSTGGKAPR where both H3K9 can be methylated and K14 can be acetylated. When we deconvoluted the relative abundance for a labeled species into its two components, differing by Lys-14 acetylation, we observed that the rate of formation of H3K9me1:1 is increased by over 50% when Lys-14 is acetylated (Fig. 8 and supplemental Table S2). No significant change was observed for H3K9me2 when Lys-14 is acetylated (supplemental Fig. S5A). When we examined the influence of H3K27 and H3K36 on each other, the formation of H3K27me1, H3K27me2, and H3K36me1 and H3K36me2 is slower when the other residue becomes progressively methylated (supplemental Table S2 and Fig. S5B). In further investigating this effect for PTM combinations with 3 methylation states, namely H3K27me1, H3K36me1 and H3K36me2, there was a significant increase in t½ with increasing methylation of Lys-36 and Lys-27 respectively (p = 0.0329, df = 11).
FIGURE 8.
Effect of acetylation on histone Lys-9 methylation. The deconvolution of the H3K9me1:1 peptides into its two components, which differ only in the acetylation state of the Lys-14 residue. Vertical bars denote S.E. Lines are not equation fitting but rather visual guides.
Kinetic Parameter Estimation from Differential Equations
In addition to calculating t½, we quantified the dynamics for each methylation state (Fig. 1B) by estimating various parameters that represented first order rate constants (day−1) in our idealized system for each residue (Table 2). This was attained by deriving a set of differential equations aimed at characterizing the dynamics of methylated residues (supplemental Fig. S2). The individual parameters derived from the relative levels are consistent with the qualitative conclusions drawn from the relative distributions. For instance, the trend that k0→1> k1→2> k2→3for H3K9, H3K27, and H4K20 is consistent with the observation that me1, me2, and me3 peptides have progressively slower rates of formation (Table 2). Finally, the contribution of unlabeled methyl groups derived from macromolecule degradation (k′0→1, k′1→2, k′2→3) was generally lower than the contribution from the labeled methyl groups (k0→1, k1→2, k2→3) across all residues. This reflects the fact that the levels of exogenous labeled methionine are greater than the levels of endogenous unlabeled methionine.
TABLE 2.
Average kinetic parameters (day−1) determined from fits of differential equations to the relative levels of various methylation species
Parameters are the rate of histone binding chromatin (α), labeled (k0→1, k1→2, k2→3) or unlabeled (k′0→1, k′1→2, k′2→3) methylation, demethylation (k3→2, k2→1, k1→0), and fragment degradation for each methylation state (μ0, μ1, μ2, μ3). All four μ parameters already account for dilution and thus represent degradation above the dilution rate. Parameters for H3K36 and H3K27 were determined with peptides in which the other residue (H3K27 and H3K36, respectively) on the same fragment was unmodified, because of the inability to localize the label on certain labeled intermediates, as detailed in Supplemental Table S1. Averages and standard deviations (parentheses) were taken for fits with an RMSD less than 125% of the minimum RMSD determined.
