Abstract
Triggering receptor expressed on myeloid cells-2 (TREM-2) is rapidly emerging as a key regulator of the innate immune response via its regulation of macrophage inflammatory responses. Here we demonstrate that proximal TREM-2 signaling parallels other DAP12-based receptor systems in its use of Syk and Src-family kinases. However, we find that the linker for activation of T cells (LAT) is severely reduced as monocytes differentiate into macrophages and that TREM-2 exclusively uses the linker for activation of B cells (LAB encoded by the gene Lat2−/−) to mediate downstream signaling. LAB is required for TREM-2-mediated activation of Erk1/2 and dampens proximal TREM-2 signals through a novel LAT-independent mechanism resulting in macrophages with proinflammatory properties. Thus, Lat2−/− macrophages have increased TREM-2-induced proximal phosphorylation, and lipopolysaccharide stimulation of these cells leads to increased interleukin-10 (IL-10) and decreased IL-12p40 production relative to wild type cells. Together these data identify LAB as a critical, LAT-independent regulator of TREM-2 signaling and macrophage development capable of controlling subsequent inflammatory responses.
Keywords: Immunology, Immunology/Innate Immunity, Receptors/Leukocyte/Lymphocyte, Signal Transduction/Adapter Proteins, Signal Transduction/Phosphotyrosine, Signal Transduction/Phosphotyrosine/SH2 Domains, Macrophage
Introduction
The triggering receptors expressed on myeloid cells (TREM)2 are a family of single immunoglobulin-like domain containing receptors expressed on a variety of innate immune cells of the myeloid lineage (1). In the mouse the TREM family genes form a loose cluster on chromosome 17 and include Trem-1, -2, -3, and -4 (pDC-TREM), an uncharacterized TREM, and TREM-like transcript (Treml-1 and -2). The human gene cluster includes Trem-1 and -2, and Treml1, -2, and -3. Of these genes the receptors encoded by Trem-1 and -2 are the best characterized to date. TREM-1 is selectively expressed by neutrophils and monocytes and is a potent amplifier of pathogenesis associated with microbial sepsis (2, 3). TREM-1 ligation on neutrophils leads to secretion of IL-8 and myeloperoxidase, and co-engagement of TREM-1 during LPS stimulation of monocytes synergistically enhances production of TNFα and monocyte chemoattractant protein-1. Moreover, inhibition of TREM-1 with soluble receptor protein, small interfering RNA, or antagonistic peptides rescues mice from microbial sepsis and can lessen the severity of experimentally induced colitis (4). In contrast, TREM-2 is expressed on murine macrophages, microglia, and osteoclasts and has been reported to play a role in the maturation and survival of human dendritic cells by inducing up-regulation of the chemokine receptor CCR7 (5).
Both TREM-1 and 2 associate with, and signal through DAP12, an immunoreceptor tyrosine-based activation motif (ITAM)-containing transmembrane adapter protein originally identified as a 16-kDa tyrosine-phosphorylated protein in NK cells functionally complexed with the non-inhibitory killer Ig-like receptors in humans and their murine counterparts within the Ly-49 gene family (6, 7). In addition to NK cells, DAP12 is expressed in a variety of other innate immune cells including granulocytes, blood monocytes, macrophages, and dendritic cells where it is non-covalently associated with a variety of receptors including signal-regulatory protein β1, myeloid DAP12-associating lectin 1, myeloid associated immunoglobulin-like receptor II, CD200 receptor-like 3, and paired immunoglobulin-like type 2 receptor β (8).
Supportive of the descriptions of TREM-1 as an amplifier of the septic response, Turnbull et al. (9) described decreased levels of plasma TNFα and IL-6, an attenuated acute phase response, and increased survival during polymicrobial sepsis in Dap12−/− mice. However, Hamerman et al. (9, 10) reported that DAP12-deficient mice were more susceptible to d-galactosamine-sensitized endotoxic shock and that macrophages from these mice showed increased TLR-induced production of TNF, IL-6, and IL-12p40. Moreover, these authors showed that bone marrow macrophages also express a TREM-2 ligand and that reconstitution of Dap12−/− macrophages with a TREM-2/DAP12 chimera restores normal inflammatory responses, suggesting that the hyper-responsiveness of Dap12−/− macrophages is due to a lack of TREM-2 signaling (10). This conclusion was confirmed by the recent demonstration of increased in vitro cytokine production in Trem-2−/− macrophages (11, 12).
Taken together these findings suggest opposing roles for TREM-1 and -2 during inflammation. Whereas TREM-1 on monocytes and neutrophils acts to amplify the septic response to facilitate the clearance of pathogens, TREM-2 on macrophages suppresses inflammatory responses perhaps to protect the host from endotoxemia and facilitate tissue repair. Consistent with a role for TREM-2 in regulation of anti-inflammatory responses, its expression is induced during allergic inflammation, and the receptor is constitutively expressed by alveolar macrophages. Moreover, alternative activation of resident peritoneal macrophages by IL-4 leads to TREM-2 expression, whereas interferon γ and LPS rapidly down-regulate cell surface expression (12). Last, soluble TREM-2 is found in the cerebrospinal fluid of patients with multiple sclerosis, and the development of experimental autoimmune encephalomyelitis in mice is exacerbated by blockade of TREM-2 and muted by administration of myeloid cells transduced with TREM-2 (13–15).
Given the profound importance of TREM-2 and DAP12 in macrophage responsiveness, it is surprising that relatively little is known about its downstream signaling mechanism in these cells. In peripheral blood monocytes, engagement of TREM-1 stimulates rapid tyrosine phosphorylation of PLCγ, intracellular calcium mobilization, and activation of the mitogen-activated protein kinase cascade (2). In macrophages, engagement of DAP12-coupled signal-regulatory protein β promotes phagocytosis by inducing the DAP12-dependent tyrosine phosphorylation of Syk, SLP-76, and the subsequent activation of mitogen-activated protein kinase (16). ITAM-mediated signals in T cells rely on the adaptor linker for activation of T cells (LAT) to translate proximal signals into downstream effects (17, 18). In contrast, DAP12 in NK cells and the Fc receptor of mast cells and monocytes use combinations of LAT and the related molecule, linker for activation in B cells (LAB), an adaptor encoded by the gene Lat2−/− that is also known as non-T cell activation linker (NTAL) (19). In T cells, mast cells, and NK cells, LAB negatively regulates signaling by disrupting the utilization of LAT. In contrast, the mechanism of TREM-mediated macrophage regulation, including its potential use of LAT and/or LAB, remains completely unknown.
