Abstract
Complex I (NQR) is a critical site of superoxide ( ) production and the major host of redox protein thiols in mitochondria. In response to oxidative stress, NQR-derived protein thiols at the 51- and 75-kDa subunits are known to be reversibly S-glutathionylated. Although several glutathionylated domains from NQR 51 and 75 kDa have been identified, their roles in the regulatory functions remain to be explored. To gain further insights into protein S-glutathionylation of complex I, we used two peptides of S-glutathionylated domain (200GAGAYICGEETALIESIEGK219 of 51-kDa protein and 361VDSDTLCTEEVFPTAGAGTDLR382 of 75-kDa protein) as chimeric epitopes incorporating a “promiscuous” T-cell epitope to generate two polyclonal antibodies, AbGSCA206 and AbGSCB367. Binding of AbGSCA206 and AbGSCB367 inhibited NQR-mediated
) production and the major host of redox protein thiols in mitochondria. In response to oxidative stress, NQR-derived protein thiols at the 51- and 75-kDa subunits are known to be reversibly S-glutathionylated. Although several glutathionylated domains from NQR 51 and 75 kDa have been identified, their roles in the regulatory functions remain to be explored. To gain further insights into protein S-glutathionylation of complex I, we used two peptides of S-glutathionylated domain (200GAGAYICGEETALIESIEGK219 of 51-kDa protein and 361VDSDTLCTEEVFPTAGAGTDLR382 of 75-kDa protein) as chimeric epitopes incorporating a “promiscuous” T-cell epitope to generate two polyclonal antibodies, AbGSCA206 and AbGSCB367. Binding of AbGSCA206 and AbGSCB367 inhibited NQR-mediated  generation by 37 and 57%, as measured by EPR spin-trapping. To further provide an appropriate control, two peptides of non-glutathionylated domain (21SGDTTAPKKTSFGSLKDFDR40 of 51-kDa peptide and 100WNILTNSEKTKKAREGVMEFL120 of 75-kDa peptide) were synthesized as chimeric epitopes to generate two polyclonal antibodies, Ab51 and Ab75. Binding of A51 did not affect NQR-mediated
generation by 37 and 57%, as measured by EPR spin-trapping. To further provide an appropriate control, two peptides of non-glutathionylated domain (21SGDTTAPKKTSFGSLKDFDR40 of 51-kDa peptide and 100WNILTNSEKTKKAREGVMEFL120 of 75-kDa peptide) were synthesized as chimeric epitopes to generate two polyclonal antibodies, Ab51 and Ab75. Binding of A51 did not affect NQR-mediated  generation to a significant level. However, binding of Ab75 inhibited NQR-mediated
generation to a significant level. However, binding of Ab75 inhibited NQR-mediated  generation by 35%. None of AbGSCA206, AbGSCB367, Ab51, or Ab75 showed an inhibitory effect on the electron transfer activity of NQR, suggesting that antibody binding to the glutathione-binding domain decreased electron leakage from the hydrophilic domain of NQR. When heart tissue homogenates were immunoprecipitated with Ab51 or Ab75 and probed with an antibody against glutathione, protein S-glutathionylation was enhanced in post-ischemic myocardium at the NQR 51-kDa subunit, but not at the 75-kDa subunit, indicating that the 51-kDa subunit of flavin subcomplex is more sensitive to oxidative stress resulting from myocardial infarction.
generation by 35%. None of AbGSCA206, AbGSCB367, Ab51, or Ab75 showed an inhibitory effect on the electron transfer activity of NQR, suggesting that antibody binding to the glutathione-binding domain decreased electron leakage from the hydrophilic domain of NQR. When heart tissue homogenates were immunoprecipitated with Ab51 or Ab75 and probed with an antibody against glutathione, protein S-glutathionylation was enhanced in post-ischemic myocardium at the NQR 51-kDa subunit, but not at the 75-kDa subunit, indicating that the 51-kDa subunit of flavin subcomplex is more sensitive to oxidative stress resulting from myocardial infarction.
Keywords: Antibodies/Epitope Mapping, Antibodies/Polyclonal, Bioenergetics/Respiratory Chain, Methods/Electron Paramagnetic Resonance EPR, Oxygen/Superoxide, Complex I, Myocardial Infarction, Protein S-Glutathionylation
Introduction
In mitochondria, the generation of  and the oxidants derived from it can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. The mitochondrial redox pool is enriched in GSH with a high physiological concentration (in the millimolar range) (1); overproduction of
and the oxidants derived from it can act as a redox signal in triggering cellular events such as apoptosis, proliferation, and senescence. The mitochondrial redox pool is enriched in GSH with a high physiological concentration (in the millimolar range) (1); overproduction of  and
and  -derived oxidants increases the ratio of GSSG3 to GSH. Moreover, the proteins of the mitochondrial electron transport chain are rich in protein thiols (2, 3). It has been documented that Complex I is the major component of the electron transport chain to host protein thiols, which comprise structural thiols involved in the ligands of iron-sulfur clusters and the reactive/regulatory thiols that are thought to function in antioxidant defense and redox signaling (4, 5). Physiologically, the Complex I-derived regulatory thiols have been implicated in the regulation of respiration, nitric oxide utilization (6, 7), and redox status of mitochondria (1–3). It has been well documented that the 51- and 75-kDa subunits of the NQR hydrophilic domain are two of the major polypeptides that host regulatory thiols in mitochondria (4, 5, 12).
-derived oxidants increases the ratio of GSSG3 to GSH. Moreover, the proteins of the mitochondrial electron transport chain are rich in protein thiols (2, 3). It has been documented that Complex I is the major component of the electron transport chain to host protein thiols, which comprise structural thiols involved in the ligands of iron-sulfur clusters and the reactive/regulatory thiols that are thought to function in antioxidant defense and redox signaling (4, 5). Physiologically, the Complex I-derived regulatory thiols have been implicated in the regulation of respiration, nitric oxide utilization (6, 7), and redox status of mitochondria (1–3). It has been well documented that the 51- and 75-kDa subunits of the NQR hydrophilic domain are two of the major polypeptides that host regulatory thiols in mitochondria (4, 5, 12).
We have reported previously that in oxidative damage to NQR, the C206 moiety of the 51-kDa subunit plays a unique role as a reactive thiol, based on the evidence of immunospin trapping with 5,5-dimethyl pyrroline N-oxide and mass spectrometry (9). With a proteomic approach, we further determined that the Cys206 of the 51-kDa subunit is involved in site-specific S-glutathionylation (12). The peptide identified as 5,5-dimethyl pyrroline N-oxide-binding and GS binding is 200GAGAYIC206GEETALIESIEGK219, which is highly conserved in the bacterial, fungal, and mammalian enzymes (90% sequence identity in the bacterial enzyme). An x-ray crystal structure of the hydrophilic domain of respiratory Complex I from Thermus thermophilus indicates that this conserved cysteine (Cys182 in T. thermophilus) is only 6 Å from the FMN, which is consistent with the role of Cys206 as a redox-sensitive thiol and with FMN serving as a source of  (10).
(10).
The polypeptide of 75 kDa is the other subunit of NQR to be involved in redox modification via S-glutathionylation (4, 5, 11, 12). Based on the LC/MS/MS analysis, S-glutathionylation of Cys367 can be induced by oxidized GSSG through protein thiol disulfide exchange (12). Cys554 and Cys727 were S-glutathionylated when the NQR of bovine heart mitochondria was oxidatively stressed by diamide (11). The GS-binding peptides identified include 361VDSDTLC367TEEVFPTAGAGTDLR382 (12), 544MLFLLGADGGC554ITR557, and 713AVTEGAHAVEEPSIC727 (11). Although the identified GS-binding domains are not conserved in the bacterial and fungal enzymes (only 27.3% sequence identity in the bacterial enzyme), they are highly conserved in the mammalian enzymes.
Based on an EPR spin-trapping study, GSSG-induced glutathionylation of NQR at the 51- and 75-kDa subunits affects the  generation activity of NQR by marginally decreasing electron leakage and increasing electron transfer efficiency (12). High dosage of GSSG or diamide-induced glutathionylation tends to decrease the catalytic function of NQR and increase enzyme-mediated
generation activity of NQR by marginally decreasing electron leakage and increasing electron transfer efficiency (12). High dosage of GSSG or diamide-induced glutathionylation tends to decrease the catalytic function of NQR and increase enzyme-mediated  generation (5, 11). Nevertheless, the catalytic role of each identified GS-binding domain remains obscure. Knowledge of the function of each GS-binding domain is imperative, if we are to better understand the redox regulation mediated by NQR.
generation (5, 11). Nevertheless, the catalytic role of each identified GS-binding domain remains obscure. Knowledge of the function of each GS-binding domain is imperative, if we are to better understand the redox regulation mediated by NQR.
Probing the functional or regulatory roles of these domains may be accomplished with antibodies against specific functional domains in the 51- or 75-kDa subunits. Peptide-based immunochemistry is an ideal approach to generating high affinity antibodies against the specific functional domains present in the NQR. The development of high titer-specific antibodies also facilitates the detection of NQR-derived S-glutathionylation ex vivo and in vivo. Furthermore, a peptide-based antibody has potential use in developing a peptide-based vaccine; therefore, a pilot study is valuable for therapeutic applications.
In the present work, we have generated four peptide-based antibodies against specific domains of NQR. Two of them are specific for one of the glutathionylated domains and one of the non-glutathionylated domains of the 51-kDa subunit; the other two are specific for the 75-kDa subunit. The effect of antibody binding on NQR-mediated  generation was analyzed by EPR spin trapping. Finally, the specific antibodies were used to immunoprecipitate the polypeptides of 51 and 75 kDa from tissue homogenates of rat hearts for detecting intrinsic cysteine S-glutathionylation and evaluating the alteration of Complex I-derived glutathionylation after myocardial infarction.
generation was analyzed by EPR spin trapping. Finally, the specific antibodies were used to immunoprecipitate the polypeptides of 51 and 75 kDa from tissue homogenates of rat hearts for detecting intrinsic cysteine S-glutathionylation and evaluating the alteration of Complex I-derived glutathionylation after myocardial infarction.
EXPERIMENTAL PROCEDURES
Reagents
Ammonium sulfate, diethylenetriaminepentaacetic acid, ubiquinone-1 (Q1), sodium cholate, deoxycholic acid, rotenone, piericidin A, manganese-superoxide dismutase (Mn-SOD), and β-nicotinamide adenine dinucleotide (reduced form, NADH) were purchased from Sigma and used as received. The 5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide (DEPMPO) spin trap was purchased from Alexis Biochemicals (San Diego, CA).
