Abstract
Plants induce immune responses against fungal pathogens by recognition of chitin, which is a component of the fungal cell wall. Recent studies have revealed that LysM receptor-like kinase 1/chitin elicitor receptor kinase 1 (LysM RLK1/CERK1) is a critical component for the immune responses to chitin in Arabidopsis thaliana. However, the molecular mechanism of the chitin recognition by LysM RLK1 still remains unknown. Here, we present the first evidence for direct binding of LysM RLK1 to chitin. We expressed LysM RLK1 fused with yeast-enhanced green fluorescent protein (LysM RLK1-yEGFP) in yeast cells. Binding studies using the solubilized LysM RLK1-yEGFP and several insoluble polysaccharides having similar structures showed that LysM RLK1-yEGFP specifically binds to chitin. Subsequently, the fluorescence microscopic observation of the solubilized LysM RLK1-yEGFP binding to chitin beads revealed that the binding was saturable and had a high affinity, with a Kd of ∼82 nm. This binding was competed by the addition of soluble glycol chitin or high concentration of chitin oligosaccharides having 4–8 residues of N-acetyl glucosamine. However, the competition of these chitin oligosaccharides is weaker than that of glycol chitin. These data suggest that LysM RLK1 has a higher affinity for chitin having a longer residue of N-acetyl glucosamine. We also found that LysM RLK1-yEGFP was autophosphorylated in vitro and that chitin does not affect the phosphorylation of LysM RLK1-yEGFP. Our results provide a new dimension to chitin elicitor perception in plants.
Keywords: Carbohydrate/Binding Protein, Immunology/Innate Immunity, Organisms/Plant, Receptors/Membrane, Receptors/Oligosaccharides, Signal Transduction/Protein Kinases
Introduction
When a plant is attacked by pathogens, it rapidly induces immune responses to stop the infection. A key step of the rapid induction of immune responses is a prompt and efficient detection of microbial invaders. In plants, this is achieved by pattern recognition receptors that recognize the conserved structures of the microbial pathogens. These conserved microbial structures are called pathogen-associated molecular patterns.
The pathogen-associated molecular patterns recognized by plants include lipopolysaccharides, peptidoglycan (PGN),3 flagellin, and bacterial elongation factor-Tu, which are derived from the bacteria (1, 2). Plants also recognize fungal pathogen-associated molecular patterns, such as chitin and ergosterol, which are components of the fungal cell wall and plasma membrane, respectively (1, 3). Recent studies have discovered the plant pattern recognition receptors participating in the perception of chitin elicitor (4–6).
A LysM motif-containing plasma membrane protein, CEBiP (chitin oligosaccharide elicitor-binding protein), participates in the perception of chitin oligosaccharides in rice (4). The CEBiP has two LysM motifs in the extracellular domain and lacks the intracellular kinase domain that is required for signal transduction. It has been demonstrated that CEBiP directly binds to chitin oligosaccharides and plays an essential role in the perception of chitin and the induction of immunity in rice (4).
On the other hand, more recently, two groups have independently reported that a LysM receptor-like kinase (RLK) plays a key role in chitin perception in Arabidopsis thaliana (5, 6). This receptor, LysM receptor-like kinase 1/chitin elicitor receptor kinase 1 (LysM RLK1/CERK1), is localized in the plasma membrane and has three LysM motifs in the extracellular domain as well as the intracellular kinase domain. Although these studies demonstrated that LysM RLK1 plays a crucial role in the immune response against fungal pathogens, the molecular mechanism of interactions of LysM RLK1 and chitin is still unknown.
The LysM motifs are known as GlcNAc-binding motifs, which occur frequently in bacterial lysins and bind to PGN (7). In plants, this motif is also present in a chitinase and directly binds to chitin or chitin oligosaccharides (8, 9).
Although LysM RLK1 has three LysM motifs in the extracellular domain, it remains unclear whether LysM RLK1 directly binds to chitin or acts via cooperation with another protein. An affinity-labeling experiment was unable to detect any specific binding protein to chitin in a membrane preparation from A. thaliana, although the same technique could detect CEBiP in plasma membrane of rice (5). Therefore, we investigated whether LysM RLK1 directly binds to chitin.
To exclude the potential effects of other plant components, we used the recombinant LysM RLK1 protein expressed in the yeast, Saccharomyces cerevisiae. Yeast is superior to Escherichia coli for the expression of recombinant membrane proteins because it has a highly regulated protein quality control system in the endoplasmic reticulum (10). In this study, we demonstrated the specific and direct binding of LysM RLK1 to chitin in vitro by binding assays using recombinant LysM RLK1 fused with yeast-enhanced green fluorescence protein (LysM RLK1-yEGFP). We also found that the full length of LysM RLK1-yEGFP was autophosphorylated without chitin and that chitin did not affect the phosphorylation of LysM RLK1-yEGFP in vitro.
EXPERIMENTAL PROCEDURES
Chitin and Chitosan Derivatives
chitin beads were purchased from New England Biolabs. Chitin oligosaccharides (degrees of polymerization = 2–6) and a chitosan oligosacchride (GlcN)6 were purchased from Seikagaku Kogyo. Colloidal chitin and colloidal chitosan were prepared as reported previously (11, 12). Glycol chitin was prepared from chitin according to the method of Yamada and Imoto (13). Reduced (GlcNAc)9, r(GlcNAc)9, was prepared from mono-N-acetylated chitononaose (GlcN)8-GlcNAc obtained by the method of Mitsutomi et al. (14). (GlcN)8-GlcNAc was reduced with sodium borohydride, and then the reduced (GlcN)8-GlcNAc was N-acetylated by a modification of the procedure of Barker et al. (15), as described previously (16).