Here we find that LAB is dramatically up-regulated during differentiation of macrophages from blood monocytes, whereas LAT expression falls. Accordingly, we find that LAB, and not LAT, couples TREM-2/DAP12 signals to downstream events in macrophages. Surprisingly, Lat2−/− macrophages showed enhanced TREM-2/DAP12-mediated phosphorylation of multiple substrates including Syk and the E3 ubiquitin ligase c-Cbl, but this result was not due to increased usage of LAT. Moreover, altered TREM-2 signaling in Lat2−/− macrophages is associated with increased IL-10 and decreased IL-12p40 production after LPS stimulation in vitro. These data suggest that by suppressing proximal TREM-2-mediated signals independently of LAT, LAB regulates the host response during inflammation by affecting functional responses of macrophages.
EXPERIMENTAL PROCEDURES
Mice, Cell Lines, and Culture of Bone Marrow-derived Macrophages (BMMΦ)
Lat2−/− mice, described previously (20), have been backcrossed onto the C57Bl/6 background for at least 10 generations. For clarity, the product of the Lat2−/− locus is here referred to as LAB. Lat2−/− and control C57BL/6 mice of the same gender and age were maintained under specific pathogen-free conditions at the National Cancer Institute-Frederick. Animal care was provided in accordance with the procedures outlined in “A Guide for the Care and Use of Laboratory Animals.” Ethical approval for the animal experimentation detailed in this article was received from the Institutional Animal Care and Use Committee at the NCI-Frederick. RAW264.7 macrophage (MΦ) cell line was maintained in RPMI 1640 medium containing 10% fetal bovine serum, 2 mm l-glutamine, penicillin/streptomycin, HEPES, nonessential amino acids, sodium pyruvate, and 2-mercaptoethanol. Immortalized MΦ cell lines were established by infecting primary bone marrow cells with the J2 recombinant retrovirus as described (21). Immortalized macrophage cell lines were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, l-glutamine, and antibiotics. BMMΦ were generated as described previously (12). The macrophage origin of the cells was confirmed by flow cytometry with antibodies against F4/80 (Caltag, Carlsbad, CA) and MAC-1, GR-1, major histocompatibility complex class II (Iab), and CD11c (BD Pharmingen). TREM-2 expression was confirmed using anti-TREM-2. There were no gross abnormalities between macrophages from WT and Lat2−/− mice.
Reagents and Antibodies
Convulxin (Cvx) was endotoxin-tested using the Limulus amebocyte lysate method and was measured at 0.078 enzyme units/ml (final concentration in stimulations). GST fusion proteins were produced and purified using established protocols. The GST-p85 SH2 fusion protein was a kind gift from Dr. Tony Pawson (University of Toronto, Toronto, Ontario, Canada), and the GST-Grb-2 SH2 (54–164) fusion protein was purchased from Santa Cruz (Santa Cruz, CA). Piceatannol and PP2 were from Calbiochem, LPS (Escherichia coli 0111:B4) was from Sigma. The following antibodies were used in these experiments: anti-LAT and anti-LAB monoclonal and polyclonal antibodies were as previously described (22, 23), anti-phosphotyrosine (clone 4G10, Upstate Biotechnology, Lake Placid, NY), anti-Syk (Novus Biologicals, Littleton, CO), anti-c-Cbl (sc-170), anti-PLCγ, anti-Csk (Santa Cruz), anti-p85, anti-GST (Upstate Biotechnology), anti-actin (Chemicon International, Temecula, CA), anti-phospho DAP12 as previously described (24), anti-phospho Erk1/2, anti- Erk1/2, anti-phosphoprotein kinase B (Cell Signaling Technology, Beverley, MA), anti-TREM-2 as previously described (Ref. 12; a kind gift from Dr. Marco Colonna, Washington University, St. Louis, MO), goat anti-rat IgG (KPL, Gaithersburg, MD), anti-human glycoprotein VI (GPVI) (HY101) as previously described (25) (a kind gift from Dr. Mark Kahn, University of Pennsylvania).
Generation of the GPVI/DAP12 Chimera
The GPVI/DAP12 chimera in pEF6 TOPO (Invitrogen) was generated by overlapping PCR from human GPVI (26) and murine DAP12 (7) cDNA templates using the following primers: hGPVI forward, GGCGCGCCACCATGAGCCCATCCCCGACCGC; mDAP12/hGPVI reverse, AGACCAGGCGTGCCAGAAACCCCGCCAGGATTAG; hGPVI/mDAP12 forward, GTTTCTGGCACGCCTGGTCTCCCGAGGTCAAG; mDAP12 reverse, GGCGCGCCTCATCTGTAATATTGCCTCTGTG; R → L DAP12 TM forward, CAACCTGGTCCTGATATGCCTCGG; R → L DAP12 TM, reverse, CCGAGGCATATGTCGACCAGGTTG. RAW cells were transfected using the Lipofectamine reagent (Invitrogen) according to the manufacturer's instructions, subcloned by limiting dilution, and characterized by flow cytometry using the anti-GPVI (HY101) antibody.
Macrophage Activation and Immunoprecipitation/GST Pulldown Assay
Cells were harvested and serum-starved for 30 min at 37 °C. 1 × 107 RAW cells were resuspended in 100 μl of DPBS and stimulated at 37 °C with 20 nm Cvx for the indicated times. MΦ/BMMΦ were resuspended in 100 μl of DPBS and placed on ice for 2 min. TREM-2 antibody was added at a concentration of 10 μg/1 × 107 cells on ice for a further 30 min. Cells were washed once with ice-cold DPBS and resuspended in 100 μl of DPBS which had been prewarmed to 37 °C. A secondary goat anti-rat antibody was added at a concentration of 10 μg/1 × 107 cells and placed at 37 °C for the indicated times. Cells were lysed with radioimmune precipitation assay lysis buffer (0.5% deoxycholic acid, 1% Triton X-100, 150 mm NaCl, 20 mm Tris (pH 8), 5 mm EDTA, 0.4 mm Na3VO4, aprotinin, leupeptin, and phenylmethylsulfonyl fluoride) or in some instances with laurylmaltoside buffer (1% laurylmaltoside in 20 mm Tris (pH 7.5), 100 mm NaCl, 10% glycerol, and protease and phosphatase inhibitors as above). Lysates were clarified by centrifugation and precipitated as described previously (27). Proteins were separated using 4–12% gels (Nupage, Invitrogen), transferred onto polyvinylidene difluoride membranes (Millipore), and analyzed by Western blot.
Raft Fractionation
Raft fractions were separated by sucrose gradient ultracentrifugation as previously described (18).
LPS Stimulations and Cytokine Determination
BMMϕ were stimulated with LPS (E. coli 0111:B4) for 24 h. Cell culture supernatants were recovered and assayed for cytokine by enzyme-linked immunosorbent assay (R&D Systems Inc., Minneapolis, MN) according to the manufacturer's protocol.