Peptide Synthesis and Purification
Peptide synthesis was performed on a Milligen/Biosearch 9600 solid-phase peptide synthesizer (Bedford MA) using Fmoc/tert-Butyl chemistry. Preloaded Fmoc-amino acids on Clear Acid resin (0.36 meq/g, Peptides International, Louisville, KY) were used for the peptide synthesis using the PyBop/HoBt coupling method. The B-cell epitope was assembled by choosing regioselective side-chain protection on Cys residues as: Cys (Trt) or Cys (Acm) essentially as described by Kaumaya et al. (13, 14). The B-cell epitope was synthesized colinearly with a “promiscuous” T-helper epitope (MVF) derived from the measles virus fusion protein (amino acids 288–305 and Table 1). Also, a MVF T-helper epitope with a four-residue linker (GPSL) was incorporated for independent folding and was assembled on the B-cell epitope as indicated in Table 1. All peptides were cleaved from the resin using global deprotection reagent B (trifluoroacetic acid:phenol:water:triisopropylsilane, 90:4:4:2). The protecting group from Cys (Trt) comes off in the global cleavage reaction. Crude peptides were purified on preparative reversed-phase high-performance liquid chromatography using a C-4 Vydac column in water (0.1% trifluoroacetic acid):acetonitrile (0.1% trifluoroacetic acid) gradient system. Pure fractions were analyzed using analytical high-performance liquid chromatography, pooled together, and lyophilized in 10% acetic acid solution. The purified peptide was hydrolyzed dry and kept at −20 °C to prevent oxidation of free sulfhydryl groups of Cys residues.
TABLE 1.
Amino acid sequence of designed peptides and their corresponding MVF fusion peptides used as immunogens
| No. | Amino acid sequencea | Sequence code | No. of amino acids | Mr |
|---|---|---|---|---|
| 1 | GAGAYICGEETALIESIEGK | pGSCA206 | 20 | 2010 |
| 2 | KLLSLIKGVIVHRLEGVEGPSLGAGAYICGEETALIESIEGK | pMVFGSCA206 | 42 | 4359 |
| 3 | VDSDTLCTEEVFPTAGAGTDLR | pGSCB367 | 22 | 2179 |
| 4 | KLLSLIKGVIVHRLEGVEGPSLVDSDTLCTEEVFPTAGAGTDLR | pMVFGSCB367 | 44 | 4519 |
| 5 | SGDTTAPKKTSFGSLKDFDR | p51 | 20 | 2139 |
| 6 | KLLSLIKGVIVHRLEGVEGPSLSGDTTAPKKTSFGSLKDEDR | pMVF51 | 42 | 4479 |
| 7 | WNILTNSEKTKKAREGVMEFL | p75 | 21 | 2495 |
| 8 | KLLSLIKGVIVHRLEGVEGPSLWNILTNSEKTKKAREGVMEFL | pMVF75 | 43 | 4835 |
a Boldface amino acids represent ■.
Peptide Immunization and Antibody Purification
For each chimeric peptide (see Table 1), two New Zealand white rabbits (6–8 weeks old, female outbred) were purchased from Harlan (Indianapolis, IN), and immunized with each chimeric peptide (1 mg) dissolved in H2O (500 μl) with 100 mg of a muramyl dipeptide adjuvant, N-acetylglucosamine-3-yl-acetyl-l-alanyl-d-isoglutamine. Peptides were emulsified (50:50) in a Montanide ISA 720 vehicle (Seppic, France). 2 ml of blood was drawn for pre-immunization sera. All rabbits were immunized subcutaneously at four spots on the back. After the first immunization, the same dose of booster injections was administered three times at 3, 6, and 9 weeks. Sera were collected by bleeding from the ear of the rabbit after each immunization for determination of antibody titers. Antibody titers were determined by ELISA. High titered sera were purified on a protein A/G-agarose column (Pierce). Eluted antibodies were concentrated and exchanged in PBS using a 100-kDa cut-off centrifuge filter units (Millipore, Bedford, MA).
Preparations of Mitochondrial Complex I
Bovine heart mitochondrial Complex I was prepared under non-reducing conditions according to the published method with modifications (15). Submitochondrial particles (SMPs) were prepared as described and used as the starting material (16), starting with 1135 g of trimmed bovine hearts with fat and connective tissues removed. The SMP preparation was suspended in 50 mm Tris-Cl buffer, pH 8.0, containing 1 mm histidine and 0.66 m sucrose (TSH), and then subjected to KCl fractionation (72 g of KCl was added to per liter of SMPs) in the presence of deoxycholate (0.3 mg/mg of protein). The supernatant thus obtained was mixed with an appropriate amount of cold water to precipitate trace amounts of cytochrome c oxidase, and then dialyzed against 10 mm Tris-Cl, pH 8.0, containing 1 mm EDTA for 6 h with one change of buffer. The dialysate was subjected to centrifugation (96,000 × g for 75 min). The pellet containing Complexes I, II, and III was homogenized in TSH buffer, and then subjected to repeated ammonium acetate fractionation in the presence of deoxycholate (0.5 mg/mg of protein). Complex I was finally resolved (39% saturation of ammonium sulfate) and separated using ammonium sulfate precipitation (35.9% saturation) in the presence of potassium cholate (0.4 mg/mg of protein). The three-subunit flavin subcomplex of Complex I containing NADH dehydrogenase was isolated from SMP under non-reducing conditions by following the established method described in a previous publication (9).
Analytical Methods
Optical spectra were measured on a Shimadzu 2401 UV-visible recording spectrophotometer. The protein concentrations of SMP and Complex I were determined by the Biuret method using bovine serum albumin as standard. The concentration of Q1 was determined by absorbance spectra from NaBH4 reduction using a millimolar extinction coefficient ϵ(275 nm-290 nm) = 12.25 mm−1cm−1 (17). To measure the electron transfer activity of Complex I, an appropriate amount of Complex I was added to an assay mixture (1 ml) containing 20 mm potassium phosphate buffer, pH 8.0, 2 mm NaN3, and 0.1 mm Q1, and 0.15 mm NADH as developed by Hatefi et al. (18). The Complex I activity was determined by measuring the decrease in absorbance at 340 nm. The specific activity of Complex I was calculated using a molar extinction coefficient ϵ340 nm = 6.22 mm−1cm−1. The purified Complex I exhibited a specific activity of ∼1.0 μmol of NADH oxidized min−1mg−1.
EPR Experiments
EPR measurements were carried out on a Bruker EMX spectrometer operating at 9.86 GHz with 100-kHz modulation frequency at room temperature. The reaction mixture was transferred to a 50-μl capillary, which was then positioned in the HS cavity (Bruker Instrument, Billerica, MA). The sample was scanned using the following parameters: center field, 3510 G; sweep width, 140 G; power, 20 milliwatts; receiver gain, 2 × 105; modulation amplitude, 1 G; time constant, 163.84 ms; and number of scans, 3. The spectral simulations were performed using the WinSim program developed at NIEHS by Duling (19). The hyperfine coupling constants used to simulate the spin adduct of DEPMPO/·OOH were isomer 1: aN = 13.14 G, aHβ = 11.04 G, aHγ = 0.96 G, and aP = 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aHβ = 12.59 G, aHγ = 3.46 G, and aP = 48.2 G (20% relative concentration) (20). The correlation coefficient of simulated spectrum is typically >0.950. Therefore, the simulated spectrum is suitable for spin quantitation (9, 21).
Immunoblotting Analysis
The reaction mixture was mixed with the Laemmli sample buffer at a ratio 4:1 (v/v), incubated at 70 °C for 10 min, and then immediately loaded onto a 4–12% Bis-Tris polyacrylamide gradient gel. Samples were run at room temperature for 55 min at 190 V. Protein bands were electrophoretically transferred to nitrocellulose membrane in 25 mm Bis-Tris, 25 mm Bicine, 0.029% (w/v) EDTA, and 10% methanol. Membranes were blocked for 1 h at room temperature in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TTBS) and 5% dry milk (Bio-Rad). The blots were then incubated overnight with generated antibodies or anti-GSH monoclonal antibody at 4 °C. Blots were then washed three times in TTBS and incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG in TTBS at room temperature. The blots were again washed twice in TTBS and twice in TBS and then visualized using ECL Western blotting Detection Reagents (Amersham Biosciences).
In Vivo Myocardial Regional Ischemia/Reperfusion Model
The procedure for the in vivo ischemia-reperfusion rat model was performed by the technique reported in the literature (8, 22, 23). Sprague-Dawley rats (∼300–350 g) were anesthetized with Nembutal administered intraperitoneally (80–100 mg/kg). After the rats were fully anesthetized, they were intubated and then ventilated with room air (1.0 ml, rate of 100 breaths/min) using a mechanical ventilator Model 683 (Harvard Apparatus, Holliston, MA). The rats then underwent a left lateral thoracotomy, the pericardium was opened, and a pericardial cradle formed to allow adequate exposure of the heart surface. The left anterior descending coronary artery was then occluded by placing a suture (6.0 nylon) around the origin of the left anterior descending coronary artery.
After 30 min of ischemia, the suture around the coronary artery was untied, allowing reperfusion to occur. Following the reperfusion, all wounds were closed and infiltrated with 0.5% xylocaine (<0.3 ml). The muscular layers and skin incisions were closed with 4-0 nylon sutures. A chest tube (2.5-cm PE 50 tubing) was inserted at the wound site and maintained in position while the animal was taken off respiratory support.
Upon spontaneous breathing, the chest tube was removed and a surgical clip applied over the withdrawal site. The animal was allowed to recover, and a physiological assessment was performed. During the recovery period the animals received supportive post-operative care as needed. Body temperature was maintained at 37 °C by a thermal heating pad. By 6 h post-operation, the animals had recovered sufficiently to eat and drink independently.
At 24-h post infarction, the rats were placed under deep anesthesia with Nembutal (200 mg/ml). Left anterior descending coronary artery was re-occluded, and Evans blue (4%) was injected from the inferior vena cava to delineate the non-ischemic myocardial tissue. Rats were then sacrificed, and the hearts were excised and placed in PBS buffer. The infarct area was identified by 2,3,5-triphenyltetrazolium chloride staining. The risk region of myocardial tissue without 2,3,5-triphenyltetrazolium chloride staining was excised and subjected to biochemical analysis.
Isolated Rat Heart Model
Global ischemia-reperfusion conditions were imposed upon isolated hearts according to published procedures (24, 25). Briefly, male Sprague-Dawley rats (∼300–350 g and 9–10 weeks old) were heparinized with heparin (500 units) and anesthetized with intraperitoneal Nembutal (30–35 mg/kg). The hearts were excised, and the aortas were cannulated and perfused at 37 °C with a constant pressure of 80 mm Hg using Krebs solution (in mm, 16.7 glucose, 120 NaCl, 25.0 NaHCO3, 2.5 CaCl2, 5.9 KCl, 0.5 EDTA, and 1.2 MgCl2) pre-bubbled with 5% CO2, 95% O2 gas. Two-side arms in the perfusion line located just proximal to the heart cannula allowed infusion of treatment agents. Hearts were subjected to control perfusion or 30-min ischemia followed by 60-min reperfusion. Hearts were then placed on ice and immediately subjected to mitochondrial preparation. Ischemic duration was 30 min. The isolated hearts were perfused for 60 min at 37 °C by the method of Langendorff at a constant pressure of 80 mm Hg with a Krebs-buffered perfusate.