Preparation of [14C]Glycol Chitin
Glycol chitosan was acetylated with [1-14C]acetic anhydride (America Radiolabeled Chemicals, Inc.) as follows. Three hundred μl of 0.04 m Na2CO3, 250 μl of methanol, and 3 μl of [1-14C]acetic anhydride (5 mCi/mmol) were added to 2 mg of glycol chitosan. After a 24-h incubation at 25 °C, the sample was passed through the Dowex 50 column. The elution evaporated three times with methanol to remove the unreacted [1-14C]acetic acid. After the evaporation, the [14C]glycol chitin was dissolved in water.
Expression and Purification of yEGFP
yEGFP was expressed in E. coli BL21 Star (DE3) (Invitrogen). First, yEGFP was cloned into pET43a(+) (Novagen, Madison, WI). To fuse yEGFP with an octahistidine tag at the C terminus and to remove an N-terminal Nus tag, the yeast recombinational cloning was carried out as reported previously (17), except that, instead of pSU0, pYES2/CT (Invitrogen) was used as a helper plasmid for the conversion of pET43a(+) into the yeast shuttle vector. The pET43a(+)-yEGFP were transformed into BL21 Star (DE3) by the standard method. The transformants were grown in the LB medium containing 100 μg/ml ampicillin to an A600 = 0.6 at 37 °C. Subsequently, isopropyl-1-thio-β-d-galactopyranoside was added at a final concentration of 1 mm, and the cells were grown at 15 °C for a further 20 h. The cells were broken by a French press, and yEGFP was purified by a nickel column as described previously (18). For the removal of imidazole, the eluted proteins were dialyzed against 200 volumes of phosphate-buffered saline containing 0.01 mm phenylmethylsulfonyl fluoride for 4 h at 4 °C with five buffer changes. The concentration of the proteins was determined by the Bradford method.
Cloning of LysM RLK1
LysM RLK1 was cloned into the expression vector, pDDGFP2 (10), to be fused with yEGFP and an octahistidine tag at the C terminus. First, the cDNA corresponding to LysM RLK1 was created by reverse transcription-PCR with SuperScript II reverse transcriptase (Invitrogen) using a gene-specific primer (5′-ATATAAATTTTCCCCCCGGCCGGACATAAGACTGA-3′) from 0.5 μg of the total RNA of A. thaliana (19). The open reading frame of LysM RLK1 was obtained after two rounds of PCR using PrimeSTAR GXL DNA Polymerase (TAKARA BIO, Ohtsu, Shiga, Japan). The first round of PCR amplified the open reading frame with the gene-specific primers 5′-ACTAGTGGATCCCCCATGAAGCTAAAGATTTCTCT-3′ and 5′-ATATAAATTTTCCCCCCGGCCGGACATAAGACTGA-3′, whereas the second round of PCR amplified it with the primers 5′-TCGACGGATTCTAGAACTAGTGGATCCCCC-3′ and 5′-AAATTGACCTTGAAAATATAAATTTTCCCC-3′ for providing this fragment with the ends homologous to pDDGFP2. Subsequently, the LysM RLK1 gene was subcloned into pDDGFP2 by the yeast recombinational cloning (10). The plasmid obtained from the yeast cells was cloned and propagated in E. coli DH5α (TOYOBO, Osaka, Japan) as we described previously (17). The sequence of insert was confirmed by a DNA sequencer (Applied Biosystems, Foster City, CA).
Expression and Purification of LysM RLK1-yEGFP Protein
The pDDGFP2 harboring LysM RLK1 (pDDGFP2-LysM RLK1) was retransformed into the S. cerevisiae strain BY2777 (MATa prb1-1122 prc1-407 pep4-3 ura3-52 leu2 trp1). Subsequently, for the expression of LysM RLK1-yEGFP, the transformants were grown in SC-Ura liquid medium containing 2% glucose, and the protein expression was induced as reported previously (10). The cells were harvested by centrifugation at 3000 × g for 5 min at 4 °C and were suspended in the lysis buffer containing 50 mm Tris-HCl (pH 7.5), 0.6 m sorbitol, 1 mm EDTA, and protease inhibitor mixture, complete EDTA-free (Roche Applied Science). After the addition of the same volume of chilled acid-washed glass beads, the cells were broken by three rounds of a Fast Prep cell disruptor (BIO 101, Vista, CA) at 5.5 ms−1 for 20 s with 2-min intervals on ice. After lysis, the lysate was centrifuged at 3000 × g for 10 min. The supernatant was further centrifuged at 21,040 × g for 1 h at 4 °C to pellet the membranes. The crude membranes were solubilized, and LysM RLK1 proteins were purified as reported previously (10), except that a detergent, lauryl dimethylamine N-oxide (LDAO), was used in solubilization and purification instead of N-dodecyl-β-d-maltoside. To remove imidazole, the eluted protein was dialyzed against 300 volumes of the binding buffer containing 50 mm Tris-HCl (pH 7.0), 0.05% LDAO, 150 mm NaCl, 5 mm MgCl2, 0.1 mm phenylmethylsulfonyl fluoride for 4 h at 4 °C with five buffer changes. The concentration of LysM RLK1-yEGFP protein was determined by comparing it with a yEGFP standard curve. The standard curve was generated by using known amounts of recombinant yEGFP purified from E. coli. 100 μl of the samples were transferred to a 96-well plate, and the yEGFP fluorescence in each well was read using a multimode plate reader (model DTX880, Beckman (Krefeld, Germany)), and the following settings were used: fluorescence intensity bottom method, 0.1-s integration time, 485-nm excitation filter, and 535-nm emission filter.