RESULTS
Direct Stimulation of DAP12 in Macrophages
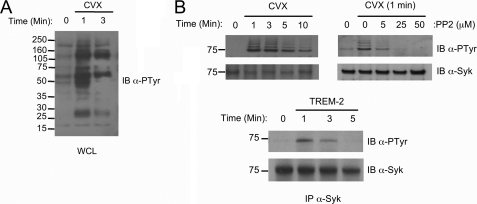
To investigate TREM-2 signaling without interference from Fc receptor or LPS effects, we generated a chimeric receptor by fusing the extracellular and transmembrane regions of GPVI to the cytoplasmic tail of DAP12 using overlapping PCR (supplemental Fig. 1A). Stimulation of the GPVI/DAP12 chimeric receptor expressed in RAW264.7 macrophages (MΦ) (supplemental Fig. 1B) with the high affinity, multivalent, GPVI-specific, ligand Cvx resulted in the rapid and transient phosphorylation of its ITAM (supplemental Fig. 1C) and multiple substrates (Fig. 1A). As in NK cells, Syk phosphorylation downstream of DAP12 was dependent on Src kinase activity (Fig. 1B). Stimulation of primary BMMϕ with antibody to TREM-2 confirmed the involvement of Syk downstream of TREM-2 in macrophages (Fig. 1B, bottom).
FIGURE 1.
Proximal TREM-2 signaling. A, whole cell lysates (WCL) from Cvx-stimulated chimera-expressing RAW cells were immunoblotted (IB) with an anti-phosphotyrosine (clone 4G10) antibody. B, top, chimera expressing RAW264.7 cells were treated for 30 min with various concentrations of the Src inhibitor PP2 as indicated (right panel) or left untreated (left panel). Cells were stimulated with Cvx for the indicated times, and Syk was immunoprecipitated, resolved via SDS-PAGE, and immunoblotted with anti-phosphotyrosine. Filters were reprobed with anti-Syk as indicated to demonstrate equal loading. B, bottom. BMMΦ of WT mice were stimulated for the times indicated with TREM-2 monoclonal antibody, and Syk phosphorylation was assayed as above.
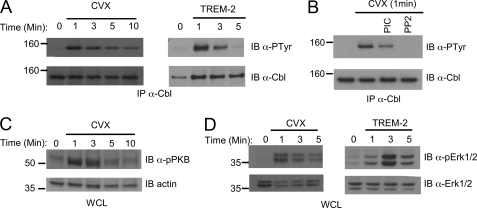
Because DAP12-dependent signaling is known to lead to phosphorylation of the E3 ubiquitin ligase c-Cbl in NK cells (27), we next determined whether c-Cbl is downstream of TREM-2 in macrophages. Using the GPVI/DAP12 chimera system, we found that c-Cbl was rapidly tyrosine-phosphorylated (Fig. 2A, left). Similarly, we found that c-Cbl is rapidly phosphorylated downstream of TREM-2 in primary macrophages (Fig. 2A, right) with peak phosphorylation occurring 1 min after stimulation. Although the phosphorylation of c-Cbl was completely inhibited by the Src-specific inhibitor PP2, it was only partially inhibited by the Syk inhibitor Piceatannol (Fig. 2B). More distally in the DAP12 pathway we found rapid and transient phosphorylation of protein kinase B/Akt, a marker of activation of the phosphatidylinositol 3-kinase pathway (Fig. 2C). In addition we detected activation of the mitogen-activated protein kinase pathway (Fig. 2D). Erk1/2 activation was again confirmed in primary BMMΦ stimulated via anti-TREM-2 (Fig. 2D, right). Thus, our results suggest that our GPVI/DAP12 chimeric receptor system accurately recapitulates endogenous DAP12 signaling, making it a powerful tool for further dissection of the TREM-2 pathway in macrophages.
FIGURE 2.
Distal TREM-2 signaling events. A, chimera expressing RAW264.7 cells (left) or BMMΦ from WT mice (right) were stimulated with 20 nm Cvx or anti-TREM-2, respectively, for the indicated times. Lysates were immunoprecipitated (IP) with anti-c-Cbl and immunoblotted with anti-phosphotyrosine. Blots were stripped and reprobed with an anti-c-Cbl. B, before stimulation with Cvx, chimera-expressing cells were treated for 30 min with 20 μg/ml piceatannol or 25 μm PP2. Lysates were immunoprecipitated with an anti-c-Cbl and immunoblotted with an anti-phosphotyrosine. Blots were stripped and reprobed with anti-c-Cbl. C, chimera expressing RAW264.7 cells were stimulated with 20 nm Cvx as in A. Lysates were immunoblotted with an anti-phosphoprotein kinase B. Reprobing with anti-actin demonstrated equal loading. WCL, whole cell lysates. D, chimera-expressing cells (left) or BMMΦ from WT mice (right) were stimulated as in A, and lysates were immunoblotted with anti-phospho-Erk1/2. Blots were stripped and reprobed with anti-Erk1/2, as indicated. Results are representative of at least two independent experiments.
LAB Expression Is Induced, and LAT Expression Is Suppressed as Monocytes Differentiate into Macrophages or Dendritic Cells
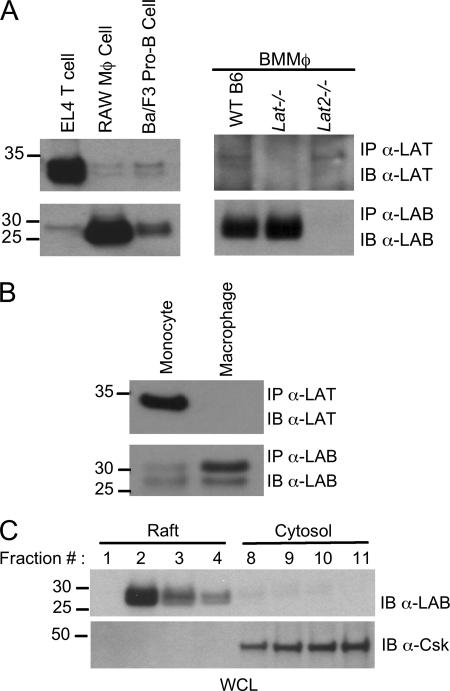
Many immune receptor systems use LAT and/or LAB to translate proximal signals into downstream effects; therefore, we examined the possibility that these proteins might be components of the TREM-2 signaling cascade and as such may be important in TREM-2-mediated regulation of macrophage TLR responses. We found that whereas LAT could easily be detected in EL4 T cells, only very low levels of the protein were detected in the RAW264.7 macrophage line or in primary BMMΦ (Fig. 3A). By comparison, we found LAB to be strongly expressed in RAW264.7 and BMMΦ relative to the Ba/F3 pro-B cell line (Fig. 3A). Bone marrow-derived macrophages expressed high levels of LAB but only barely detectable levels of LAT (Fig. 3A, left). Moreover, our analysis demonstrated no compensatory increases in either LAT or LAB when the other adaptor was deleted (Fig. 3A, right panel). In agreement with previous reports, we found that purified human monocytes do express LAT (Fig. 3B and Ref. 19). However, upon differentiation of these cells into macrophages in vitro LAT was down-regulated, and LAB expression increased (Fig. 3B). As expected, LAB localization was largely restricted to the detergent-insoluble lipid raft fractions of the plasma membrane, consistent with a possible role in TREM-2 signaling, whereas the control cytosolic c-Src kinase (Csk) was found in the cytosol fractions (Fig. 3C).