RESULTS
 Generation Mediated by NQR as Measured by EPR Spin Trapping with DEPMPO
Generation Mediated by NQR as Measured by EPR Spin Trapping with DEPMPO
Complex I (NQR) is one of the major sources of oxygen free radical generation in mitochondria. To obtain direct evidence for  production mediated by NQR, we employed the EPR spin-trapping technique to measure
production mediated by NQR, we employed the EPR spin-trapping technique to measure  generation unambiguously. Of the available spin traps, DEPMPO is ideal for quantitating
generation unambiguously. Of the available spin traps, DEPMPO is ideal for quantitating  production by NQR based on the following advantages: (a) DEPMPO is 40-fold more sensitive than the cytochrome c assay for the detection of
production by NQR based on the following advantages: (a) DEPMPO is 40-fold more sensitive than the cytochrome c assay for the detection of  (26), (b) DEPMPO traps
(26), (b) DEPMPO traps  with an efficiency of 60–70% (26), and (c) the
with an efficiency of 60–70% (26), and (c) the  adduct of DEPMPO/·OOH is more stable than that of 5,5-dimethyl pyrroline N-oxide/·OOH (20).
adduct of DEPMPO/·OOH is more stable than that of 5,5-dimethyl pyrroline N-oxide/·OOH (20).
When isolated NQR (0.1 mg/ml) was incubated with DEPMPO (20 mm) in PBS, and the reaction was initiated by the addition of NADH (0.5 mm), a multiline EPR spectrum was produced that was characteristic of DEPMPO/·OOH (Fig. 1A, solid line) based on the hyperfine coupling constants (isomer 1: aN = 13.14 G, aHβ = 11.04 G, aHγ = 0.96 G, and aP = 49.96 G (80% relative concentration); isomer 2: aN = 13.18 G, aHβ = 12.59 G, aHγ = 3.46 G, and aP = 48.20 G (20% relative concentration) (9, 20)) obtained from computer simulation (Fig. 1A, dashed line). Trapping of NQR-mediated  by DEPMPO was thus highly specific and suitable for quantitative analysis.
by DEPMPO was thus highly specific and suitable for quantitative analysis.
FIGURE 1.
EPR spin trapping of  generated from NQR in the presence of DEPMPO. A, the computer simulation (dashed line) superimposed on the experimental spectrum (solid line) obtained using NQR (0.1 mg/ml), DEPMPO (20 mm), diethylenetriaminopentaacetic acid (1 mm), and NADH (0.5 mm) in PBS. The experimental spectrum was recorded after signal averaging 3 scans at room temperature. B, the same as A, except that rotenone (25 μm) was added to the mixture before the reaction was initiated by NADH. C, the same as A, except that ubiquinone-1 (Q1, 200 μm) was added to the mixture before the reaction was initiated by NADH. D, the same as C, except that rotenone (25 μm) was added to the mixture before the reaction was initiated by NADH. E, the same as A (or C) except that the NQR was omitted from the system. The spin quantitation for each spectrum was obtained by double integration of the simulation spectrum. The instrumental settings are described under “Experimental Procedures.” F, the same as C, except that DEPMPO was omitted from the system. G, the same as D, except that DEPMPO was omitted from the system. H, the same as G except that rotenone was replaced with piericidin A (250 nm) in the system. I, the EPR spectrum was recorded by the reaction mixture containing 2-(N,N-diethylamino)diazenolate 2-oxide (DEANO; 3 mm), Q1H2 (0.2 mm), and diethylenetriaminepentaacetic acid (1 mm) in 50 mm phosphate buffer, pH 7.4. The instrumental settings for F–I are: center field, 3525 G; sweep width, 20 G; power, 20 milliwatts; receiver gain, 2 × 105; modulation amplitude, 1 G; time of conversion, 163.84 ms; time constant, 327.68 ms; and number of scans, 5.
generated from NQR in the presence of DEPMPO. A, the computer simulation (dashed line) superimposed on the experimental spectrum (solid line) obtained using NQR (0.1 mg/ml), DEPMPO (20 mm), diethylenetriaminopentaacetic acid (1 mm), and NADH (0.5 mm) in PBS. The experimental spectrum was recorded after signal averaging 3 scans at room temperature. B, the same as A, except that rotenone (25 μm) was added to the mixture before the reaction was initiated by NADH. C, the same as A, except that ubiquinone-1 (Q1, 200 μm) was added to the mixture before the reaction was initiated by NADH. D, the same as C, except that rotenone (25 μm) was added to the mixture before the reaction was initiated by NADH. E, the same as A (or C) except that the NQR was omitted from the system. The spin quantitation for each spectrum was obtained by double integration of the simulation spectrum. The instrumental settings are described under “Experimental Procedures.” F, the same as C, except that DEPMPO was omitted from the system. G, the same as D, except that DEPMPO was omitted from the system. H, the same as G except that rotenone was replaced with piericidin A (250 nm) in the system. I, the EPR spectrum was recorded by the reaction mixture containing 2-(N,N-diethylamino)diazenolate 2-oxide (DEANO; 3 mm), Q1H2 (0.2 mm), and diethylenetriaminepentaacetic acid (1 mm) in 50 mm phosphate buffer, pH 7.4. The instrumental settings for F–I are: center field, 3525 G; sweep width, 20 G; power, 20 milliwatts; receiver gain, 2 × 105; modulation amplitude, 1 G; time of conversion, 163.84 ms; time constant, 327.68 ms; and number of scans, 5.
That the DEPMPO/·OOH adduct arose from the trapping of  was confirmed by the addition of manganese-superoxide dismutase (Mn-SOD, 200 units/ml) to the reaction system (data not shown); upon its addition, adduct formation was completely prevented. In the absence of NQR, no DEPMPO/·OOH adduct was detected (Fig. 1E), indicating the enzymatic dependence of the DEPMPO adduct formation. When native NQR was replaced with heat-denatured (70 °C for 5 min) NQR, the formation of DEPMPO/·OOH was inhibited (data not shown), suggesting that electron transfer activity in the enzyme is required for
was confirmed by the addition of manganese-superoxide dismutase (Mn-SOD, 200 units/ml) to the reaction system (data not shown); upon its addition, adduct formation was completely prevented. In the absence of NQR, no DEPMPO/·OOH adduct was detected (Fig. 1E), indicating the enzymatic dependence of the DEPMPO adduct formation. When native NQR was replaced with heat-denatured (70 °C for 5 min) NQR, the formation of DEPMPO/·OOH was inhibited (data not shown), suggesting that electron transfer activity in the enzyme is required for  generation. The amount of DEPMPO/·OOH greatly decreased when NADH was omitted from the system (data not shown), supporting the role of NADH as the direct electron donor for NQR-mediated
generation. The amount of DEPMPO/·OOH greatly decreased when NADH was omitted from the system (data not shown), supporting the role of NADH as the direct electron donor for NQR-mediated  production.
production.
The enzymatic activity of NQR is sensitive to rotenone, which is assumed to inhibit electron transfer activity through blocking the Q-binding site. Pretreatment of NQR with rotenone (25 μm) inhibited ∼80% of the electron transfer activity from NADH to Q1, but failed to inhibit the  production mediated by NQR (97.8 ± 2.2% superoxide generation activity (SGA) remained, Fig. 1B), implying that the flavin subcomplex (Fp) controls NQR-mediated
production mediated by NQR (97.8 ± 2.2% superoxide generation activity (SGA) remained, Fig. 1B), implying that the flavin subcomplex (Fp) controls NQR-mediated  production in the absence of Q1 (9).
production in the absence of Q1 (9).
In the presence of the electron acceptor Q1 (200 μm), the generation of  by NADH-energized NQR was enhanced nearly 2-fold (195.3 ± 9.7% SGA of control, Fig. 1C) under the same assay condition of EPR spin trapping, suggesting two possibilities: (a)
by NADH-energized NQR was enhanced nearly 2-fold (195.3 ± 9.7% SGA of control, Fig. 1C) under the same assay condition of EPR spin trapping, suggesting two possibilities: (a)  generation by NQR, under the conditions of enzyme turnover in the presence of NADH and Q1, is mediated mainly by the Fp subcomplex and ubisemiquinone and (b) ∼50% of the NQR-mediated
generation by NQR, under the conditions of enzyme turnover in the presence of NADH and Q1, is mediated mainly by the Fp subcomplex and ubisemiquinone and (b) ∼50% of the NQR-mediated  generation was derived from the source of ubisemiquinone-1 under the conditions of enzyme turnover; presumably the formation of ubisemiquinone-1 was caused by incomplete reduction of Q1, which is mediated by the Q-binding domain.
generation was derived from the source of ubisemiquinone-1 under the conditions of enzyme turnover; presumably the formation of ubisemiquinone-1 was caused by incomplete reduction of Q1, which is mediated by the Q-binding domain.
Rotenone is proposed as a specific inhibitor of NQR through blocking the Q-binding site of the enzyme. Pretreatment of NQR with rotenone (25 μm) significantly inhibited  generation by ∼35–45% (117.3 ± 10.2% SGA of control, Fig. 1D) under the conditions of enzyme turnover, thus confirming that the source of ubisemiquinone-1 mediated by the Q-binding site controlled most of the NQR-mediated
generation by ∼35–45% (117.3 ± 10.2% SGA of control, Fig. 1D) under the conditions of enzyme turnover, thus confirming that the source of ubisemiquinone-1 mediated by the Q-binding site controlled most of the NQR-mediated  production.
production.
An organic radical with a multiline spectrum was detected under the conditions of enzyme turnover (Fig. 1C, arrow). The EPR signal was subsequently enhanced in the presence of rotenone (Fig. 1D, arrow) or piericidin A (data not shown). Presumably, the detected organic radical was derived from incomplete reduction of Q1 and mediated by the ubiquinone-binding site of NQR (27). To test this hypothesis, the EPR experiments were conducted in the absence of spin trap, resulting in the detection of a nine-line spectrum during the steady state of enzyme turnover conditions at room temperature (Fig. 1F). The intensity of this EPR signal increased proportional to the dosage of Q1 (supplemental Fig. S1). The EPR signal was totally dependent on the presence of Complex I, NADH, and Q1 in the reaction system (data not shown) and was subsequently enhanced by rotenone (by 81.4 ± 12.7%, n = 3, Fig. 1G) or piericidin A (by 72.7 ± 6.3%, n = 3, Fig. 1H). To confirm that the detected spectrum was the Q1-derived ubisemiquinone (Q1·− or SQ1), we incubated ubiquinol-1 (Q1H2, 0.2 mm) with 2-(N,N-diethylamino)diazenolate 2-oxide (DEANO; NO donor, 3 mm, Fig. 1I) and obtained its EPR spectrum. The hyperfine splitting constants for the known spectrum of Q1·− based on the computer simulation are aH (3H) = 2.02 G and aH (2H) = 0.99 G (line width = 0.305 G).