| H3K4 | H3K18 | H4R3 | H1.4K26 | H3K36 | H3K79 | H3K9 | H3K27 | H4K20 | |
|---|---|---|---|---|---|---|---|---|---|
| α | 0.6948 (0.0016) | 1.4611 (0.2633) | 1.9590 (1.1634) | 2.8488 (1.0027) | 0.2539 (0.0750) | 0.7088 (0.0977) | 0.9453 (0.2226) | 0.3848 (0.0540) | 0.8118 (0.1372) |
| k0→1 | 0.0939 (0.0016) | 0.0052 (0.0008) | 0.0117 (0.0015) | 0.0223 (0.0010) | 3.4092 (0.4868) | 0.1186 (0.0077) | 3.8746 (0.1482) | 7.0139 (0.0720) | 3.4327 (0.1048) |
| k1→2 | – | – | – | – | 5.4056 (0.0277) | 0.2177 (0.0099) | 2.2240 (0.0038) | 4.1676 (0.0070) | 1.4813 (0.0123) |
| k2→3 | – | – | – | – | – | – | 0.2676 (0.0003) | 0.2329 (0.0014) | 0.0000 (0.0000) |
| k′0→1 | 0.0200 (0.0007) | 0.0023 (0.0004) | 0.0019 (0.0006) | 0.0045 (0.0006) | 0.3104 (0.0520) | 0.0447 (0.0055) | 0.4320 (0.0229) | 1.0496 (0.0198) | 0.2212 (0.0186) |
| k′1→2 | – | – | – | – | 0.3323 (0.0194) | 0.0004 (0.0050) | 0.2043 (0.0016) | 0.0302 (0.0097) | 0.0039 (0.0118) |
| k′2→3 | – | – | – | – | – | – | 0.0531 (0.0007) | 0.0852 (0.0048) | 0.1603 (0.0133) |
| k1→0 | 0.0009 (0.0052) | 0.4943 (0.0188) | 0.1249 (0.1279) | 0.0401 (0.0422) | 1.2228 (0.4952) | 0.0125 (0.0308) | 0.2480 (0.2283) | 0.0016 (0.0084) | 0.0061 (0.0274) |
| k2→1 | – | – | – | – | 0.2510 (0.0455) | 0.0008 (0.0018) | 0.1020 (0.0016) | 0.0635 (0.0022) | 0.0703 (0.0038) |
| k3→2 | – | – | – | – | – | – | 0.0000 (0.0009) | 0.1680 (0.0101) | 6.9116 (1.2521) |
| μ0 | 0.0001 (0.0003) | 0.7704 (0.2633) | 1.2807 (1.1787) | 2.2330 (1.0385) | 3.2997 (1.7333) | 0.0171 (0.1193) | 0.6811 (1.1148) | 1.8727 (1.4989) | 0.7828 (0.8591) |
| μ1 | 0.0031 (0.0119) | 0.2834 (0.2406) | 0.1208 (0.0906) | 0.0374 (0.0315) | 2.6177 (1.2729) | 0.0221 (0.0447) | 0.7617 (0.2429) | 0.0047 (0.0396) | 0.0075 (0.0346) |
| μ2 | – | – | – | – | 0.3448 (0.0929) | 0.0061 (0.0476) | 0.0008 (0.0032) | 0.0000 (0.0007) | 0.0017 (0.0080) |
| μ3 | – | – | – | – | – | – | 0.0001 (0.0007) | 0.0003 (0.0045) | 0.2931 (0.4585) |
DISCUSSION
We performed heavy methyl SILAC with the goal of quantifying methylation rates for distinct histone residues. One advantage of this approach is the unbiased and direct detection of histone PTMs, in contrast to earlier 15N labeling methods that require an a priori knowledge of which proteins are modified (28). The versatility of heavy methyl SILAC has been demonstrated in studies that differentiate histone demethylation and histone turnover as possibilities to account for reduced levels of H3K9me3 (29). Whereas a pulse-chase analysis determines the fates of only the histones modified and synthesized during the pulse, it requires an additional media transfer step, is less robust toward noise in label detection, and does not provide significantly more information than a continuous pulse labeling approach with regard to steady state kinetics.
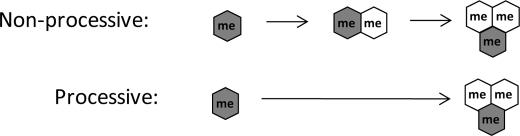
In addition to detection of the unlabeled and fully labeled methyl species, our experiments revealed the presence of labeled progressively methylated “intermediates” for the di- and trimethylated states. Such “intermediates” could link specific methyltransferases with a specific intermediate state. For instance, the formation of H3K9me3 can be catalyzed by Suv39H1, G9a, and SETDB1 (9). Yet, the three methyltransferases have different mechanisms. X-ray crystallographic analysis and in vitro assays have revealed that G9a (30) and the SET (31) domain act in a processive manner, where the enzyme can remain bound to the histone while subsequent methylations occur. In contrast, Suv39h1 is believed to act non-processively (32), where the enzyme must rebind to the histone after each round of methylation. We anticipate that the processive and non-processive mechanisms lead to different labeled intermediates (Scheme 1), which can be detected and resolved by our MS technique. Furthermore, the quantification of labeled intermediates at each residue allowed fitting to idealized differential equations and provided a more complete picture of how histone methylation is actively regulated in vivo. We observed some striking patterns in the MS data that warrant further discussion.