Analysis of the Proteins by In-gel Fluorescence
Protein samples were mixed with SDS sample buffer and then incubated at 45 °C for 3 min. After incubation, the samples were analyzed by SDS-PAGE. The GFP fluorescence was measured using a Typhoon 9200 fluorescence imager (GE Healthcare). The SDS-polyacrylamide gel was scanned using the green laser (532 nm) excitation source, and the resulting fluorescence emitted (526 nm, short pass filter) was recorded using a PMT (a value for sensitivity) of 800 V at a resolution of 100 μm. The in-gel fluorescence analysis was followed by CBB staining, using CBB G-250.
Ligand Specificity Assay
The binding specificity of LysM RLK1 protein for various ligands was examined by a polysaccharide binding assay, which was performed as reported previously (20, 21) with minor modifications. Thirty pmol of purified LysM RLK1-yEGFP or yEGFP (negative control) was mixed with 200 μg of insoluble polysaccharides, chitin beads, colloidal chitin, colloidal chitosan, or PGN derived from Staphylococcus aureus (Sigma) in a total volume of 200 μl of binding buffer incubated for 30 min by rotating end over end. The samples were centrifuged at 15,000 × g for 15 min at 4 °C. The pellets were washed three times with the ice-cold binding buffer and mixed with SDS sample buffer. The eluted proteins were electrophoresed, and the binding proteins were analyzed by in-gel fluorescence as described earlier. The proteins bound to the polysaccharides were quantified by ImageJ software (National Institutes of Health).
Detection of Chitin Bead-LysM RLK1 Interaction by Fluorescence Microscopy
Aliquots (1 μl) of chitin beads (50% slurry) were suspended in 50 μl of the binding buffer containing various concentrations (3.9, 7.8, 15.6, 31, 63, 125, 250, and 500 nm) of LysM RLK1-yEGFP or yEGFP. Subsequently, the suspensions were incubated for 20 min. In the case of the competition assay, glycol chitin, glycol chitosan, a chitosan oligosaccharide (GlcN)6 and various lengths of chitin oligosaccharides were incubated with 250 nm proteins before the addition of the chitin beads. The LysM RLK1 or yEGFP bound to the chitin were directly visualized using a fluorescence microscope (model BX-51; Olympus, Tokyo, Japan) with a fluorescence mirror unit, U-MWIB3 (Olympus). The images were taken with a Nikon Coolpix 4500 digital camera (Nikon, Tokyo, Japan), whose shutter speed and aperture were fixed at ⅛ and F3.1, respectively. The fluorescence intensities of the edges of the beads were analyzed by ImageJ software. The dissociation constant, Kd was determined by nonlinear regression using the GraphPad Prism software (GraphPad Software Inc.).
[14C]Glycol Chitin Binding Assay
Purified 40 pmol of LysM RLK1-yEGFP was bound to nickel-chelating Sepharose and suspended in 100 μl of the binding buffer. It was then incubated with 0.25, 0.5, 2.5, 5, 12.5, and 25 μg/ml [14C]glycol chitin with or without a 100-fold excess of unlabeled glycol chitin for 30 min at room temperature. The samples were washed three times with 5 ml of ice-cold binding buffer on GF/C glass microfiber filters (Whatman, Maidstone, UK). After the nickel-chelating Sepharoses on the filters were dried, the samples were quantified by scintillation counting. In the case of the competition assay, glycol chitin, glycol chitosan, and various lengths of chitin oligosaccharides were incubated with LysM RLK1-yEGFP on nickel-chelating Sepharose before the addition of the glycol chitin.
Phosphorylation Assay
The phosphorylation of LysM RLK1 was analyzed as previously described (22). Briefly, various concentrations of LysM RLK-yEGFP or yEGFP were incubated with or without 500 μm r(GlcNAc)9 or 25 μg/ml glycol chitin at 25 °C for 30 min in 20 μl of kinase assay buffer containing 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 0.1% LDAO, 10 mm MgCl2, 1 mm dithiothreitol, 50 μm unlabeled ATP, and 370 kBq of [γ-32P]ATP (111 TBq/mmol; PerkinElmer Life Sciences). The reactions were terminated by adding SDS sample buffer and boiled for 3 min, and the samples were separated by SDS-PAGE. The phosphorylated proteins were visualized by autoradiography after the gels were dried under the vacuum.
RESULTS
Expression and Purification of LysM RLK1-yEGFP
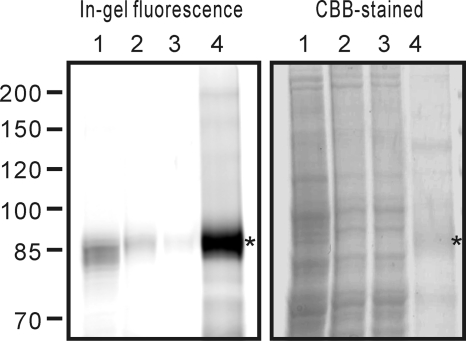
LysM RLK1 was expressed fused with yEGFP and octahistidine tag at the C terminus in S. cerevisiae. The use of yEGFP allows easy and specific detection of the target protein without antibodies and makes optimization of the protein expression and purification easy. We expressed the LysM RLK1-yEGFP fusion protein in S. cerevisiae strain, BY2777, following the procedure described previously (10). After the induction, we could detect the yEGFP fluorescence on the plasma membrane of the cells by a fluorescence microscope (data not shown). The membrane proteins were then analyzed by the in-gel fluorescence after SDS-PAGE, and an ∼94 kDa protein band corresponding to the LysM RLK1-yEGFP was detected in the membrane fraction (Fig. 1, left, lane 1).