FIGURE 3.
LAT and LAB expression in monocytes and macrophages. Equal amounts of protein derived from lysates of EL4 T cells, RAW264.7, Ba/F3 B cells, WT, Lat−/−, and Lat2−/− BMMΦ (A) or human monocytes or macrophages differentiated with M-CSF (B) were immunoprecipitated with anti-LAT or anti-LAB antibodies as indicated. The corresponding immunoprecipitates (IP) were subject to immunoblotting (IB) with anti-LAT or anti-LAB monoclonal antibodies. C, detergent-insoluble raft fractions were isolated by sucrose-density gradient ultracentrifugation from RAW264.7 lysates. Raft fractions (1–4) and non-raft fractions (9–11) were immunoblotted with an anti-LAB antibody or anti-Csk antibody. Results are representative of at least two independent experiments. WCL, whole cell lysates.
LAB Is a Substrate for Tyrosine Phosphorylation and Interacts with Grb-2 in the TREM-2 Pathway
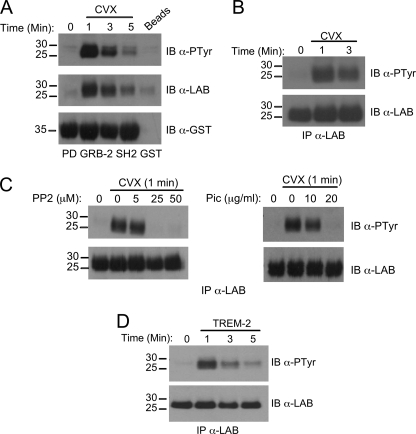
In Cvx-stimulated whole cell lysates of chimera expressing RAW264.7 MΦ cells, a strong tyrosine-phosphorylated band migrating at the appropriate size for LAB was present (Fig. 1A). LAB has a total of nine tyrosine phosphorylation sites that are conserved between human and mouse proteins, five of which fall into the consensus for Grb-2 binding (22). We, therefore, determined whether the observed protein was associated with Grb-2 after stimulation of the GPVI/DAP12 chimera. Accordingly, in Grb-2 SH2 domain pulldown assays there was a phosphorylated band reactive with a LAB monoclonal antibody and bound to Grb-2 SH2 after Cvx treatment (Fig. 4A). Direct immunoprecipitation confirmed that LAB was in fact a substrate for tyrosine phosphorylation downstream of the GPVI/DAP12 chimera (Fig. 4B). The dose-dependent inhibition of LAB phosphorylation by inhibitors of either Src or Syk placed it downstream of these kinases within the signaling cascade (Fig. 4C). Importantly, LAB phosphorylation was not observed in chimera expressing RAW264.7 treated with LPS or in control RAW264.7 treated with Cvx (data not shown).
FIGURE 4.
LAB is tyrosine-phosphorylated downstream of TREM-2. A–C, chimera expressing RAW264. 7 cells were stimulated for the indicated times with 20 nm Cvx. A, pulldown assays were performed on postnuclear lysates using Grb-2 SH2 domain fusion protein then immunoblotted (IB) with an anti-phosphotyrosine. The same blot was stripped and reprobed with anti-LAB antibody followed by anti-GST. B, Cvx-stimulated lysates were immunoprecipitated (IP) with anti-LAB antibody and immunoblotted with an anti-phosphotyrosine. The same blot was stripped and reprobed with an anti-LAB. C, before stimulation with Cvx, samples were treated for 30 min with various concentrations of the PP2 or piceatannol, as indicated. D, BMMΦ from WT mice were stimulated for the times indicated by TREM-2 cross-linking. Lysates were immunoprecipitated with anti-LAB antibody and immunoblotted with an anti-phosphotyrosine. The same blots were stripped and reprobed with an anti-LAB antibody. Results are representative of several independent experiments.
Thus, we next sought to confirm the direct involvement of LAB in TREM-2 signaling in primary bone marrow macrophages. To this end we stimulated primary macrophages with anti-TREM-2 antibody, immunoprecipitated LAB, and immunoblotted with antiphosphotyrosine. These experiments confirmed the rapid and transient phosphorylation of LAB in response to TREM-2 engagement (Fig. 4D) and established LAB as a prominent, tyrosine-phosphorylated intermediary in macrophages.
LAB Is Present in a Complex with p85 That Includes c-Cbl
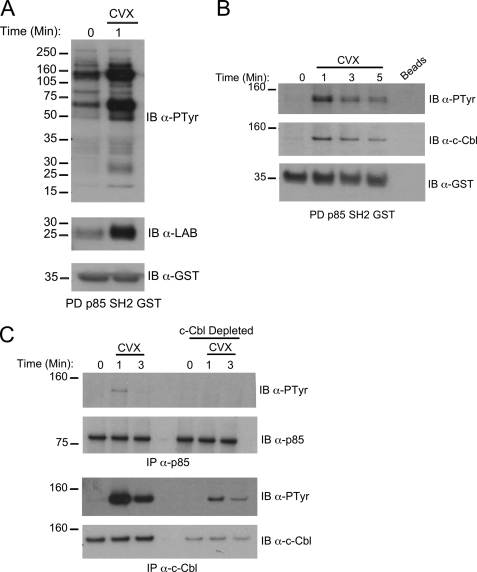
c-Cbl can recruit the p85 adaptor subunit of phosphatidylinositol 3-kinase (PI3K) in macrophages/osteoclasts and positively influence PI3K activation (28). Moreover, LAB has been demonstrated to interact with c-Cbl in the monocytic cell line THP-1. Therefore, we examined the possibility that there is a complex formed between LAB, c-Cbl, and p85 downstream of TREM-2/DAP12. Indeed we observed the inducible association of tyrosine-phosphorylated LAB with a p85 SH2 domain fusion protein (Fig. 5A). A number of other tyrosine-phosphorylated bands were prominent in the Cvx stimulated p85 pulldown assays, one of which we subsequently identified as c-Cbl (Fig. 5B). Consistent with a c-Cbl/p85 co-association, a tyrosine-phosphorylated band corresponding to the approximate size of c-Cbl was observed in p85 immunoprecipitates, and this band was eliminated by c-Cbl depletion (Fig. 5C). Thus, LAB nucleates a complex that contains both phosphatidylinositol 3-kinase and c-Cbl.
FIGURE 5.