SCHEME 1.
Consequences of processive and non-processive methylation. Non-processive, rather than processive, methylation from me1:0 to me3:2 should result in the observed appearance of the me2:1 peptide because of the detachment and rebinding of the methyltransferase after the first round of methylation to me2.
Methylation Rates of Active and Silencing Marks
H3K27me3:3 and H4K20me3:3 have the slowest rates of formation among the histone PTMs examined. Interestingly, H3K27me3 (33) and H4K20me3 (34) are both silencing marks. The longer t½ of these PTMs may reflect the inheritance of the epigenetic mark across mitotic cycles. For instance, H3K27me3 is maintained by the EED-EZH2 PcG complex and enriched on the inactivated X chromosome in stem cells (35). In embryonic stem cells, H3K27me3 localization rises to 50% maximal levels 1 day after differentiation, and then drops back to 50% at ∼5 days after reaching the maximal abundance. These findings are broadly consistent with our value of t½ for H3K27me3 in HeLa cells.
Another histone PTM associated with gene silencing is H4K20me3, which is localized to pericentric heterochromatin in Drosophila (34). Although H3K9me3 and H3K27me1 (36) have the same cytological localization as H4K20me3, our study found both of those PTMs have shorter half-maximal times than H4K20me3. Thus, H4K20me3 may represent a more stable PTM for pericentric heterochromatin formation. It is tempting to hypothesize that in bivalent chromatin domains containing both activating and silencing marks (37), the mark that is epistatic to the other may have a slower rate of formation. The techniques described in this report, combined with middle-down MS sequencing to detect simultaneously multiple PTMs on the same H3 polypeptide fragment, could test this hypothesis (19).
In contrast to H3K27me3 and H4K20me3, H3K9me1 has among the fastest half-maximal times. From the sites examined, we found that active marks are formed significantly faster than silencing marks. This suggests that cells require more fine temporal control over which genes are activated than which genes are silenced. Such a relation was proposed in a recent review, with the logic that silencing marks would be more easily perpetuated as epigenetic inheritance with slower turnovers (38). Although there might be exceptions, we can speculate the epigenetic features of certain PTMs based on their rate of formation. H3K18me1 is not known presently to be an active or silencing mark, but the PTM has a half-maximal time slower than all other me1 sites with gene activating function and similar to overall H3. Based on this trend, we hypothesize that H3K18me1 acts as a silencing mark. Additionally, because H3K18ac is highly correlated with active genes (39), our speculation that H3K18me1, which should be an antagonistic PTM to H3K18ac, acts as a silencing mark is not unreasonable, but this hypothesis requires further biological experiments to prove. Such behavior might also implicate H3K18me1 and H3K18ac1 and PTMs on other sites to act as “binary switches” on the same residue, similar to H3K9me3 and H3K9ac (40).
It is important to note that our current data on histone methylation rates are averaged over the entire genome. The more rapid histone exchange over specific genomic regions such as cis-regulatory elements (41) could influence the abundance of chromatin-bound histones. For instance, we predict that more rapid histone exchange leads to faster rates of formation of the labeled methylated species.