FIGURE 1.
Expression and purification of LysM RLK1. LysM RLK1-yEGFP overexpressed in yeast membrane was solubilized and purified by a nickel column. The samples were separated on a 6% SDS gel and analyzed by in-gel fluorescence (left) and CBB staining (right). Lane 1, crude membrane containing LysM RLK1-yEGFP; lane 2, solubilized membrane proteins by 1% LDAO; lane 3, flow-through of the nickel column; lane 4, eluted proteins from the nickel column. *, LysM RLK1 yEGFP.
Subsequently, to select an adequate detergent for solubilization of LysM RLK1-yEGFP, several kinds of detergents were examined including LDAO, cholic acid, deoxycholic acid, CHAPS, and Triton X-100 at 1% (w/v). The amounts of solubilized LysM RLK1 were determined by measuring the florescence of the solubilized proteins with a plate reader as described previously (10). We found that the LDAO was the most effective detergent for solubilizing LysM RLK1-yEGFP from the membranes (data not shown).
The membrane proteins were then solubilized by incubating with 1% LDAO, and LysM RLK1-yEGFP protein was purified by the nickel column chromatography. The samples were analyzed by in-gel fluorescence and CBB-stained after the electrophoresis (Fig. 1). We had observed a slight shift in band size of LysM RLK1-yEGFP in the crude membrane fraction on SDS-PAGE (Fig. 1, left, lane 1). Not only the LysM RLK1-yEGFP protein but also other proteins in the crude membrane fraction migrated faster than that in the LDAO-solubilized fraction (Fig. 1, right, lane 1). Membrane contaminant might affect the band patterns because we could detect the non-shifted band of LysM RLK1-yEGFP after the removal of the LDAO-insoluble fraction by ultracentrifuge (Fig. 1, lane 2). The amount of LysM RLK1-yEGFP protein was determined by comparison with the yEGFP standard curve, because the CBB staining is poor and inconsistent for many membrane protein samples (10). We obtained about 0.3–0.4 mg of LysM RLK1-yEGFP protein per liter of culture after the purification.
Ligand Specificity Assay
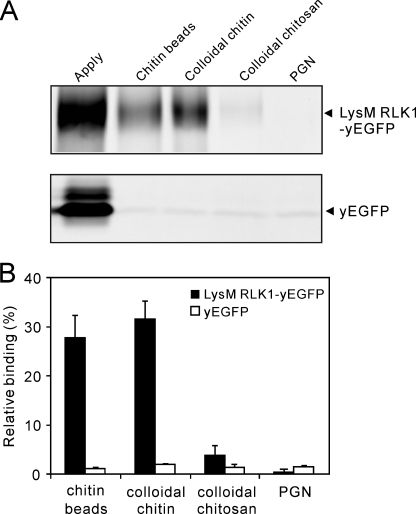
To study the biochemical interactions between LysM RLK1 and chitin, we first examined the binding of LysM RLK1-yEGFP to insoluble chitin, such as chitin beads and colloidal chitin, and other insoluble polysaccharides, colloidal chitosan, and PGN, containing a common GlcNAc backbone. These insoluble polysaccharides were incubated with the same amount of LysM RLK1-yEGFP protein (Fig. 2A, Apply lane). Subsequently, the proteins bound to the polysaccharides were analyzed by in-gel fluorescence after SDS-PAGE. The LysM RLK1-yEGFP was largely specific for chitin. It strongly bound the chitin beads and colloidal chitin (Fig. 2, A (top) and B). However, we could detect no or little binding of the LysM RLK1-yEGFP to colloidal chitosan and PGN (Fig. 2, A (top) and B). As a negative control, these polysaccharides were also incubated with the yEGFP protein. However, we could detect little binding of yEGFP to any polysaccharides (Fig. 2, A (bottom) and B). Thus, these data indicate that LysM RLK1 specifically and directly binds to chitin.
FIGURE 2.
LysM RLK1 specifically bound to chitin but not to chitosan and PGN. A, binding of LysM RLK1-yEGFP or yEGFP to various polysaccharides was analyzed by in-gel fluorescence. 30 pmol of purified LysM RLK1-yEGFP (top) or yEGFP (bottom) were incubated with 200 μg of chitin beads, colloidal chitin, colloidal chitosan, and PGN. After the polysaccharides were washed with the buffer, the proteins bound to polysaccharides were eluted and electrophoresed on a 6% SDS gel for LysM RLK1 and a 12% gel for yEGFP. B, the amounts of the proteins bound to polysaccharides were analyzed by ImageJ. The experiment was repeated three times. The percentage of binding protein rate was calculated from the ratio of band fluorescence of the proteins eluted from polysaccharides and the applied proteins.