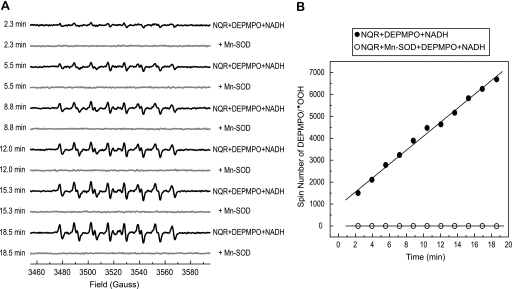
The formation of the DEPMPO/·OOH mediated by NQR was time-dependent and completely attenuated in the presence of Mn-SOD. As shown in Fig. 2A, the intensity of DEPMPO/·OOH gradually increased with time. Each spectrum obtained from the time course was simulated, and the spin quantitation of each simulated spectrum was obtained from double integration. The spin number of DEPMPO/·OOH was plotted versus time as indicated in Fig. 2B, showing the linear part of reaction of NQR-mediated  generation was occurred within 20 min.
generation was occurred within 20 min.
FIGURE 2.
Time dependence of DEPMPO/·OOH formation from the reaction of NQR, DEPMPO, and NADH. A, the components in the system were the same as those described in the legend of Fig. 1A without Mn-SOD or with Mn-SOD addition (1 unit/μl, spectra in gray). The instrumental settings were the same as those described under “Experimental Procedures” except the time of conversion was 81.92 ms and the number of scans was one. B, the spectra (in black) in A were simulated using the parameters of DEPMPO/·OOH. The spin number of each simulated spectrum was calculated based on the area obtained by double integration.
Generation of Antibodies against the Epitope of the Glutathione-binding Domains of NQR
In a previous study, we identified a specific tryptic peptide, 200GAGAYIC206GEETALIESIEGK219, that is involved in the GS-binding domain of the 51-kDa subunit of NQR (12). Cys206 is also identified as the site of specific protein radical formation resulting from oxygen free-radical attack (9). To gain insight into the functional role of the identified GS-binding domain, we generated a polyclonal antibody against this domain.
Based on the published three-dimensional structure of the hydrophilic domain of NQR from T. thermophilus (10), we identified a 20-residue peptide sequence (amino acids 200–219), which is highly conserved in the bovine protein, exhibiting a helix-turn-helix conformation (amino acids 208–217) as indicated in supplemental Fig. S2A. We have previously demonstrated that the design, synthesis, and immunological and structural characterization of such motifs can be achieved successfully (28). The 20-residue peptide pGSCA206 sequence in bovine protein, 200GAGAYIC206GEETALIESIEGK219, was designed as a B cell epitope (Table 1). The peptide of pMVFGSCA206 was synthesized as a chimeric construct incorporating a T-cell epitope consisting of an 18-residue promiscuous T-helper measles virus (MVF sequence: 288KLLSLIKGVIVHRLEGVE305), linked via a 4-residue linker (GPSL), and pGSCA206 B cell epitope on a Milligen/Biosearch 9600 solid-phase peptide synthesizer as described under “Experimental Procedures.” The crude peptide was purified to homogeneity by reverse phase high-performance liquid chromatography and fully characterized by matrix-assisted laser desorption ionization mass spectroscopy, which gave the exact mass unit [(M+H)+ 4359.41].
The other domain involved in GS binding was identified as the tryptic peptide 361VDSDTLC367TEEVFPTAGAGTDLR382 of the 75-kDa polypeptide of NQR. The predicted structure of this domain is a random coil-turn-helix as indicated in supplemental Fig. S2B. With the same strategy as above, we designed the B-cell epitope as 354LKDLLNKVDSDTLC367TEEVF372 (pGSCB367 and Table 1) and the chimeric construct with the MVF T cell epitope as KLLSLIKGVIVHRLEGVEGPSLLKDLLNKVDSDTLCTEEVF, or pMVFGSCB367 (Table 1). The purified pMVFGSCB367 was characterized with matrix-assisted laser desorption ionization mass spectroscopy, which gave the same mass [(M+H)+ 4519.61] as calculated [(M+H)+ 4519.49].
Four New Zealand white rabbits (6–8 weeks old) were immunized with the immunogens pMVFGSCA206 and pMVFGSCB367, respectively. Sera were collected, and antibodies were purified as described under “Experimental Procedures.” The antibodies thus generated are termed AbGSCA206 and AbGSCB367.
Immunological Specificity of Antibodies
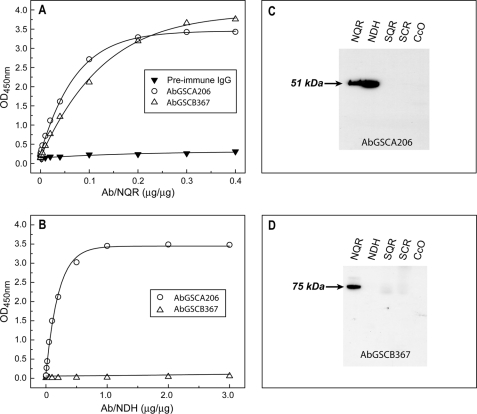
Fig. 3 shows the purified antibody response to the purified mitochondrial Complex I (NQR) and its flavin protein subcomplex (NDH). The immunological cross-reactivity of antibodies was analyzed by ELISA (Fig. 3, A and B) using NQR and its Fp subcomplex, NDH, as antigens. When each antigen, at a fixed protein concentration (see the figure legend), was titrated with various amounts of antibody preparation, the AbGSCA206 antibodies reacted at a high titer, with antigens containing the 51-kDa FMN-binding subunit, including NQR (Fig. 3A) and NDH (Fig. 3B). There was practically no binding detected between pre-immune IgG and the antigens used. The protein concentration of NQR used for ELISA was 11 times higher than that of NDH, because the amount of Fp subcomplex present in NQR was ∼8.3%.
FIGURE 3.
Immunological specificity of the antibodies AbGSCA206 and AbGSCB367 analyzed by ELISA and Western blotting. A and B, the antigens, 400 μg of NQR in A and 40 μg of NDH in B, were coated on the ELISA plate and then reacted with various amounts of purified AbGSCA208 (○), AbGSCB367 (△), and pre-immune IgG (▼). A goat anti-rabbit IgG-horseradish peroxidase conjugate was then added. The binding of AbGSCA206 and AbGSCB367 to the antigens was quantified by measuring the optical density at 450 nm of the diimine product resulting from two-electron oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) using a TMB Peroxidase ELISA Substrate Kit (Bio-Rad). C and D, SDS-PAGE was carried out according to the procedure described under “Experimental Procedures.” The isolated enzymes, NQR (40 μg), NDH (4 μg), SQR (40 μg), SCR (40 μg), and CcO (40 μg) were immunoblotted with AbGSCA206 in C and AbGSCB367 in D, respectively.
As indicated in Fig. 3, A and B, the AbGSCB367 antibodies react at a high titer with the antigen of NQR but not with the antigen of NDH due to the absence of the 75-kDa polypeptide in the Fp subcomplex of NQR. These results confirmed the immunological specificity of antibodies AbGSCA206 and AbGSCB367.
The immunological specificity of antibodies was further characterized by a Western blot (Fig. 3, C and D) using the mitochondrial electron transfer complex, including NQR, NDH, SQR (Complex II), QCR (Complex III), and CcO (Complex IV). As indicated in Fig. 3 (C and D), AbGSCA206 binds specifically to the 51-kDa subunit of NQR and NDH (Fig. 3C), whereas AbGSCB367 only binds specifically to the 75-kDa subunit of NQR (Fig. 3D). As expected, there was no binding observed to SQR, QCR, or CcO by either antibodies of AbGSCA206 or antibodies of AbGSCB367.
Immunoinhibition of NQR-mediated  Generation by AbGSCA206 and AbGSCB367
Generation by AbGSCA206 and AbGSCB367
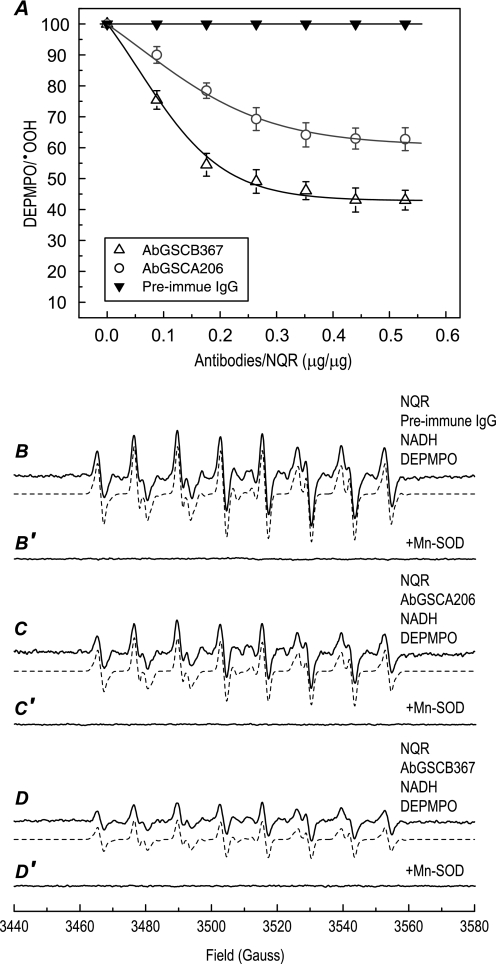
Fig. 4A (open circles) shows the effect of AbGSCA206 binding on the  generation activity of NQR. Mediation of
generation activity of NQR. Mediation of  by NQR was induced by NADH in the absence of Q1 and measured by EPR spin trapping with DEPMPO. The production of DEPMPO/·OOH adduct was mainly controlled by the Fp subcomplex or the hydrophilic domain of NQR under the above assay conditions. When the isolated NQR was incubated with various amounts of antibodies, the
by NQR was induced by NADH in the absence of Q1 and measured by EPR spin trapping with DEPMPO. The production of DEPMPO/·OOH adduct was mainly controlled by the Fp subcomplex or the hydrophilic domain of NQR under the above assay conditions. When the isolated NQR was incubated with various amounts of antibodies, the  generation activity of NQR was decreased as the amounts of antibodies were increased. A maximum inhibition of ∼37% was observed with 353 μg of antibody per mg of NQR (Figs. 4A, 4B, 4C, and 6), indicating that the binding of AbGSCA206 with the epitope that is involved in the GS-binding domain (amino acids 200–219) of the 51-kDa subunit marginally decreased the electron leakage mediated by the Fp subcomplex or hydrophilic domain. The binding of the antibody AbGSCA206 did not affect the electron transfer activity from NADH to Q1 catalyzed by NQR, indicating that the identified GS-binding domain from the 51-kDa subunit is not essential for the catalytic function of NQR.
generation activity of NQR was decreased as the amounts of antibodies were increased. A maximum inhibition of ∼37% was observed with 353 μg of antibody per mg of NQR (Figs. 4A, 4B, 4C, and 6), indicating that the binding of AbGSCA206 with the epitope that is involved in the GS-binding domain (amino acids 200–219) of the 51-kDa subunit marginally decreased the electron leakage mediated by the Fp subcomplex or hydrophilic domain. The binding of the antibody AbGSCA206 did not affect the electron transfer activity from NADH to Q1 catalyzed by NQR, indicating that the identified GS-binding domain from the 51-kDa subunit is not essential for the catalytic function of NQR.