LAB is present in a complex with p85 and c-Cbl after TREM-2 stimulation. A, GST pulldown (PD) assays from Cvx-stimulated RAW264.7 cell lysates were performed using a p85 SH2 domain GST fusion protein. Protein complexes were resolved by SDS-PAGE and immunoblotted (IB) with anti-phosphotyrosine (top), anti-LAB antibody (middle), or anti-GST (bottom). B, after Cvx stimulation for the indicated times lysates were precipitated with empty beads (lane 5) or GST-p85 SH2 (lanes 1–4). Complexes were resolved as above and immunoblotted with anti-phosphotyrosine (top), anti-c-Cbl (middle), or anti-GST (bottom). C, Cvx-stimulated lysates were subjected to three rounds of immunodepletion with anti-c-Cbl. Cbl-depleted lysates were subsequently immunoprecipitated with either anti-p85 (top two panels) or anti-c-Cbl (bottom two panels). Immunoblots were probed with anti-phosphotyrosine and then stripped and reprobed with anti-p85 or anti-c-Cbl as appropriate. Results are representative of at least two independent experiments.
LAB Suppresses TREM-2 Signaling in Macrophages Independently of the Regulation of LAT Usage
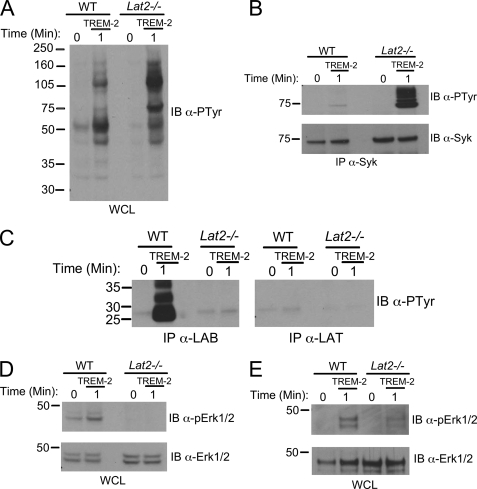
We next examined the effects of LAB deficiency on TREM-2 signaling by deriving immortalized MΦ cell lines from wild type C57Bl/6 mice (WT) and Lat2−/− (the gene encoding LAB/NTAL) mice. These lines expressed comparable levels of TREM-2 on their surface (data not shown); however, surprisingly, we observed enhanced tyrosine phosphorylation of a number of substrates in lysates from TREM-2-stimulated Lat2−/− cells compared with WT cells (Fig. 6A). Notably, immunoprecipitation of Syk from WT and Lat2−/− cells revealed that TREM-2-dependent phosphorylation of this kinase was dramatically increased in cells lacking LAB (Fig. 6B). In addition, total levels of Syk protein were slightly up-regulated in the absence of LAB. Taken together, these data highlight LAB as a major negative regulator of proximal TREM-2 signaling in macrophages. Consistent with the relative lack of LAT protein expression in macrophages, we could not detect its TREM-2-inducible phosphorylation, even in Lat2−/− BMMϕ, whereas significant LAB phosphorylation in WT BMMϕ was observed under the same conditions (Fig. 6C). Further strengthening our contention that LAT does not contribute to TREM-2 signaling in macrophages, Erk1/2 phosphorylation was not intact in the Lat2−/− MΦ cell line or in Lat2−/− primary BMMΦ (Fig. 6, D and E, respectively). Taken together, these data suggest that in macrophages LAB is a potent negative regulator of TREM-2-induced proximal tyrosine phosphorylation via a mechanism independent of LAT but is required for efficient activation of Erk.
FIGURE 6.
LAB regulates TREM-2 signaling in macrophages independently of LAT. A, immortalized MΦ from WT or Lat2−/− mice were stimulated by TREM-2 cross-linking for the times indicated. Equal amounts of protein were immunoblotted (IB) with anti-phosphotyrosine. WCL, whole cell lysates. B, lysates as in A were immunoprecipitated (IP) with anti-Syk and immunoblotted with an anti-phosphotyrosine. The same blot was stripped and reprobed with an anti-Syk. Results are representative of at least three independent experiments. C, equal amounts of protein from TREM-2-stimulated lysates of BMMΦ from WT or Lat2−/− mice were immunoprecipitated with anti-LAB or anti-LAT antibodies. Blots were probed with anti-phosphotyrosine, and both panels shown here are from the same exposure. D, immortalized MΦ from WT and Lat2−/− mice were stimulated with TREM-2 and immunoblotted with an anti-phospho Erk1/2 antibody. The same blot was stripped and reprobed with anti- Erk1/2. Results are representative of at least two independent experiments. E, BMMΦ derived from WT or Lat2−/− mice were assayed for Erk1/2 activation as in D.
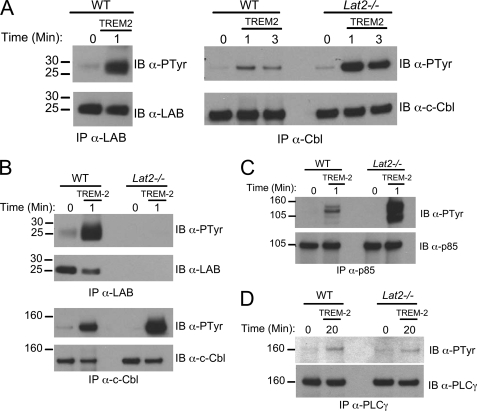
Enhanced c-Cbl Phosphorylation in the Absence of LAB
To examine the effect of LAB deficiency on c-Cbl phosphorylation, we utilized our MΦ cell lines from WT and Lat2−/− mice. We found that TREM-2 cross-linking caused much higher levels of c-Cbl tyrosine phosphorylation in LAB-deficient cells as compared with WT macrophages (Fig. 7A). Consistent with this result we also observed increased Cvx-induced c-Cbl phosphorylation after small interfering RNA knockdown of LAB in GPVI/DAP12 chimera-expressing RAW264.7 cells (data not shown). Last, we detected significantly increased TREM-2-mediated c-Cbl phosphorylation in primary BMMϕ from Lat2−/− null mice (Fig. 7B). Consistent with the c-Cbl·p85·LAB complex we detected above (see Fig. 5), we also found a marked increase in TREM-2-mediated tyrosine phosphorylation of the p85 co-immunoprecipitating band we had previously determined to be c-Cbl (Fig. 7C), whereas the absence of LAB appears to have no affect on PLCγ phosphorylation (Fig. 7D). Together these findings suggest that the altered TREM-2 signaling of Lat2−/− macrophages leads to higher c-Cbl phosphorylation and increased association of c-Cbl with the p85 subunit of phosphatidylinositol 3-kinase.
FIGURE 7.