Binding of LysM RLK1 to Chitin Beads
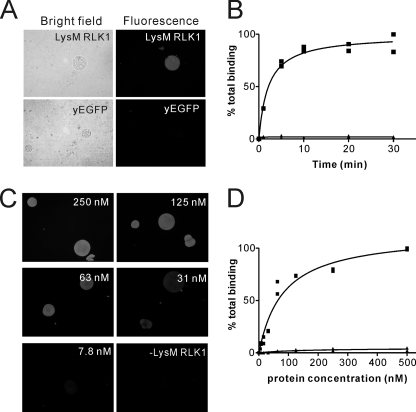
To investigate the binding property of LysM RLK1 to chitin, we subsequently directly visualized the binding of LysM RLK1 to chitin beads. Chitin beads are fine chitin particles (50–250 μm), and recent studies have used them in the bioassay or binding assay instead of chitin (21, 23). We incubated LysM RLK1-yEGFP with the chitin beads and directly observed their interaction using a fluorescence microscope. When 500 nm LysM RLK1-yEGFP was incubated with the chitin beads, strong green florescence was detected on the chitin beads (Fig. 3A). However, no fluorescence was detected on the beads when they were incubated with yEGFP (Fig. 3A). The protein binding was completed within 20 min of the start of incubation (Fig. 3B). Accordingly, all of the subsequent binding studies were performed at 20 min after the addition of chitin beads.
FIGURE 3.
Fluorescence microscopic visualization of the interaction of LysM RLK1 and chitin beads. A, 500 nm LysM RLK1-yEGFP or yEGFP was incubated with chitin beads, and then the interactions of LysM RLK1-yEGFP or yEGFP with chitin beads were observed by the fluorescence microscope. After 20-min incubations, the strong yEGFP fluorescence was observed on the chitin beads incubated with LysM RLK1-yEGFP, but no fluorescence was detected on the beads incubated with yEGFP. B, the time course of the interaction of LysM RLK1-yEGFP and chitin beads was examined. The samples were observed after 1, 5, 10, 20, and 30 min of the incubation. The amount of the proteins bound to chitin beads was analyzed by ImageJ. C and D, the various concentrations of LysM RLK1-yEGFP (3.9, 7.8, 15.6, 31, 63, 125, 250, or 500 nm) were incubated with chitin beads. Only the beads that were incubated without or with 250, 125, 63, 31, or 7.8 nm LysM RLK1-yEGFP are shown (C). The amount of the proteins bound to chitin beads was analyzed by ImageJ (D). ■, LysM RLK1-yEGFP; ▴, yEGFP. These experiments were performed twice with similar results.
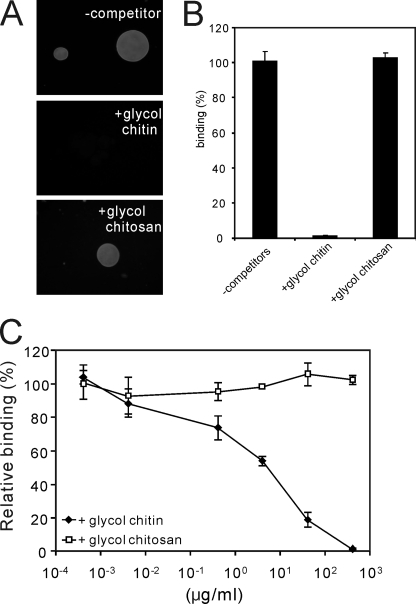
To determine whether the binding was saturable, as would be expected for a receptor-ligand interaction, the chitin beads were incubated with various concentrations of LysM RLK1-yEGFP (Fig. 3C). We found that the binding was saturable, and the dissociation constant (Kd) of the proteins was ∼82 nm (Fig. 3, C and D). Furthermore, fluorescence of chitin beads was completely diminished by the addition of an excess of glycol chitin but not glycol chitosan (400 μg/ml), which are known as water-soluble chitin and chitosan derivatives (13), respectively (Fig. 4, A and B). The competition by glycol chitin and glycol chitosan was assessed in experiments in which increasing amounts of them competed with LysM RLK1-yEGFP for binding to chitin beads. Glycol chitin competed for the binding to chitin beads with a half-maximal inhibitory concentration (IC50) of 4 μg/ml (Fig. 4C). These data show that LysM RLK1 specifically and directly binds to chitin.
FIGURE 4.
Competition of glycol chitin and glycol chitosan for the binding of LysM RLK1-yEGFP to chitin beads. A, 250 nm LysM RLK1-yEGFP incubated with 400 μg/ml glycol chitin and glycol chitosan and then chitin beads was added to the solution and incubated for 20 min. After the incubation, the beads were directly observed by a fluorescence microscope, and the fluorescence images are shown. B, the amount of proteins bound to chitin beads was analyzed by ImageJ. C, competition of the binding of LysM RLK1-yEGFP to chitin beads by glycol chitin and glycol chitosan was analyzed by the addition of increasing concentrations (400, 40, 4, 0.4, 0.04, and 0.004 μg/ml) of them. The experiments were repeated three times with similar results. The relative percentage binding was calculated by comparison to the fluorescence intensity of beads in the absence of competitors (100% binding).