FIGURE 4.
Immunoinhibition of NQR-mediated  generation activity by AbGSCA206 and AbGSCB367 as assayed by EPR spin-trapping with DEPMPO. A, titration of NQR-mediated
generation activity by AbGSCA206 and AbGSCB367 as assayed by EPR spin-trapping with DEPMPO. A, titration of NQR-mediated  generation by antibodies of AbGSCA206 (○), AbGSCB367 (△), and pre-immune IgG (▼). Isolated NQR (0.91 mg/ml) was incubated with various amounts of antibodies at 4 °C for 12 h. The
generation by antibodies of AbGSCA206 (○), AbGSCB367 (△), and pre-immune IgG (▼). Isolated NQR (0.91 mg/ml) was incubated with various amounts of antibodies at 4 °C for 12 h. The  generation mediated by NQR was initiated by NADH (0.5 mm) in the presence of DEPMPO (20 mm), and subsequently measured by EPR as described under “Experimental Procedures.” B–D, the computer simulation (dashed line) superimposed on the experimental EPR spectra (solid line) obtained from the reaction systems containing NQR (0.91 mg/ml), antibodies (0.36 mg/ml, pre-immune IgG in B, AbGSCA206 in C, and AbGSCB367 in D), diethylenetriaminepentaacetic acid (1 mm), DEPMPO (20 mm), and NADH (0.5 mm). B′, C′, and D′, same as B, C, and D except that Mn-SOD (1 unit/μl) was present in the reaction mixture.
generation mediated by NQR was initiated by NADH (0.5 mm) in the presence of DEPMPO (20 mm), and subsequently measured by EPR as described under “Experimental Procedures.” B–D, the computer simulation (dashed line) superimposed on the experimental EPR spectra (solid line) obtained from the reaction systems containing NQR (0.91 mg/ml), antibodies (0.36 mg/ml, pre-immune IgG in B, AbGSCA206 in C, and AbGSCB367 in D), diethylenetriaminepentaacetic acid (1 mm), DEPMPO (20 mm), and NADH (0.5 mm). B′, C′, and D′, same as B, C, and D except that Mn-SOD (1 unit/μl) was present in the reaction mixture.
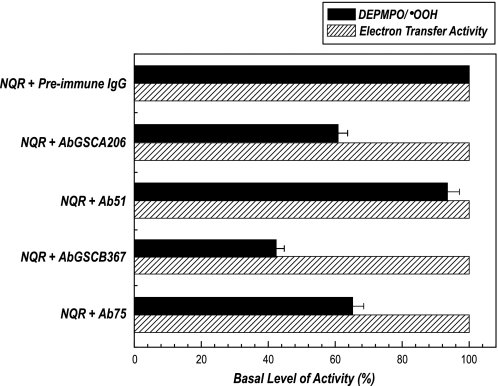
FIGURE 6.
Effect of antibodies Ab51, AbGSCA206, Ab75, and AbGSCB367 on the  generation and electron transfer activity mediated by NQR. Isolated NQR (0.91 mg/ml) was incubated with preimmune IgG or Ab51 or AbGSCA206 or Ab75 or AbGSCB367 (0.36 mg/ml) at 4 °C for 12 h. The
generation and electron transfer activity mediated by NQR. Isolated NQR (0.91 mg/ml) was incubated with preimmune IgG or Ab51 or AbGSCA206 or Ab75 or AbGSCB367 (0.36 mg/ml) at 4 °C for 12 h. The  generation activity (black bars) was analyzed by EPR spin trapping with DEPMPO in the absence of Q1, and the electron transfer activity (dashed bars) of Q1-stimulated NADH oxidation was assayed at room temperature. The spin quantitation for each spectrum was obtained by double integration of the simulation spectrum. The instrumental settings are described under “Experimental Procedures.”
generation activity (black bars) was analyzed by EPR spin trapping with DEPMPO in the absence of Q1, and the electron transfer activity (dashed bars) of Q1-stimulated NADH oxidation was assayed at room temperature. The spin quantitation for each spectrum was obtained by double integration of the simulation spectrum. The instrumental settings are described under “Experimental Procedures.”
To test the role of the GS-binding domain from the 75-kDa subunit in the NQR-derived SGA, a fixed amount of isolated NQR was incubated with various amounts of AbGSCB367, and the  generation mediated by NQR was assayed by EPR spin trapping with DEPMPO. As indicated in Fig. 4A (open triangle), production of DEPMPO/·OOH decreased as the dosage of AbGSCB367 increased. A maximum reduction of 57% in the
generation mediated by NQR was assayed by EPR spin trapping with DEPMPO. As indicated in Fig. 4A (open triangle), production of DEPMPO/·OOH decreased as the dosage of AbGSCB367 increased. A maximum reduction of 57% in the  generation was observed with 440 μg of AbGSCB367 per mg of NQR (Figs. 4A, 4B, 4D, and 6), suggesting that the binding of AbGSCB367 with the epitope involved in the GS-binding domain of the 75-kDa subunit significantly decreased the electron leakage for
generation was observed with 440 μg of AbGSCB367 per mg of NQR (Figs. 4A, 4B, 4D, and 6), suggesting that the binding of AbGSCB367 with the epitope involved in the GS-binding domain of the 75-kDa subunit significantly decreased the electron leakage for  production mediated by the hydrophilic domain of NQR.
production mediated by the hydrophilic domain of NQR.
Note that the formation of DEPMPO/·OOH mediated by NQR+AbGSCA206 or NQR+AbGSCB367 is time-dependent and completely attenuated in the presence of Mn-SOD as indicated in the supplemental Fig. S3 and Fig. 4 (C′ and D′). The signal intensity at the specific time point adopted (8.4 min in the data of Fig. 3) to quantify  production is thus within the linear part of the reaction.
production is thus within the linear part of the reaction.
Generation of Antibodies against the 51-kDa FMN-binding and 75-kDa Subunits of the NQR
The use of pre-immune IgG has provided a good experimental control for the study. However, pre-immune IgG does not bind to NQR. To provide an additional experimental control, we have designed B-cell epitopes of the 51- and 75-kDa subunits, respectively, which are 21SGDTTAPKKTSFGSLKDFDR40 (p51 in Table 1), corresponding to the N-terminal amino acid residues 1–20 of the 51-kDa mature protein, and 100WNILTNSEKTKKAREGVMEFL120 (p75 in Table 1), corresponding to amino acid residues 77–97 of the 75-kDa mature protein. The predicted structure of p51 contains a major β-sheet conformation (residues 22GDTTAPKK29). The structure of p75 contains a major α-helix conformation (supplemental Fig. S2C). The chimeric constructs of the fusion peptides used as immunogens were KLLSLIKGVIVHRLEGVEGPSLSGDTTAPKKTSFGSLKDEDR (mass unit = 4479.61, or pMVF51 in Table 1) and KLLSLIKGVIVHRLEGVEGPSLWNILTNSEKTKKAREGVMEFL (mass unit = 4834.79, or pMVF75 in Table 1). The polyclonal antibodies thus generated were named Ab51 and Ab75.
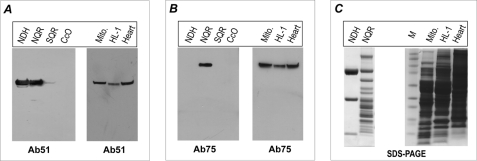
The immunological specificities of Ab51 and Ab75 were characterized by an ELISA (supplemental Fig. S4) using NQR and Western blot (Figs. 5A and 5B) using the mitochondrial electron transfer complex, including NQR, NDH, SQR, and CcO. As indicated in Figs. 5A and 5B, Ab51 binds specifically to the 51-kDa subunit of NQR and NDH (Fig. 5A), whereas Ab75 binds specifically only to the 75-kDa subunit of NQR (Fig. 5B, left-side panel). As expected, there is no binding observed to SQR or CcO by either antibodies of Ab51 or antibodies of Ab75. The antibodies were further used to probe the 51- and 75-kDa subunits in isolated mitochondria of the rat heart, rat myocardial tissues, and the mouse cardiac myocyte cell line HL-1, where the antibodies bind specifically to the 51- and 75-kDa subunits of NQR, as shown in Fig. 5 (A and B, right-side panel). Taken together, these results demonstrated that the designed antibodies against B-cell epitopes, p51 and p75, are highly specific with high sensitivity, thus suitable for in vivo, ex vivo, or in vitro studies.
FIGURE 5.
Immunological specificity of the antibody Ab51 and Ab75 analyzed by immunoblotting. The antigens, 4 μg of NDH, 40 μg of NQR, 40 μg of SQR, 40 μg of CcO, and isolated mitochondria (Mito., 100 μg) from rat heart, cardiac myocytes (HL-1, 100 μg), and tissue homogenates of rat heart (Heart, 100 μg), were probed with antibodies Ab51 in A and Ab75 in B. C, SDS-PAGE with Coomassie Blue staining of NDH (8 μg), NQR (30 μg), isolated mitochondria (100 μg), HL-1 lysate (100 μg), and rat heart tissue homogenates (100 μg). M represents a molecular weight marker.
Immunoinhibition of NQR-mediated  Generation by Ab75 but Not by Ab51
Generation by Ab75 but Not by Ab51
The effect of antibodies binding to NQR on the enzymatic functions was determined. Binding of Ab51 or Ab75 to NQR did not significantly alter the electron transfer activity of NQR (Fig. 6). However, binding of Ab51 to NQR slightly reduced the mediation of  generation (by ∼7%, versus ∼37% reduction by AbGSCA206 binding) as shown by EPR spin trapping in the absence of Q1. Binding of Ab75 to NQR inhibited
generation (by ∼7%, versus ∼37% reduction by AbGSCA206 binding) as shown by EPR spin trapping in the absence of Q1. Binding of Ab75 to NQR inhibited  generation by ∼35% (versus ∼57% inhibition by AbGSCB367 binding) as indicated in Fig. 6. The formation of DEPMPO/·OOH mediated by NQR+Ab75 is time-dependent and completely inhibited by Mn-SOD (supplemental Fig. S3). The signal intensity at the specific time point to quantify
generation by ∼35% (versus ∼57% inhibition by AbGSCB367 binding) as indicated in Fig. 6. The formation of DEPMPO/·OOH mediated by NQR+Ab75 is time-dependent and completely inhibited by Mn-SOD (supplemental Fig. S3). The signal intensity at the specific time point to quantify  production is thus within the linear part of the reaction.
production is thus within the linear part of the reaction.