Enhanced c-Cbl phosphorylation in the absence of LAB. A, immortalized MΦ from WT and Lat2−/− mice were stimulated by TREM-2 cross-linking for the indicated times. Lysates were immunoprecipitated (IP) with an anti-LAB polyclonal antibody (left) or an anti-c-Cbl antibody (right). Immunoblots (IB) were probed with anti-phosphotyrosine, anti-LAB, or anti-c-Cbl as indicated. B, BMMΦ from the indicated strain were stimulated, immunoprecipitated, and immunoblotted as in A. C, lysates from TREM-2-stimulated immortalized MΦ were immunoprecipitated with an anti-p85 and immunoblotted with an anti-phosphotyrosine. The same blot was stripped and reprobed with an anti-p85. D, lysates from TREM-2-stimulated immortalized MΦ were immunoprecipitated with an anti-PLCγ and immunoblotted with an anti-phosphotyrosine. The same blot was stripped and reprobed with an anti-PLCγ. Results are representative of several independent experiments.
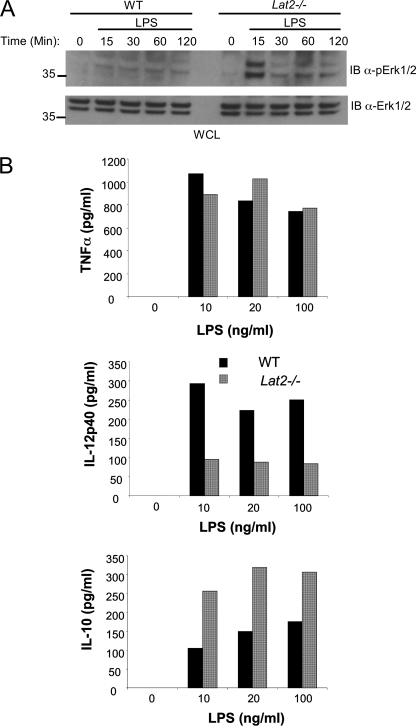
LAB Differentially Regulates the Production of IL-10 and IL-12p40
Recent studies have suggested that through its interaction with an endogenous ligand expressed on bone marrow-derived macrophages, TREM-2 suppresses TLR-mediated production of TNFα, IL-6, and IL-12p40 (9–12). The altered TREM-2 signaling we found in Lat2−/− macrophages prompted us to investigate LPS-induced responses in these cells. In contrast to the reduced levels of TREM-2-induced Erk phosphorylation we detected in the absence of LAB, LPS-stimulated activation of Erk1/2 was increased in Lat2−/− BMMΦ compared with WT cells (Fig. 8A). Engagement of TLR-5 by flagellin also resulted in increased Erk phosphorylation in Lat2−/− macrophages relative to WT cells (data not shown).
FIGURE 8.
LAB differentially regulates production of IL-10 and IL-12p40 by macrophages. A, BMMΦ from WT and Lat2−/− mice were stimulated for the indicated times with LPS, and phospho-Erk was assessed by immunoblotting (IB) whole cell lysates. B, BMMΦ from WT and Lat2−/− mice were stimulated for 24 h with various concentrations of LPS as indicated. Supernatants were quantified for IL-10, IL-12p40, and TNFα by enzyme-linked immunosorbent assay. Black bars represent WT, and gray bars represent Lat2−/−. Results are representative of at least three independent experiments with a pool of three mice in each.
Consistent with increased LPS-induced Erk activation, LPS-induced IL-10 production by Lat2−/− BMMΦ was also increased compared with WT cells. In contrast, production of IL-12p40 was substantially decreased in the absence of LAB. Interestingly, TNFα production by Lat2−/− macrophages was comparable with that of WT cells, suggesting that the enhanced TREM-2 signaling of these cells results in skewing of LPS-induced cytokine production patterns rather than the global increases reported in Dap12−/− or Trem-2−/− cells (9–12) (Fig. 8B). Similar results for IL-10 and TNFα production from WT and Lat2−/− macrophages were found using the LINCOplex system (supplemental Fig. 2). These assays also demonstrated that Lat2−/− macrophages produce lower amounts of IL-1β (supplemental Fig. 2) and normal IL-6 (data not shown) levels when compared with WT cells, confirming suppression of LPS-induced pro-inflammatory cytokines in Lat2−/− cells. To further assess the effects of Lat2−/− mutation on macrophage function, we primed cells with interferon γ before LPS. Priming of macrophages failed to overcome the skewed cytokine production in Lat2−/− cells (data not shown).
DISCUSSION
Preferential Use of LAB in Macrophages
We and others have described the importance of LAT downstream of DAP12 in NK cell activation, suggesting it may also be involved in TREM-2-mediated signaling (29). Moreover, LAT phosphorylation has been demonstrated after ligation of ITAM-coupled Fc receptors in human monocytes (19). Although we could detect LAT expression in human monocytes, we found that its expression was markedly down-regulated upon in vitro differentiation into macrophages. Similarly, we could detect only weak LAT expression in the RAW264.7 MΦ cell line or in primary murine BMMΦ. In contrast, the LAT homologous protein, LAB, was poorly expressed in monocytes, and its expression was dramatically up-regulated upon differentiation to macrophages.
Similar to LAT, LAB contains a membrane proximal palmitoylation motif and has a cytoplasmic tail with several highly conserved tyrosine residues that, upon phosphorylation, facilitate binding of Grb-2. However unlike LAT, LAB does not bind directly to PLCγ, suggesting that LAB-mediated signaling is qualitatively distinct from signaling cassettes dependent on LAT (22, 30, 31). Thus, whereas LAT may be important in blood monocytes and NK cells, LAB is the primary adaptor protein expressed in macrophages. Because murine bone marrow macrophages express predominately TREM-2 rather than TREM-1, our findings suggest that adaptor cassettes may be intentionally regulated in concordance with the TREM receptor systems (data not shown). This coordinated regulation may assure the functional coupling of TREM-1 on monocytes specifically to LAT, whereas the TREM-2 of macrophages signals through LAB.
Negative Regulation of TREM-2 Signaling by LAB Independent of the Use of LAT
The utilization of LAB by TREM-2 is intriguing given recent studies suggesting that LAB can negatively regulate ITAM-driven signaling (20, 32). In LAB-deficient Mast cells, FcϵRI-stimulated calcium mobilization, degranulation, and cytokine production is enhanced. Notably, mast cells express both LAT and LAB, and the inhibitory role of LAB has been suggested to result from its competition with LAT for position within lipid rafts. Consistent with this view, in the absence of LAB, more LAT is partitioned into lipid rafts, where it is more highly phosphorylated and consequently more efficient at propagating downstream signaling.
Here we show that LAB is a potent negative regulator of proximal TREM-2 signaling through a LAT-independent mechanism. Evidence in support of this conclusion is two-fold. First, there is no compensatory increase in LAT expression, nor were we able to detect TREM-2-mediated LAT phosphorylation even in Lat2−/− macrophages. Second, Erk1/2 phosphorylation via TREM-2 was diminished in the absence of LAB, whereas we would expect mitogen-activated protein kinase activation to be intact, if not elevated, if LAT was able to deliver a competent signal in these cells (20). Thus, from our data it appears that LAB negatively modulates proximal TREM-2 signaling via mechanisms other than through competition with LAT. Moreover, our data suggest that LAB is critical for efficient TREM-2-mediated activation of Erk.