Synergistic and Antagonistic Influences of Multiple PTMs
In addition to finding different methylation rates between active and silencing marks, we found that the formation of H3K9me1:1 is increased when Lys-14 is acetylated. The behavior is consistent with our hypothesis that active marks are formed faster than silencing marks and with the finding that demethylation of H3K4 by LSD1 is inhibited by acetylation of H3K9, H3K14, and H3K18 (42). Because levels of H3K9me1 are a product of both methylation and demethylation, a decrease in demethylation due to H3K14ac could account for the increase in H3K9me1:1 levels. Further supporting evidence was found in a past report using a continuous pulse labeling with [3H]methionine in alfalfa histone H3.1 and H3.2 (10). The author proposed that increased acetylation relaxes the nucleosome to increase access for methyltransferases, although we found such a model unlikely given the spatial location of the H3 tails relative to the nucleosome (43).
In contrast to the synergistic effect of H3K14ac on H3K9me1, we believe that H3K27 and H3K36 may have antagonistic effects toward each other. In particular, the presence of another PTM on the same peptide with an opposite epigenetic effect may render the doubly modified peptide slower to form. Middle Down MS sequencing could reveal the effects of multiple PTMs on methylation rate (44). The histone code hypothesis proposes that combinatorial patterns of multiple histone PTMs lead to different transcriptional outputs (45). Thus, differences in peptide methylation rates observed due to PTMs on different residues may agree with such a model.
Insights from Kinetic Modeling
It is important to emphasize that the “rate constants” derived from our modeling do not reflect the activity of a specific methyltransferase or demethylase, but rather the average concerted activity of all the enzymes in vivo. Actual histone methylation dynamics are likely more complicated than our idealized system. For instance, we assumed sequential methylation and demethylation. In addition to the earlier examples for processive and non-processive methyltransferase, DIM-5 is a methyltransferase that sequentially methylates H3K9 to produce H3K9me2 and then H3K9me3 (46). However, ASH1 is known to trimethylate H3K9 from an unmodified state (22). Sequential methylation may likely not be the most accurate way to model such behavior. Although higher order differential equations could more accurately describe the enzymatically catalyzed methylation and demethylation, at least for overall H3 methylation, past studies found that turnover could indeed be modeled as a first order reaction (12).
Despite these caveats, the rates calculated from the relative levels are consistent with and provide a quantitative explanation for the half-maximal times from the relative distribution and qualitative inspection of the MS data. For instance, H3K36me2 has a faster half-maximal time than H3K9me2. Our modeling efforts reveal that this behavior is due to the greater rate of dimethylation (k1→2 and k′1→2), demethylation (k2→1), and degradation (μ2) of H3K36me2 than H3K9me2 (Table 2). Stated differently, H3K36me2 is more rapidly formed and reformed after demethylation than H3K9me2, indicating that H3K36me2 has faster turnover than H3K9me2.
Similar logic applies within a residue, such as H3K9. H3K9me1 is methylated faster (k0→1 and k′0→1) than H3K9me2 (k1→2 and k′1→2), which is methylated faster than H3K9me3 (k2→3 and k′2→3). Although H3K9me1 and H3K9me2 have similar rates of demethylation (k1→0 and k2→1) and degradation (μ1 and μ2), both have faster rates than H3K9me3 (k3→2 and μ3). The net effect is that increasing methylation states on H3K9 have slower turnover. Additionally, for H3K79, there appears to be little detectable demethylation (k1→0 and k2→1). Interestingly, there is currently no known demethylase for H3K79 (47). In contrast, although no known demethylase exists for H4K20 (47), our modeling identified a non-zero demethylation rate (k2→1 and k3→2) for this site.
In summary, the quantification of all labeled intermediates for each methylation state at each residue allowed fitting of the data to idealized differential equations; these data provide a more complete picture of how actively regulated the histone PTMs are in vivo. Application of the findings and methods of this report provide a platform for further investigation into the dynamic regulation of histone PTMs under varying physiological conditions and over various genomic loci, an understudied yet highly informative part of histone biology and epigenetics.