Protein S-glutathionylation of the NQR 51-kDa FMN Binding Subunit and 75-kDa Subunit in the Post-ischemic Heart
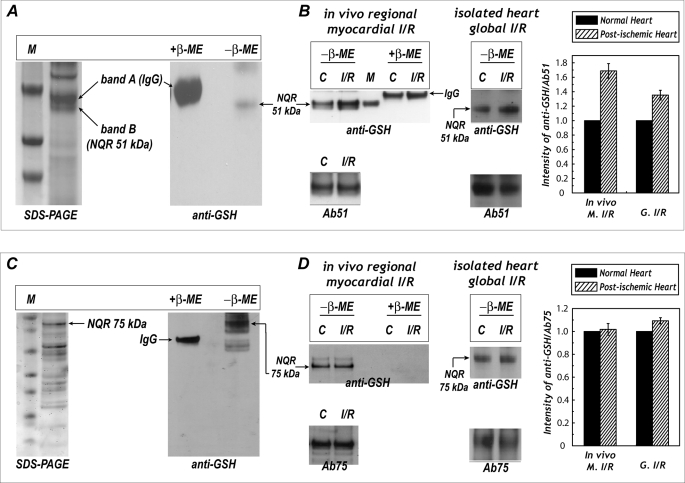
To test whether protein S-glutathionylation of NQR 51- and 75-kDa subunits is biologically and physiologically relevant, antibodies of Ab51 and Ab75 were used to immunoprecipitate the 51- and 75-kDa proteins from rat heart mitochondria, followed by immunoblotting with a monoclonal antibody against GSH. As indicated in Fig. 7 (A and C), we detected a signal of S-glutathionylation on the 51- and 75-kDa subunits (right-side panels) of NQR, and the signals of protein S-glutathionylation were subsequently eliminated in the presence of β-ME. In the SDS-PAGE of Fig. 7A (left-side panel), two major protein bands (band A and band B) with a molecular mass near 50 kDa were examined with LC/MS/MS. With this technique, 17.1% of the amino acid sequence of the rat heart NQR-51-kDa subunit was unambiguously identified on the gel band B of the lower side (supplemental Fig. S5). As shown in the table of supplemental Fig. S5, MS/MS spectra of tryptic peptides from the NQR 51-kDa subunit of the rat heart included 41IFTNLYGR48 (m/z = 492.522+), 112YLVVNADEGEPGTCK126 (m/z = 826.242+), 112YLVVNADEGEPGTCKDR128 (m/z = 641.793+), 175EAYEAGLIGK184 (m/z = 525.702+), 200GAGAYICGEETALIESIEGK219 (m/z = 1034.762+), 406PAEIDSLWEISK417 (m/z = 694.652+), and 441HFRPELEDR449 (m/z = 400.502+). The gel band A of the upper side was confirmed to be immunoglobulin G (IgG) as verified by LC/MS/MS analysis (data not shown).
FIGURE 7.
Protein S-glutathionylation of the 51-kDa subunit of NQR in myocardial tissue homogenates and the effect of post-ischemic injury. A, mitochondrial preparation (1 mg) from rat heart was subjected to immunoprecipitation with Ab51 antibody and subsequently subjected to SDS-PAGE (left-side panel) and immunoblotting with anti-GSH monoclonal antibody (right-side panel). M denotes a molecular weight marker standard. B, myocardial tissue homogenates (300 μg) were obtained from the infarction region (left-side panel, in vivo regional I/R rat heart) or isolated heart subjected to global I/R injury (right-side panel). Tissue homogenates were immunoprecipitated with Ab51 and then probed with anti-GSH antibody and Ab51. M denotes the molecular weight marker obtained from S-glutathionylated NDH at the 51-kDa FMN-binding subunit (12), C denotes non-ischemic tissue (left-side panel) or normal control rat heart (right-side panel). C, Ab75 was immobilized to Affi-Gel 10 in accordance with the product literature. Mitochondrial preparation (1 mg) from rat heart was subjected to immunoprecipitation with immobilized Ab75. Proteins obtained from immunoprecipitation were subjected to SDS-PAGE (left-side panel) and probed with anti-GSH monoclonal antibody (right-side panel). D, same as B, except that Ab75 was used for immunoprecipitation. The density ratio of the blotting signals between anti-GSH and Ab51 (in B) or Ab75 (in D) was quantitated by using ImageJ.
Likewise, the protein band with a molecular mass near 77 kDa (the calculated molecular mass of mature rat 75 kDa is 76860.16) in the SDS-PAGE of Fig. 7C (left-side panel) was further subjected to proteolytic digestion with trypsin, and examined by LC/MS/MS. We identified 33% of the amino acid sequence of the 75-kDa subunit (supplemental Fig. S6). MS/MS spectra of tryptic peptides from the NQR-75-kDa subunit of the rat heart revealed twenty different molecular ions as shown in the table of supplemental Fig. S6.
Immobilized antibodies of Ab51 and Ab75 were further used to immunoprecipitate the NQR 51- and 75-kDa polypeptides from tissue homogenates of the post-ischemic heart, followed by immunoblotting with anti-GSH monoclonal antibody. The detected NQR-derived S-glutathionylation on the 51-kDa subunit was enhanced by 60.8 ± 5.7% (in vivo occlusion/reperfusion model, n = 5) and 35.3 ± 2.7% (isolated heart global I/R model, n = 5) in the post-ischemic heart (Fig. 7B). However, S-glutathionylation of the NQR 75-kDa subunit was not significantly increased (Fig. 7D).
DISCUSSION
Superoxide Generation Mediated by NQR
In the current investigation, we have provided direct evidence for NQR-mediated  generation using EPR spin trapping. We have demonstrated, with a combination of EPR spin trapping and immunochemistry, the catalytic role of the 51-kDa subunit, the 75-kDa subunit, and one of their GS-binding domains in the
generation using EPR spin trapping. We have demonstrated, with a combination of EPR spin trapping and immunochemistry, the catalytic role of the 51-kDa subunit, the 75-kDa subunit, and one of their GS-binding domains in the  generation activity of NQR.
generation activity of NQR.
In the absence of ubiquinone (Q1 in this study), we hypothesized that NQR mediates  production mainly through the hydrophilic domain of the enzyme, and this hypothesis was strongly supported by the failure of rotenone (or piericidin A) to inhibit
production mainly through the hydrophilic domain of the enzyme, and this hypothesis was strongly supported by the failure of rotenone (or piericidin A) to inhibit  generation (Fig. 1B). One major site of
generation (Fig. 1B). One major site of  generation is widely recognized to be the cofactor FMN near the site of NADH oxidation on the 51-kDa subunit (9, 29–31). Here
generation is widely recognized to be the cofactor FMN near the site of NADH oxidation on the 51-kDa subunit (9, 29–31). Here  generation mediated by NQR is likely determined by a bimolecular reaction of O2 and FMNH2 or the formation of the FMNH· intermediate.
generation mediated by NQR is likely determined by a bimolecular reaction of O2 and FMNH2 or the formation of the FMNH· intermediate.
The second site associated with NQR-mediated  generation has been proposed to be the site of ubiquinone reduction, presumably due to the formation of an unstable semiquinone radical that is the source of
generation has been proposed to be the site of ubiquinone reduction, presumably due to the formation of an unstable semiquinone radical that is the source of  . Under conditions of enzyme turnover in the presence of Q1, NQR-mediated
. Under conditions of enzyme turnover in the presence of Q1, NQR-mediated  generation was enhanced 2-fold as shown by EPR spin trapping with DEPMPO (Fig. 1C). More than 80% of the enhanced
generation was enhanced 2-fold as shown by EPR spin trapping with DEPMPO (Fig. 1C). More than 80% of the enhanced  generation was inhibited by rotenone, thus supporting (but not necessarily proving) an
generation was inhibited by rotenone, thus supporting (but not necessarily proving) an  generation mechanism involving the reduction of ubiquinone to an unstable semiquinone. The formation of an unstable semiquinone under enzyme turnover conditions is likely mediated through the ubiquinone-binding domain of Complex I (27). In support of this mechanism, we have detected the formation of a stable SQ1 under enzyme turnover conditions (or steady state) in the absence of spin trap; the signal of SQ1 was enhanced by rotenone or piericidin A. Presumably, binding of rotenone or piericidin A to the Q-binding site of NQR decreases the formation of the unstable semiquinone and subsequently increases the formation of stable SQ1 at steady state, thus reducing
generation mechanism involving the reduction of ubiquinone to an unstable semiquinone. The formation of an unstable semiquinone under enzyme turnover conditions is likely mediated through the ubiquinone-binding domain of Complex I (27). In support of this mechanism, we have detected the formation of a stable SQ1 under enzyme turnover conditions (or steady state) in the absence of spin trap; the signal of SQ1 was enhanced by rotenone or piericidin A. Presumably, binding of rotenone or piericidin A to the Q-binding site of NQR decreases the formation of the unstable semiquinone and subsequently increases the formation of stable SQ1 at steady state, thus reducing  generation mediated by the Q-binding site of NQR.
generation mediated by the Q-binding site of NQR.
Ohnishi et al. (32) have reported the presence of three distinct semiquinone species with different spin relaxation times in Complex I in situ based on a study using rat heart SMP. The SQ1 detected during steady state at room temperature can be logically assigned to be SQNs (slow relaxing) and SQNx (very slow relaxing), because the signal was not quenched by rotenone or piericidin A. The unstable semiquinone as a source of  is likely to be SQNf (fast relaxing), which is highly sensitive to rotenone or piericidin A (32).
is likely to be SQNf (fast relaxing), which is highly sensitive to rotenone or piericidin A (32).