Competition of LysM RLK1 Binding to Chitin Beads by Chitin Oligosaccharides
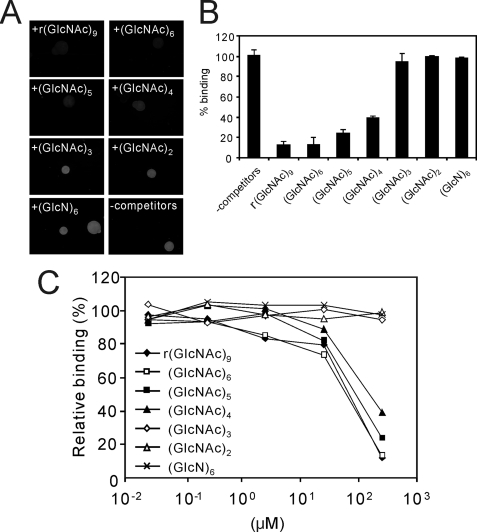
It is known that the biological activity of the soluble chitin oligosaccharides depends on their degree of polymerization. Therefore, we tested the chitin oligosaccharides of different lengths for their efficiency to compete for the binding of LysM RLK1 to chitin beads. After 250 μm concentrations of the various lengths of chitin oligosaccharides were incubated with 250 nm LysM RLK1-yEGFP, the chitin beads were added to this solution, and the binding of LysM RLK1-yEGFP to chitin beads was analyzed by the fluorescence microscopy. We found that the competition of chitin oligosaccharides occurred in a size-dependent manner. r(GlcNAc)9 (an analog of (GlcNAc)8), (GlcNAc)6, and (GlcNAc)5 strongly competed with LysM RLK1-yEGFP for >75% of the binding to the chitin beads (Fig. 5, A and B). Although (GlcNAc)6 strongly competed the binding, (GlcN)6 did not compete for the binding. This finding also indicates that the LysM RLK1 specifically binds chitin oligosaccharides. (GlcNAc)4 also competed with it for about 60% of the binding (Fig. 5, A and B). However, (GlcNAc)3 and (GlcNAc)2 did not show noticeable competition with regard to the binding of LysM RLK1-yEGFP to the chitin beads (Fig. 5, A and B).
FIGURE 5.
Competition of chitin oligosaccharides and a chitosan oligosaccharide (GlcN)6 for the binding of LysM RLK1-yEGFP to chitin beads. A, 250 nm LysM RLK1-yEGFP was incubated with a 250 μm concentration of various chitin oligosaccharides and a chitosan oligosaccharide (GlcN)6 and subsequently, the chitin beads were added to the solution and incubated for 20 min. After the incubation, the beads were directly observed by a fluorescence microscope. B, the amounts of LysM RLK1-yEGFP bound to chitin beads were analyzed by ImageJ. C, competition of the binding of LysM RLK1 to chitin beads by chitin and chitosan oligosaccharides was analyzed by the addition of increasing concentrations (500, 50, 5, 0.5, and 0.05 μm) of them. The experiments were repeated three times with similar results. The relative percentage binding was calculated in the same way as in Fig. 4.
We also assessed the competition by the serial diluted chitin oligosaccharides. Even larger chitin oligosaccharides could weakly compete for the binding of LysM RLK1-yEGFP to chitin beads with an IC50 of about 100 μm (Fig. 5C).
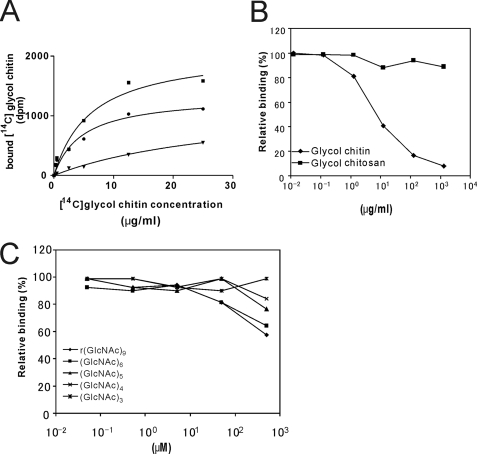
Binding of 14C-Labeled Glycol Chitin to LysM RLK1-yEGFP
14C-Labeled glycol chitin was used to confirm the binding of chitin to LysM RLK1. The purified LysM RLK1-yEGFP was bound to nickel-chelating Sepharose and suspended in 100 μl of binding buffer containing the various concentrations of [14C]glycol chitin with or without a 100-fold excess of unlabeled glycol chitin. The binding of [14C]glycol chitin to LysM RLK1-yGFP was saturable, and a Kd value was 3.5 μg/ml (Fig. 6A). We also performed the competition assay with the serial diluted glycol chitin, glycol chitosan, and oligosaccharides of various lengths. Glycol chitin but glycol chitosan competed for the binding of [14C]glycol chitin to LysM RLK1-yEGFP with an IC50 of 10 μg/ml (Fig. 6B). However, even larger chitin oligosaccharides could weakly compete for the binding of it at a higher concentration, such as 500 μm (Fig. 6C).
FIGURE 6.
Binding of [14C]glycol chitin to LysM RLK1-yEGFP. A, 40 pmol of LysM RLK1-yEGFP bound to nickel-chelating Sepharose was incubated with 0.025, 0.5, 2.5, 5, 12.5, and 25 μg/ml at room temperature for 30 min in the absence of (■, total binding) or presence of (▾, nonspecific binding) 100-fold unlabeled glycol chitin. To determine the specific binding (●), nonspecific binding was subtracted from total binding. B, competition of the binding of [14C]glycol chitin to LysM RLK1-yEGFP by the unlabeled glycol chitin. 40 pmol of LysM RLK1-yEGFP bound to nickel-chelating Sepharose was incubated with various concentrations of glycol chitin and glycol chitosan (0.125, 1.25, 12.5, 125, and 1250 μg/ml) and then incubated with 12.5 μg/ml [14C]glycol chitin at room temperature for 30 min. C, competition of the binding of [14C]glycol chitin to LysM RLK1-yEGFP by chitin oligosaccharides. 40 pmol of LysM RLK1-yEGFP bound to nickel-chelating Sepharose was incubated with various concentrations of chitin oligosaccharides (0.05, 0.5, 5, 50, and 500 μm) and then incubated with 12.5 μg/ml [14C]glycol chitin at room temperature for 30 min. The relative percentage binding was calculated by comparison with the dpm in the absence of competitors (100% binding). The experiments were repeated three times (A) or twice (B and C), and the averages of the results are shown.