Supplementary Material
Acknowledgments
We thank Joshua Bloom, R. Scott McIsaac, Fred M. Hughson, Thomas Gregor, John D. Storey, and Joshua D. Rabinowitz for helpful discussion, Robert Endres for supplying an early MATLAB code, and all members of the Garcia laboratory for technical assistance and enlightening conversations.
This work was supported by Princeton University, the National Science Foundation Grant CBET-0941143, and an American Society for Mass Spectrometry research award sponsored by the Waters Corporation (to B. A. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Data, Table S1, and Figs. S1–S5.
- PTM
- post-translational modifications
- MS
- mass spectrometry
- ChIP
- chromatin immunoprecipitation
- SILAC
- stable isotope labeling by amino acids in cell culture
- RMSD
- root mean-squared difference.
REFERENCES
- 1.Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2.Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 3.Rice J. C., Briggs S. D., Ueberheide B., Barber C. M., Shabanowitz J., Hunt D. F., Shinkai Y., Allis C. D. (2003) Mol. Cell 12, 1591–1598 [DOI] [PubMed] [Google Scholar]
- 4.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Cell 125, 483–495 [DOI] [PubMed] [Google Scholar]
- 5.Klose R. J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. (2006) Nature 442, 312–316 [DOI] [PubMed] [Google Scholar]
- 6.Noma K., Allis C. D., Grewal S. I. (2001) Science 293, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 7.Wang H., Huang Z. Q., Xia L., Feng Q., Erdjument-Bromage H., Strahl B. D., Briggs S. D., Allis C. D., Wong J., Tempst P., Zhang Y. (2001) Science 293, 853–857 [DOI] [PubMed] [Google Scholar]
- 8.Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Green R. D., Kouzarides T. (2009) Nat. Struct. Mol. Biol. 16, 449–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y., Whetstine J. R. (2007) Mol. Cell 25, 1–14 [DOI] [PubMed] [Google Scholar]
- 10.Waterborg J. H. (1993) J. Biol. Chem. 268, 4918–4921 [PubMed] [Google Scholar]
- 11.Byvoet P., Shepherd G. R., Hardin J. M., Noland B. J. (1972) Arch. Biochem. Biophys. 148, 558–567 [DOI] [PubMed] [Google Scholar]
- 12.Thomas G., Lange H. W., Hempel K. (1975) Eur. J. Biochem. 51, 609–615 [DOI] [PubMed] [Google Scholar]
- 13.Pesavento J. J., Yang H., Kelleher N. L., Mizzen C. A. (2008) Mol. Cell. Biol. 28, 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McManus K. J., Biron V. L., Heit R., Underhill D. A., Hendzel M. J. (2006) J. Biol. Chem. 281, 8888–8897 [DOI] [PubMed] [Google Scholar]
- 15.Scharf A. N., Barth T., Imhof A. (2009) Nucleic Acids Res. 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morillon A., Karabetsou N., Nair A., Mellor J. (2005) Mol. Cell 18, 723–734 [DOI] [PubMed] [Google Scholar]
- 17.Ong S., Mittler G., Mann M. (2004) Nat. Methods 1, 1–8 [DOI] [PubMed] [Google Scholar]
- 18.Trojer P., Li G., Sims R. J., 3rd, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D., Wang Y. H., Reinberg D. (2007) Cell 129, 915–928 [DOI] [PubMed] [Google Scholar]
- 19.Garcia B. A., Pesavento J. J., Mizzen C. A., Kelleher N. L. (2007) Nat. Methods 4, 487–489 [DOI] [PubMed] [Google Scholar]
- 20.Garcia B. A., Mollah S., Ueberheide B. M., Busby S. A., Muratore T. L., Shabanowitz J., Hunt D. F. (2007) Nat. Protoc. 2, 933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeRoy G., Weston J. T., Zee B. M., Young N. L., Plazas-Mayorca M. D., Garcia B. A. (2009) Mol. Cell. Proteomics 8, 2432–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beisel C., Imhof A., Greene J., Kremmer E., Sauer F. (2002) Nature 419, 857–862 [DOI] [PubMed] [Google Scholar]