So how might LAB regulate TREM signaling in a cell that lacks LAT? In Lat2−/− macrophages, the dramatic enhancement of Syk phosphorylation together with increased Syk protein levels suggests that LAB acts proximally in the TREM-2 pathway perhaps via regulation of Syk itself. The E3-ubiquitin ligase c-Cbl has a well established role as a negative regulator of protein-tyrosine kinases including Src and Syk, both early components of the TREM-2 pathway (33, 34). Indeed by virtue of its interaction with LAB in TREM-2-stimulated macrophages, c-Cbl would be placed in a prime position to regulate proximal signaling events such as the activation of Syk. Thus, our data suggest that upon engagement of TREM-2, LAB is phosphorylated by receptor-associated Syk. Phosphorylated LAB then acts as the docking site for Grb-2, facilitating the activation of Erk1/2 and the recruitment of c-Cbl. c-Cbl in turn down-regulates proximal TREM-2 signals including the activation of Syk. Such a model is supported by previous data suggesting that in monocytes and B cells LAB can interact with c-Cbl and is itself ubiquitinated in these cells (31).
Anti-inflammatory Responses of Lat2−/− Macrophages
Macrophages developing in culture express both TREM-2 and an as yet unidentified TREM-2 ligand. Thus, it has been suggested that this presumably low affinity, cis-interaction delivers a tonic TREM-2 signal that in wild type mice reduces their responses to subsequent TLR stimulation (10, 12). In addition, it has been demonstrated that TLR stimulation, when delivered together with potent Erk activation via the ITAM FcγRI γ chain, results in an alternative state of macrophage activation. Characteristics of alternatively activated macrophages include relatively high production of IL-10 and relatively low production of IL-12p40; TNFα levels are comparable with classically activated macrophages (35). Indeed, we found that LAB−/− macrophages do exhibit an apparent alternative activation profile when stimulated with LPS in vitro. The higher production of IL-10 by LPS-stimulated LAB−/− macrophages is consistent with the increased Erk1/2 activation we detect after stimulation by either TLR4 (Fig. 8) or TLR5 (data not shown) (36). Although the exact mechanism responsible for enhanced TLR-mediated Erk activation in Lat2−/− macrophages remains unknown, one possibility is that in WT macrophages, continuous TREM-2 signaling may prevent efficient subsequent activation by TLR. In Lat2−/− cells TREM-2 cannot efficiently activate Erk, resulting in a resting pool of Erk1/2 poised for activation by TLR-mediated signaling pathways. Nonetheless, it appears that accentuated TLR-mediated Erk activation results in production of IL-10 at the expense of IL-12p40 in Lat2−/− macrophages.
IL-10 is a critical regulator in murine models of acute endotoxic shock through the suppression of multiple macrophage functions including the LPS-induced production of TNFα, IL-1, IL-6, and IL-12 (37–39). Therefore, although our data do not definitively implicate LAB-mediated regulation specifically of TREM-2 as the cause of the altered macrophage phenotype, our findings support such a model. We propose that deregulated TREM-2 signaling in Lat2−/− mice results in altered macrophage cytokine responses during LPS-priming, shifting the cytokine balance from IL-12 toward IL-10 production, weaker priming, and reduced subsequent inflammatory responses. These data are in accordance with the reported higher IL-10 production of TREM-2-transduced myeloid cells (40).
The skewed cytokine responses of Lat2−/− macrophages we report here contrast with a recent study suggesting enhanced TREM-1-induced TNFα and IL-8 production in monocyte cell lines where LAB expression had been suppressed by small interfering RNA (41). These authors also detected increased TREM-1 induced Erk1/2 activation in LAB knockdown monocytes, whereas we find clear reductions in TREM-2-medated Erk activation in Lat2−/− macrophages. Given our finding of dramatic differences in LAT and LAB expression in monocytes relative to macrophages, we suggest that these discrepancies may not be due to inherent differences in the specific receptors engaged; TREM-1 in the previous work versus TREM-2 here. Instead, we suggest that the observed differences reflect our use of macrophages rather than pro-monocytic cell lines, the former being exclusively dependent on LAB. Contrasts such as these, however, underscore the complexity inherent in TREM signaling where both receptors and intracellular signaling components are regulated during myeloid differentiation. The data we present here suggest that this complex regulation is likely critical for the specific tuning of monocyte and macrophage responses during inflammatory responses.
Supplementary Material
Acknowledgments
We thank Dr. Howard Young and Della Reynolds for the production of macrophage cell lines and S. Anderson and P. Schwartzberg for critical reading of the manuscript. In addition, we thank Dr. Mark Kahn for the gift of HY101 antibody.
This work was supported, in whole or in part, by the National Institutes of Health (Intramural Research Programs, NCI, Center for Cancer Research, and NIAID).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- TREM
- triggering receptor expressed on myeloid cells
- BMMΦ
- bone marrow-derived macrophages
- Cvx
- convulxin
- DAP12
- DNAX-activating protein of 12 kDa
- IL
- interleukin
- ITAM
- immune receptor tyrosine-based activation motif
- LAB
- linker for activation of B cells
- LAT
- linker for activation of T cells
- LPS
- lipopolysaccharide
- NTAL
- non-T cell activation linker
- TNFα
- tumor necrosis factor α
- GPVI
- human glycoprotein VI
- PLC
- phospholipase C
- WT
- wild type
- GST
- glutathione S-transferase
- Erk
- extracellular signal-regulated kinase
- DPBS
- Dulbecco's phosphate-buffered saline
- TLR
- Toll-like receptor.