Phosphorylation of LysM RLK1
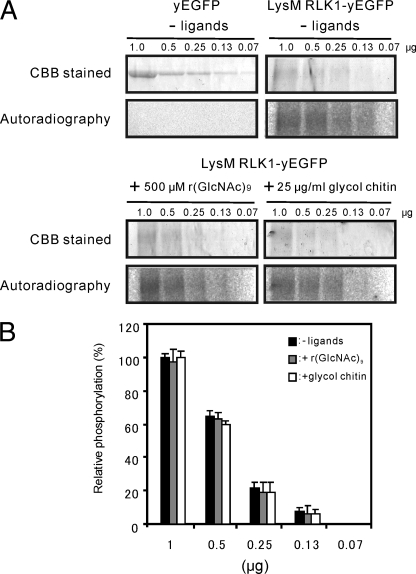
Many animal receptor kinases exhibit ligand-dependent autophosphorylation (24). To determine whether the early events in chitin perception of plants share this mechanism, we carried out an in vitro kinase assay. Various concentrations of LysM RLK1-yEGFP and yEGFP were assayed for autophosphorylation. The samples were electrophoresed and analyzed by autoradiography. The bands corresponding to LysM RLK1-yEGFP were detected in a dose-dependent manner without chitin, but those of yEGFP were not detected (Fig. 7A). Furthermore, to examine whether its ligand, chitin, affects the phosphorylation state of LysM RLK1, LysM RLK1-yEGFP was incubated with 500 μm r(GlcNAc)9 or 25 μg/ml glycol chitin and assayed for autophosphorylation. However, neither of them affected the phosphorylation of LysM RLK1-yEGFP (Fig. 7, A and B). These results demonstrate that the LysM RLK1 was autophosphorylated without chitin and that the addition of chitin does not affect the phosphorylation state of it in vitro.
FIGURE 7.
Kinase activity of the full length of LysM RLK1-yEGFP in a dose-dependent manner. A, various concentrations of LysM RLK1-yEGFP and yEGFP were subjected to an autophosphorylation assay without chitin, or various concentrations of LysM RLK1-yEGFP were also subjected to an autophosphorylation assay with 500 μm r(GlcNAc)9 or 25 μg/ml glycol chitin. The samples were electrophoresed on a 6% SDS gel for LysM RLK1 and a 12% gel for yEGFP. After the SDS-PAGE, the gels were CBB-stained, dried under the vacuum, and analyzed by autoradiography. The gels were exposed to an imaging plate for the same time period. B, the relative amount of phosphorylated LysM RLK1-yEGFP bands were determined by ImageJ and calculated by comparison with the band intensity in 1 μg of LysM RLK1-yEGFP without chitin (100% phosphorylation). The experiments were repeated twice, and similar results were obtained.
DISCUSSION
Recent studies have identified a plant receptor, LysM RLK1, playing an essential role in defense responses to chitin in A. thaliana (5, 6). Although these studies revealed that the mutations in LysM RLK1 abolished the defense responses against chitin, the molecular mechanisms of the perception of chitin by LysM RLK1 were not described. In this study, we showed that LysM RLK1 specifically and directly binds to chitin.
We first examined whether LysM RLK1 binds specifically and directly to chitin, using various polysaccharides, chitin, chitosan, and PGN, which has a common backbone, GlcNAc. Interestingly, although these polysaccharides have similar structures, we could detect the strong binding of LysM RLK1-yEGFP only to chitin, chitin beads, and colloidal chitin and not colloidal chitosan and PGN, at least under our experimental conditions (Fig. 2). This result demonstrates that LysM RLK1 may recognize the acetyl group of N-acetylglucosamine residues of chitin, and it may be inhibited by the bulky peptide group cross-linked to N-acetyl-muramic acid residues of PGN.
The PGN is a pathogen-associated molecular pattern derived from bacteria. Therefore, this result is consistent with a previous study showing that the mutations in the LysM RLK1 gene did not affect the defense response against bacteria (6). Moreover, a previous study also suggested that the recognition site of chitin is different from that of PGN in the leaf of A. thaliana (2). The LysM motifs are known as the PGN binding motif, and it is known that there are four other putative LysM RLKs in A. thaliana (25). Hence, we speculate that some of them may be involved in PGN perception in A. thaliana.
To examine the binding property of LysM RLK1, we next directly visualized the binding of LysM RLK1 to chitin beads. The binding studies on many chitin-binding proteins were performed using powdered chitin or colloidal chitin (26, 27). However, these methods required a large amount of protein, and it was difficult to detect a small amount of proteins bound to chitin because the binding proteins were quantified by measuring the UV absorbance at 280 nm. However, the direct observation by fluorescence microscopy revealed the binding of LysM RLK-yEGFP to chitin beads in only 50 μl of the sample with high sensitivity. Moreover, fluorescence of bound proteins could also be monitored without the removal of unbound proteins. Therefore, this method is considered to be better than the conventional methods.
The binding of LysM RLK1 to chitin beads was completed within 20 min of the start of incubation and the half-maximal binding is about 3 min. This is unexpectedly slow for simple ligand binding. This result shows that the interaction of LysM RLK1 and chitin is not a simple ligand binding reaction, and this interaction may be accompanied with a conformational change of receptor protein as often seen in other receptor-ligand interactions (28, 29). Therefore, it takes several minutes to reach the equilibrium.