Immunoinhibition of  Generation by Antibodies against the 51-kDa Subunit
Generation by Antibodies against the 51-kDa Subunit
Binding of AbGSCA206 to intact NQR inhibited SGA by ∼37% but did not affect the electron transfer activity (ETA) of the enzyme complex. This result supports the concept of Cys206 as the redox-sensitive thiol (9, 10). The redox GS-binding domain hosting Cys206 is highly conserved among the enzymes from bacteria, fungi, and mammals. Therefore, the redox domain of the NQR 51-kDa polypeptide is likely responsible for modulating electron leakage for  production. An x-ray crystal structure of the hydrophilic domain of respiratory Complex I from T. thermophilus indicates that this conserved cysteine (Cys182 in T. thermophilus) is only 6 Å from the FMN. FMN serves as a source of
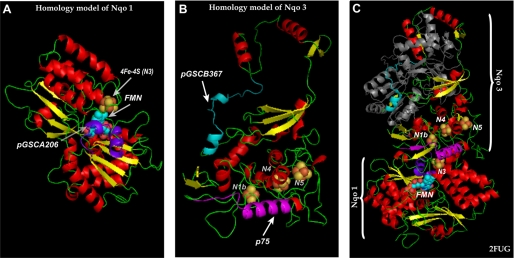
production. An x-ray crystal structure of the hydrophilic domain of respiratory Complex I from T. thermophilus indicates that this conserved cysteine (Cys182 in T. thermophilus) is only 6 Å from the FMN. FMN serves as a source of  , thus the binding of AbGSCA206 likely triggers a conformational change in this domain and decreases the electron leakage from FMNH2. The homology model (amino acids 38–444 in bovine protein precursor and shown in the Fig. 8A) is built via MODELLER ver. 8 (33), using the structure of hydrophilic domain of respiratory Complex I of T. thermophilus (2FUG) as template with a sequence identity of 46%. Binding of AbGSCA206 clearly induced a shielding effect on FMN, thus preventing molecular oxygen from accessing FMN and electron leakage for
, thus the binding of AbGSCA206 likely triggers a conformational change in this domain and decreases the electron leakage from FMNH2. The homology model (amino acids 38–444 in bovine protein precursor and shown in the Fig. 8A) is built via MODELLER ver. 8 (33), using the structure of hydrophilic domain of respiratory Complex I of T. thermophilus (2FUG) as template with a sequence identity of 46%. Binding of AbGSCA206 clearly induced a shielding effect on FMN, thus preventing molecular oxygen from accessing FMN and electron leakage for  production.
production.
FIGURE 8.
Three-dimensional structure indicating the locations of pGSCA206 in the NQR 51-kDa subunit and pGSCB367/p75 in the NQR 75-kDa subunit. A, homolog structure (amino acids 38–444 in bovine protein) of the 51-kDa subunit using the crystal structure of T. thermophilus (2FUG) as a template. An arrow shows the domain of pGSCA206 with a helix-turn-helix conformation, denoted by the purple ribbon. B, homolog structure (amino acids 31–408 in bovine protein) of the 75-kDa subunit. Arrows point to the domains of pGSCB367 (random coil turn helix) and p75 (α-helix), denoted by cyan and magenta ribbons, respectively. C, the folds of 51-kDa (Nqo1) and 75-kDa (Nqo3) subunits from the structure of hydrophilic domain of respiratory Complex I of T. thermophilus (2FUG). Fe-S centers are shown as red spheres for iron atoms and yellow spheres for sulfur atoms with clusters named in white. FMN is shown in cyan spheres. The domains of pGSCA206 (in A), pGSCB367 (in B), and p75 (in B) based on sequence alignment are shown in purple, cyan, and magenta ribbons. The gray ribbon in C denotes amino acid residues 403–767 from Nqo3; the conservation of this partial sequence is too low in mammalian protein to be built in the homolog model (in B).
The B-cell epitope of Ab51 antibody is located on the 20 amino acid residues at the N terminus of the NQR 51-kDa subunit. The sequence is not conserved (15% sequence similarity) in Complex I of T. thermophilus. However, the binding of Ab51 had little or no effect on the electron transfer or  generation activities of NQR, indicating that the N-terminal domain of the 51-kDa subunit is not essential for the catalytic functions of NQR.
generation activities of NQR, indicating that the N-terminal domain of the 51-kDa subunit is not essential for the catalytic functions of NQR.
Immunoinhibition of  Generation by Antibodies against the 75 kDa Subunit
Generation by Antibodies against the 75 kDa Subunit
Binding of AbGSCB367 to NQR inhibited  generation by up to 57% under the assay conditions without Q1. Furthermore, binding the Ab75 antibody to NQR resulted in the inhibition of
generation by up to 57% under the assay conditions without Q1. Furthermore, binding the Ab75 antibody to NQR resulted in the inhibition of  generation by 35%. These results suggest that both glutathionylated and non-glutathionylated domains of the 75-kDa subunit can regulate electron leakage for
generation by 35%. These results suggest that both glutathionylated and non-glutathionylated domains of the 75-kDa subunit can regulate electron leakage for  production by the hydrophilic domain of NQR, which may further imply an essential role for the 75-kDa subunit in the catalytic function of NQR-mediated
production by the hydrophilic domain of NQR, which may further imply an essential role for the 75-kDa subunit in the catalytic function of NQR-mediated  generation.
generation.
The homology model (amino acids 31–408 in bovine protein precursor, indicated in the color ribbon of Fig. 8B) is built using the structure of T. thermophilus Complex I (2FUG) as a template with 36.2% identity and 42.5% homology. The B-cell epitope (100WNILTNSEKTKKAREGVMEFL120) of Ab75 shares 42.8% sequence homology (91MVVDTLSDVVREAQAGMVEFT111 in T. thermophilus). The predicted conformation of p75 based on the homology model is dominated by an α-helix (magenta helix denoted by p75 in Fig. 8B). A similar structure with a β-sheet-α-turn-helix (denoted by magenta color in Fig. 8C) is observed in T. thermophilus. Together with the x-ray structure, the homology model is likely to logically explain the prevention of electron leakage for  generation by Ab75. The nearest distances between the p75 major α-helix and the iron-sulfur clusters are 8.4 Å (to N1b), 10.1Å (to N5), and 10.7 Å (to N4) based on the x-ray structure of bacterial protein (Fig. 8C). Therefore, three mechanisms are likely involved as follows. (i) Binding of Ab75 is likely to stabilize the conformation of the protein matrix surrounding the iron-sulfur clusters, thus enhancing electron transfer efficiency. (ii) The second possible mechanism involved is that Ab75 binding may protect the protein matrix from molecular oxygens accessing the iron-sulfur clusters, including N1b, N4, and N5 in bovine protein, therefore minimizing the electron leakage from iron-sulfur clusters. The above mechanism is proposed based on the hypothesis that electron leakage can occur at the iron-sulfur clusters (34). (iii) Ab75 binding could have induced the conformational change of flavin subcomplexes of NQR and decreased electron leakage from FMN.
generation by Ab75. The nearest distances between the p75 major α-helix and the iron-sulfur clusters are 8.4 Å (to N1b), 10.1Å (to N5), and 10.7 Å (to N4) based on the x-ray structure of bacterial protein (Fig. 8C). Therefore, three mechanisms are likely involved as follows. (i) Binding of Ab75 is likely to stabilize the conformation of the protein matrix surrounding the iron-sulfur clusters, thus enhancing electron transfer efficiency. (ii) The second possible mechanism involved is that Ab75 binding may protect the protein matrix from molecular oxygens accessing the iron-sulfur clusters, including N1b, N4, and N5 in bovine protein, therefore minimizing the electron leakage from iron-sulfur clusters. The above mechanism is proposed based on the hypothesis that electron leakage can occur at the iron-sulfur clusters (34). (iii) Ab75 binding could have induced the conformational change of flavin subcomplexes of NQR and decreased electron leakage from FMN.
Based on the homology structure (Fig. 8B), the domain of pGSCB367 is surface-exposed, and its conformation is mostly conserved in the Nqo3 of T. thermophilus except for an extra very short helix present in the homology structure of bovine protein (compare the cyan ribbon between Fig. 8B and C). The closest distance to the iron-sulfur clusters is relatively long, ∼23 Å (from the second helix to N1b). Binding of AbGSCB367 to the 75-kDa subunit of NQR likely triggers long range conformational changes in the 75-kDa subunit to decrease the approach of O2. Overall, the significant inhibitory effect of AbGSCB367 binding highly supports the notion that the GS-binding domain of NQR is closely related to the regulation of enzyme-mediated  generation (12).
generation (12).
Protein S-Glutathionylation of Complex I
Protein S-glutathionylation of Complex I has been demonstrated in the systems of purified enzyme (4, 12) and isolated mitochondria (5, 11). In most cases, the detected S-glutathionylation of the complex was induced by GSSG (4, 12) or by oxidative stress in vitro (5, 11). Intrinsic S-glutathionylation has been detected in the isolated mitochondrial Complex II (8). However, intrinsic S-glutathionylation has not been detected in the isolated Complex I; presumably GS binding was progressively dissociated from Complex I during purification. In this investigation, we have, for the first time, detected the intrinsic S-glutathionylation of Complex I from myocardial tissue homogenates and isolated mitochondria through the use of two peptide-based antibodies, Ab51 and Ab75. These results provide direct evidence to support the physiological relevance of Complex I-derived S-glutathionylation.
Intrinsic GSH binding to Complex I likely plays the role of an antioxidant defense under normal physiological conditions, because S-glutathionylation of Complex I may reduce  generation via the mechanisms of (i) reduction of O2 accessibility to FMN as seen in the case of AbGSCA206 binding to NQR and (ii) induction of conformational change in NQR to decrease electron leakage as seen in the case of AbGSCB367 binding to NQR.
generation via the mechanisms of (i) reduction of O2 accessibility to FMN as seen in the case of AbGSCA206 binding to NQR and (ii) induction of conformational change in NQR to decrease electron leakage as seen in the case of AbGSCB367 binding to NQR.
Post-ischemic reperfusion resulted in a significant enhancement of S-glutathionylation at the 51-kDa polypeptide in the rat heart (Fig. 7B). Enhancing oxidation of GSH to GSSG resulting from increasing oxidative stress during myocardial infarction is likely one of the causative factors. Quantitative determination of GSH and GSSG levels in the mitochondrial preparation was determined by enzymatic recycling method (36), indicating elevation of GSSG level (from 0.33 ± 0.03 nmol/mg of protein to 0.69 ± 0.11 nmol/mg of protein, n = 6) and reduction of GSH/GSSG ratio (from 8.9 ± 0.9 to 3.6 ± 0.6, n = 6) in the post-ischemic myocardium (data not shown). Reduction of GSH/GSSG ratio (from 36.0 ± 2.1 to 25.7 ± 3.2, n = 3) was also detected in the post-ischemic myocardium, but significant higher GSH/GSSG ratio is normally observed in the cytosol of myocardium. Therefore, the difference is likely due to (i) the concentration gradient between the cytosol and the mitochondria or (ii) a significant amount of oxidant stress had occurred during mitochondrial preparation. It is worth noting that depletion of GSH level in myocardium is normally occurred during myocardial infarction (decreased from 1.91 ± 0.15 nmol/mg of non-ischemic tissue to 1.37 ± 0.13 nmol/mg of post-ischemic tissue, n = 3), thus leading to a lower GSH/GSSG ratio. Accumulation of GSSG in mitochondria is unlikely unless re-reduction of GSSG is inhibited. More likely, NQR-mediated generation of  in the presence of GSH will produce other forms of more reactive glutathionylating species such as GS-thiyl radical, GS-OH (35), or even the NQR-derived protein thiyl radical (9), and these glutathionylating species may enhance S-glutathionylation at the 51-kDa subunit during myocardial infarction. In light of this mechanism, our recent research progress shows that the formation protein thiyl radical and GS-thiyl radical intermediates are involved in the mechanism of protein S-glutathionylation of NQR.4
in the presence of GSH will produce other forms of more reactive glutathionylating species such as GS-thiyl radical, GS-OH (35), or even the NQR-derived protein thiyl radical (9), and these glutathionylating species may enhance S-glutathionylation at the 51-kDa subunit during myocardial infarction. In light of this mechanism, our recent research progress shows that the formation protein thiyl radical and GS-thiyl radical intermediates are involved in the mechanism of protein S-glutathionylation of NQR.4
However, at the 75-kDa polypeptide in the rat heart, S-glutathionylation was not significantly increased by post-ischemic infarction (Fig. 7D). These results imply that the 51-kDa subunit of Complex I is more sensitive to oxidative stress caused by myocardial infarction. Analysis of the ETA of Complex I in situ from tissue homogenates indicated a significant impairment of NQR activity in the rat heart after ischemia-reperfusion (50.8 ± 2.6% of ETA remained in the infarct region and 71.5 ± 6.2% of ETA remained in the isolated heart subjected to global ischemia and reperfusion). Exposure of mitochondria to tert-butyl hydroperoxide or excess NO donor resulted in increased NQR-SSG and decreased ETA (5). In mitochondria, incubation with the thiol oxidant diamide glutathionylated only the 75-kDa subunit of Complex I and resulted in damage to its ETA, but reversal of glutathionylation did not restore NQR activity (11), suggesting a non-essential role for glutathionylation in diamide-induced oxidative injury to ETA. In contrast, in vitro S-glutathionylation of NQR with a low dosage of GSSG (1–3 mm) led to both 51- and 75-kDa S-glutathionylation, marginal enhancement of the electron transfer efficiency, and a decrease in the electron leakage by NQR (12). Therefore, glutathionylation of NQR in vivo is likely important in preventing oxidative damage to the enzyme through recycling back to free thiols the thiyl radicals and sulfenic acids formed on the cysteine residues, as suggested by Hurd and coworkers (11).