- 23.Osley M. A. (1991) Annu. Rev. Biochem. 60, 827–861 [DOI] [PubMed] [Google Scholar]
- 24.Worcel A., Han S., Wong M. L. (1978) Cell 15, 969–977 [DOI] [PubMed] [Google Scholar]
- 25.Sirotkin A. M., Edelmann W., Cheng G., Klein-Szanto A., Kucherlapati R., Skoultchi A. I.(1995) Proc. Natl. Acad. Sci. U. S. A. 92, 6434–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T. (2000) Nature 408, 877–881 [DOI] [PubMed] [Google Scholar]
- 27.Djondjurov L. P., Yancheva N. Y., Ivanova E. C. (1983) Biochemistry 22, 4095–4102 [DOI] [PubMed] [Google Scholar]
- 28.Oda Y., Huang K., Cross F. R., Cowburn D., Chait B. T. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 6591–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fodor B. D., Kubicek S., Yonezawa M., O'Sullivan R. J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. (2009) Genes Dev. 20, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patnaik D., Chin H. G., Estève P., Benner J., Jacobsen S. E., Pradhan S. (2004) J. Biol. Chem. 279, 53248–53258 [DOI] [PubMed] [Google Scholar]
- 31.Trievel R. C., Beach B. M., Dirk L. M., Houtz R. L., Hurley J. H. (2002) Cell 111, 91–103 [DOI] [PubMed] [Google Scholar]
- 32.Chin H. G., Patnaik D., Estève P., Jacobsen S. E., Pradhan S. (2006) Biochemistry 45, 3272–3284 [DOI] [PubMed] [Google Scholar]
- 33.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 34.Schotta G., Lachner M., Sarma K., Ebert A., Sengupta R., Reuter G., Reinberg D., Jenuwein T. (2004) Genes Dev. 18, 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plath K., Fang J., Mlynarczyk-Evans S. K., Cao R., Worringer K. A., Wang H., de la Cruz C. C., Otte A. P., Panning B., Zhang Y. (2003) Science 300, 131–135 [DOI] [PubMed] [Google Scholar]
- 36.Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. (2003) Mol. Cell 12, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 37.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 38.Lee B. M., Mahadevan L. C. (2009) J. Cell. Biochem. 108, 22–34 [DOI] [PubMed] [Google Scholar]
- 39.Kurdistani S. K., Tavazoie S., Grunstein M. (2004) Cell 117, 721–733 [DOI] [PubMed] [Google Scholar]
- 40.Fischle W., Wang Y., Allis C. D. (2003) Nature 425, 475–479 [DOI] [PubMed] [Google Scholar]
- 41.Mito Y., Henikoff J. G., Henikoff S. (2007) Science 315, 1408–1411 [DOI] [PubMed] [Google Scholar]
- 42.Forneris F., Binda C., Dall'Aglio A., Fraaije M. W., Battaglioli E., Mattevi A. (2006) J. Biol. Chem. 281, 35289–35295 [DOI] [PubMed] [Google Scholar]
- 43.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 44.Boyne M. T., Garcia B. A., Li M., Zamdborg L., Wenger C. D., Babai S., Kelleher N. L. (2009) J. Proteome Res. 8, 374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 46.Tamaru H., Zhang X., McMillen D., Singh P. B., Nakayama J., Grewal S. I., Allis C. D., Cheng X., Selker E. U. (2003) Nat. Genetics 34, 75–79 [DOI] [PubMed] [Google Scholar]
- 47.Lan F., Shi Y. (2009) Science in China Series C: Life Sciences 52, 311–322 [DOI] [PubMed] [Google Scholar]
- 48.Bell O., Wirbelauer C., Hild M., Scharf A. N., Schwaiger M., MacAlpine D. M., Zilbermann F., van Leeuwen F., Bell S. P., Imhof A., Garza D., Peters A. H., Schübeler D. (2007) EMBO J. 26, 4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.