REFERENCES
- 1.Ford J. W., McVicar D. W. (2009) Curr. Opin. Immunol. 21, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouchon A., Dietrich J., Colonna M. (2000) J. Immunol. 164, 4991–4995 [DOI] [PubMed] [Google Scholar]
- 3.Bouchon A., Facchetti F., Weigand M. A., Colonna M. (2001) Nature 410, 1103–1107 [DOI] [PubMed] [Google Scholar]
- 4.Schenk M., Bouchon A., Seibold F., Mueller C. (2007) J. Clin. Invest. 117, 3097–3106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchon A., Hernández-Munain C., Cella M., Colonna M. (2001) J. Exp. Med. 194, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanier L. L., Corliss B. C., Wu J., Leong C., Phillips J. H. (1998) Nature 391, 703–707 [DOI] [PubMed] [Google Scholar]
- 7.Mason L. H., Willette-Brown J., Anderson S. K., Gosselin P., Shores E. W., Love P. E., Ortaldo J. R., McVicar D. W. (1998) J. Immunol. 160, 4148–4152 [PubMed] [Google Scholar]
- 8.Whittaker G. C., Anderson S. K., McVicar D. W. (2007) UCSD-Nature Molecule Pages doi:10.1038/mp.a000750.01 [Google Scholar]
- 9.Turnbull I. R., McDunn J. E., Takai T., Townsend R. R., Cobb J. P., Colonna M. (2005) J. Exp. Med. 202, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamerman J. A., Tchao N. K., Lowell C. A., Lanier L. L. (2005) Nat. Immunol. 6, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamerman J. A., Jarjoura J. R., Humphrey M. B., Nakamura M. C., Seaman W. E., Lanier L. L. (2006) J. Immunol. 177, 2051–2055 [DOI] [PubMed] [Google Scholar]
- 12.Turnbull I. R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. (2006) J. Immunol. 177, 3520–3524 [DOI] [PubMed] [Google Scholar]
- 13.Piccio L., Buonsanti C., Cella M., Tassi I., Schmidt R. E., Fenoglio C., Rinker J., 2nd, Naismith R. T., Panina-Bordignon P., Passini N., Galimberti D., Scarpini E., Colonna M., Cross A. H. (2008) Brain 131, 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piccio L., Buonsanti C., Mariani M., Cella M., Gilfillan S., Cross A. H., Colonna M., Panina-Bordignon P. (2007) Eur. J. Immunol. 37, 1290–1301 [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K., Rochford C. D., Neumann H. (2005) J. Exp. Med. 201, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi A., Ohnishi H., Okazawa H., Nakazawa S., Ikeda H., Motegi S., Aoki N., Kimura S., Mikuni M., Matozaki T. (2004) J. Biol. Chem. 279, 29450–29460 [DOI] [PubMed] [Google Scholar]
- 17.Zhang W., Sommers C. L., Burshtyn D. N., Stebbins C. C., DeJarnette J. B., Trible R. P., Grinberg A., Tsay H. C., Jacobs H. M., Kessler C. M., Long E. O., Love P. E., Samelson L. E. (1999) Immunity 10, 323–332 [DOI] [PubMed] [Google Scholar]
- 18.Zhang W., Trible R. P., Samelson L. E. (1998) Immunity 9, 239–246 [DOI] [PubMed] [Google Scholar]
- 19.Tridandapani S., Lyden T. W., Smith J. L., Carter J. E., Coggeshall K. M., Anderson C. L. (2000) J. Biol. Chem. 275, 20480–20487 [DOI] [PubMed] [Google Scholar]
- 20.Zhu M., Liu Y., Koonpaew S., Granillo O., Zhang W. (2004) J. Exp. Med. 200, 991–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blasi E., Radzioch D., Merletti L., Varesio L. (1989) Cancer Biochem. Biophys. 10, 303–317 [PubMed] [Google Scholar]
- 22.Janssen E., Zhu M., Zhang W., Koonpaew S., Zhang W. (2003) Nat. Immunol. 4, 117–123 [DOI] [PubMed] [Google Scholar]
- 23.Zhang W., Sloan-Lancaster J., Kitchen J., Trible R. P., Samelson L. E. (1998) Cell 92, 83–92 [DOI] [PubMed] [Google Scholar]
- 24.Mason L. H., Willette-Brown J., Taylor L. S., McVicar D. W. (2006) J. Immunol. 176, 6615–6623 [DOI] [PubMed] [Google Scholar]
- 25.Chen H., Locke D., Liu Y., Liu C., Kahn M. L. (2002) J. Biol. Chem. 277, 3011–3019 [DOI] [PubMed] [Google Scholar]
- 26.Zheng Y. M., Liu C., Chen H., Locke D., Ryan J. C., Kahn M. L. (2001) J. Biol. Chem. 276, 12999–13006 [DOI] [PubMed] [Google Scholar]
- 27.McVicar D. W., Taylor L. S., Gosselin P., Willette-Brown J., Mikhael A. I., Geahlen R. L., Nakamura M. C., Linnemeyer P., Seaman W. E., Anderson S. K., Ortaldo J. R., Mason L. H. (1998) J. Biol. Chem. 273, 32934–32942 [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki T., Sanjay A., Neff L., Tanaka S., Horne W. C., Baron R. (2004) J. Biol. Chem. 279, 17660–17666 [DOI] [PubMed] [Google Scholar]
- 29.Chiesa S., Mingueneau M., Fuseri N., Malissen B., Raulet D. H., Malissen M., Vivier E., Tomasello E. (2006) Blood 107, 2364–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janssen E., Zhu M., Craven B., Zhang W. (2004) J. Immunol. 172, 6810–6819 [DOI] [PubMed] [Google Scholar]
- 31.Brdicka T., Imrich M., Angelisová P., Brdicková N., Horváth O., Spicka J., Hilgert I., Lusková P., Dráber P., Novák P., Engels N., Wienands J., Simeoni L., Osterreicher J., Aguado E., Malissen M., Schraven B., Horejsí V. (2002) J. Exp. Med. 196, 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volná P., Lebduska P., Dráberová L., Símová S., Heneberg P., Boubelík M., Bugajev V., Malissen B., Wilson B. S., Horejsí V., Malissen M., Dráber P. (2004) J. Exp. Med. 200, 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andoniou C. E., Lill N. L., Thien C. B., Lupher M. L., Jr., Ota S., Bowtell D. D., Scaife R. M., Langdon W. Y., Band H. (2000) Mol. Cell. Biol. 20, 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ota S., Hazeki K., Rao N., Lupher M. L., Jr., Andoniou C. E., Druker B., Band H. (2000) J. Biol. Chem. 275, 414–422 [DOI] [PubMed] [Google Scholar]
- 35.Mosser D. M. (2003) J. Leukocyte Biol. 73, 209–212 [DOI] [PubMed] [Google Scholar]
- 36.Lucas M., Zhang X., Prasanna V., Mosser D. M. (2005) J. Immunol. 175, 469–477 [DOI] [PubMed] [Google Scholar]
- 37.Berg D. J., Kühn R., Rajewsky K., Müller W., Menon S., Davidson N., Grünig G., Rennick D. (1995) J. Clin. Invest. 96, 2339–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Waal Malefyt R., Abrams J., Bennett B., Figdor C. G., de Vries J. E. (1991) J. Exp. Med. 174, 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiorentino D. F., Zlotnik A., Mosmann T. R., Howard M., O'Garra A. (1991) J. Immunol. 147, 3815–3822 [PubMed] [Google Scholar]
- 40.Takahashi K., Prinz M., Stagi M., Chechneva O., Neumann H. (2007) PLoS Med. 4, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tessarz A. S., Weiler S., Zanzinger K., Angelisová P., Horejsí V., Cerwenka A. (2007) J. Immunol. 178, 1991–1999 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.