CONCLUSION
The new information resulting from this study includes delineation of mechanism of  production by NQR, demonstration of immunochemical approach to decrease enzyme-mediated
production by NQR, demonstration of immunochemical approach to decrease enzyme-mediated  generation, identification of evidence of NQR-derived intrinsic S-glutathionylation in vivo, and work showing how this redox modification is altered by myocardial infarction. The use of EPR spin trapping facilitates direct measurement of NQR-mediated
generation, identification of evidence of NQR-derived intrinsic S-glutathionylation in vivo, and work showing how this redox modification is altered by myocardial infarction. The use of EPR spin trapping facilitates direct measurement of NQR-mediated  generation. With this technique, we demonstrated that both FMN- and ubiquinone-binding moieties were involved in enzyme-mediated
generation. With this technique, we demonstrated that both FMN- and ubiquinone-binding moieties were involved in enzyme-mediated  production under conditions of enzyme turnover. With the use of our unique peptide-based antibodies that block
production under conditions of enzyme turnover. With the use of our unique peptide-based antibodies that block  generation, we have established that two of the glutathione-binding domains and one non-glutathionylated domain are involved in
generation, we have established that two of the glutathione-binding domains and one non-glutathionylated domain are involved in  production by NQR. The approach used in this study of engineering conformational peptides to glutathione-binding domains to elicit anti-peptide antibodies may pave the way for the development of novel therapeutic approaches to suppress oxidative stress.
production by NQR. The approach used in this study of engineering conformational peptides to glutathione-binding domains to elicit anti-peptide antibodies may pave the way for the development of novel therapeutic approaches to suppress oxidative stress.
Supplementary Material
Acknowledgments
We thank Drs. Liwen Zhang and Kari B. Green-Church (CCIC, Mass Spectrometry and Proteomics Facility, The Ohio State University) for analysis in mass spectrometry.
This work was supported, in whole or in part, by National Institutes of Health Grants HL83237 (to Y.-R. C.) and HL63744 (to J. L. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
J. Chen and Y.-R. Chen, unpublished observation.
- GSSG
- oxidized glutathione
- Ab
- antibody
- MVF
- measles virus fusion
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- NQR
- NADH ubiquinone reductase, or mitochondrial Complex I
- NDH
- NADH dehydrogenase or flavin protein subcomplex of Complex I

- superoxide anion radical
- SMP
- submitochondrial particle
- Q1
- ubiquinone-1
- SQ1/Q1·−
- Q1-derived ubisemiquinone
- DEPMPO
- 5-diethoxylphosphoryl-5-methyl-1-pyrroline N-oxide
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- PBS
- phosphate-buffered saline
- β-ME
- β-mercaptoethanol
- I/R
- ischemia/reperfusion
- SGA
- superoxide generation activity
- SOD
- superoxide sisumtase
- ELISA
- enzyme-linked immunosorbent assay
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- TBS
- Tris-buffered saline
- Fp
- flavin subcomplex
- GS
- glutathione.
REFERENCES
- 1.Costa N. J., Dahm C. C., Hurrell F., Taylor E. R., Murphy M. P. (2003) Antioxid. Redox Signal. 5, 291–305 [DOI] [PubMed] [Google Scholar]
- 2.Hurd T. R., Filipovska A., Costa N. J., Dahm C. C., Murphy M. P. (2005) Biochem. Soc. Trans. 33, 1390–1393 [DOI] [PubMed] [Google Scholar]
- 3.Hurd T. R., Costa N. J., Dahm C. C., Beer S. M., Brown S. E., Filipovska A., Murphy M. P. (2005) Antioxid. Redox Signal. 7, 999–1010 [DOI] [PubMed] [Google Scholar]
- 4.Beer S. M., Taylor E. R., Brown S. E., Dahm C. C., Costa N. J., Runswick M. J., Murphy M. P. (2004) J. Biol. Chem. 279, 47939–47951 [DOI] [PubMed] [Google Scholar]
- 5.Taylor E. R., Hurrell F., Shannon R. J., Lin T. K., Hirst J., Murphy M. P. (2003) J. Biol. Chem. 278, 19603–19610 [DOI] [PubMed] [Google Scholar]
- 6.Beltrán B., Orsi A., Clementi E., Moncada S. (2000) Br. J. Pharmacol. 129, 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementi E., Brown G. C., Feelisch M., Moncada S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7631–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y. R., Chen C. L., Pfeiffer D. R., Zweier J. L. (2007) J. Biol. Chem. 282, 32640–32654 [DOI] [PubMed] [Google Scholar]
- 9.Chen Y. R., Chen C. L., Zhang L., Green-Church K. B., Zweier J. L. (2005) J. Biol. Chem. 280, 37339–37348 [DOI] [PubMed] [Google Scholar]
- 10.Sazanov L. A., Hinchliffe P. (2006) Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 11.Hurd T. R., Requejo R., Filipovska A., Brown S., Prime T. A., Robinson A. J., Fearnley I. M., Murphy M. P. (2008) J. Biol. Chem. 283, 24801–24815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C. L., Zhang L., Yeh A., Chen C. A., Green-Church K. B., Zweier J. L., Chen Y. R. (2007) Biochemistry 46, 5754–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dakappagari N. K., Lute K. D., Rawale S., Steele J. T., Allen S. D., Phillips G., Reilly R. T., Kaumaya P. T. (2005) J. Biol. Chem. 280, 54–63 [DOI] [PubMed] [Google Scholar]
- 14.Sundaram R., Lynch M. P., Rawale S. V., Sun Y., Kazanji M., Kaumaya P. T. (2004) J. Biol. Chem. 279, 24141–24151 [DOI] [PubMed] [Google Scholar]
- 15.Galante Y. M., Hatefi Y. (1979) Arch. Biochem. Biophys. 192, 559–568 [DOI] [PubMed] [Google Scholar]
- 16.Vinogradov A. D., King T. E. (1979) Methods Enzymol. 55, 118–127 [DOI] [PubMed] [Google Scholar]
- 17.Redfearn E. R., Whittaker P. A. (1966) Biochim. Biophy.s Acta 118, 413–418 [DOI] [PubMed] [Google Scholar]
- 18.Hatefi Y. (1978) Methods Enzymol. 53, 11–14 [DOI] [PubMed] [Google Scholar]
- 19.Duling D. R. (1994) J. Magn. Reson. B 104, 105–110 [DOI] [PubMed] [Google Scholar]
- 20.Frejaville C., Karoui H., Tuccio B., Le Moigne F., Culcasi M., Pietri S., Lauricella R., Tordo P. (1995) J. Med. Chem. 38, 258–265 [DOI] [PubMed] [Google Scholar]
- 21.Vásquez-Vivar J., Kalyanaraman B., Martásek P., Hogg N., Masters B. S., Karoui H., Tordo P., Pritchard K. A., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9220–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X., He G., Chen Y. R., Pandian R. P., Kuppusamy P., Zweier J. L. (2005) Circulation 111, 2966–2972 [DOI] [PubMed] [Google Scholar]
- 23.Guo Y., Wu W. J., Qiu Y., Tang X. L., Yang Z., Bolli R. (1998) Am. J. Physiol. 275, H1375–H1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zweier J. L., Kuppusamy P., Williams R., Rayburn B. K., Smith D., Weisfeldt M. L., Flaherty J. T. (1989) J. Biol. Chem. 264, 18890–18895 [PubMed] [Google Scholar]
- 25.Wang P., Zweier J. L. (1996) J. Biol. Chem. 271, 29223–29230 [DOI] [PubMed] [Google Scholar]
- 26.Roubaud V., Sankarapandi S., Kuppusamy P., Tordo P., Zweier J. L. (1997) Anal. Biochem. 247, 404–411 [DOI] [PubMed] [Google Scholar]
- 27.Gong X., Xie T., Yu L., Hesterberg M., Scheide D., Friedrich T., Yu C. A. (2003) J. Biol. Chem. 278, 25731–25737 [DOI] [PubMed] [Google Scholar]
- 28.Kobs-Conrad S., Lee H., DiGeorge A. M., Kaumaya P. T. (1993) J. Biol. Chem. 268, 25285–25295 [PubMed] [Google Scholar]
- 29.Kussmaul L., Hirst J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin A., Brandt U. (2005) J. Biol. Chem. 280, 30129–30135 [DOI] [PubMed] [Google Scholar]
- 31.Kudin A. P., Bimpong-Buta N. Y., Vielhaber S., Elger C. E., Kunz W. S. (2004) J. Biol. Chem. 279, 4127–4135 [DOI] [PubMed] [Google Scholar]
- 32.Magnitsky S., Toulokhonova L., Yano T., Sled V. D., Hägerhäll C., Grivennikova V. G., Burbaev D. S., Vinogradov A. D., Ohnishi T. (2002) J. Bioenerg. Biomembr. 34, 193–208 [DOI] [PubMed] [Google Scholar]
- 33.Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi S. T., Ohnishi T., Muranaka S., Fujita H., Kimura H., Uemura K., Yoshida K., Utsumi K. (2005) J. Bioenerg. Biomembr. 37, 1–15 [DOI] [PubMed] [Google Scholar]
- 35.Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 36.Rahman I., Kode A., Biswas S. K. (2006) Nat. Protocols 1, 3159–3165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.