This binding was saturable, and the dissociation constant (Kd) was ∼82 nm (Fig. 3, C and D). These values might be different from that of those in vivo, because we used soluble LysM RLK1-yEGFP that was not anchored. A previous study reported that the dimerization of RLK or its anchoring is important for high affinity interaction of the receptor and its ligand (30). However, the Kd value of LysM RLK1 for chitin beads was lower than that of other chitin-binding proteins to chitin beads (21), and this binding was eliminated by the excess amount of 400 μg/ml glycol chitin but glycol chitosan. Therefore, we conclude that LysM RLK1 directly and specifically binds to chitin.
We also examine the direct binding of [14C]glycol chitin with LysM RLK1-yEGFP. The binding of [14C]glycol chitin to LysM RLK1-yGFP was saturable, and the Kd value was 3.5 μg/ml (Fig. 6A). Previous research studied elicitor activity of A. thaliana by using glycol chitin in a concentration of about 0.05% (500 μg/ml) (31, 32). Moreover, chitin was previously shown to induce expression of the gene, such as lectin-like protein or zinc finger protein, in A. thaliana (33). Its half-maximal effective concentration (EC50) was more than 1 μg/ml (33). Therefore, the Kd value may be in the physiological range.
Subsequently, we tested the chitin oligosaccharides of different lengths for their efficiency to compete with the binding of LysM RLK1 to chitin beads. Two hundred fifty μm of the r(GlcNAc)9, (GlcNAc)6, and (GlcNAc)5 strongly competed for the binding of LysM RLK1-yEGFP to chitin beads (Fig. 5, A and B). On the other hand, (GlcNAc)3 and (GlcNAc)2 did not show noticeable competition with regard to the binding of LysM RLK1-yEGFP to the chitin beads (Fig. 5, A and B). The biological activity of the chitin oligomers was observed to depend on their degree of polymerization. A previous study showed that the larger chitin oligosaccharides (degree of polymerization = 6–8) had the most effective activity in A. thaliana (33). However, smaller chitin oligosaccharides (degree of polymerization = 2–5) had a little biological activity (33). The competition efficiency of the various lengths of chitin oligosaccharides beads was consistent with the previous data, except that the (GlcNAc)5 had a relatively higher competition activity in our experiment. This might be due to the difference in the efficiency of the length of the chitin oligosaccharides on the binding and biological activity.
In the competition assay of Fig. 5, although the larger chitin oligosaccharides (degree of polymerization = 4–8) competed for the binding of LysM RLK1-yEGFP to chitin beads at their higher concentration of 250 μm (Fig. 5B), the competition of these larger chitin oligosaccharides is also weaker than that of glycol chitin. IC50 of r(GlcNAc)9 and (GlcNAc)6 is about 100 μm in Fig. 5B, and that is also about 180 and 120 μg/ml, respectively, in weight per volume concentration units. This value is much higher than that of glycol chitin (IC50 = 4 μg/ml).
In the competition assay of Fig. 6, chitin oligosaccharides also weakly competed for the binding of [14C]glycol chitin to LysM RLK1-yEGFP (Fig. 6, B and C). The IC50 values in the competition assay of Fig. 6 are different from those in competition assays in Figs. 4 and 5. The discrepancy of the IC50 may be caused by the use of different assays. However, in both cases, the competition of chitin oligosaccharides is much weaker than that of glycol chitin. These data show that LysM RLK1 has a higher affinity for chitin polymer than chitin oligosaccharides.
It is proposed that a single LysM motif binds 4–5 residues of N-acetyl glucosamine (9, 34). It is also known that CEBiP, which directly binds (GlcNAc)8 in rice, has two extracellular LysM motifs (4). On the other hand, LysM RLK1 has three LysM extracellular motifs. Therefore, it may have the binding capacity for 12–15 residues of N-acetyl glucosamine. CEBiP may be a main binding protein for (GlcNAc)8 in rice because a CEBiP RNA interference plant lost the (GlcNAc)8 binding activity (4). It is also shown that binding protein(s) or receptor(s) for (GlcNAc)8 in rice binds (GlcNAc)8 with about 2000- and 10,000-fold higher affinities than (GlcNAc)5 and (GlcNAc)4, respectively (35). Therefore, CEBiP may bind (GlcNAc)8 with both of its two LysM motifs, and the binding of CEBiP to chitin oligosaccharides having fewer than 8 residues would be unstable. In our competition assay, even relatively larger chitin oligosaccharides, such as r(GlcNAc)9 or (GlcNAc)6, weakly competed the binding of LysM RLK1-yEGFP to chitin beads or [14C]glycol chitin. In addition, we also examined the direct binding of [14C]r(GlcNAc)9 to LysM RLK1-yEGFP, but we could not detect the binding of it to LysM RLK1-yEGFP at a few μm range (data not shown). These results are also consistent with the previous result, which failed to detect any specific binding protein to (GlcNAc)8 in a membrane preparation from A. thaliana (5). It may be because LysM RLK1 recognizes chitin oligosaccharides with more than 8 residues of N-acetyl glucosamine with its three LysM motifs that LysM RLK1 very weakly binds to chitin oligosaccharides having 8 or fewer N-acetyl glucosamine residues. Previous studies showed that (GlcNAc)8 binds to various plant membranes with a high affinity and is enough for the activation of innate immunity in plants (33, 35–37). However, it is unknown how the plant membrane recognizes chitin oligosaccharides having more than 8 residues and induces the innate immunity because it is difficult to obtain enough of these large chitin oligosaccharides for the assays.
Although LysM RLK1 weakly binds chitin oligosaccharides having 8 or fewer N-acetyl glucosamine residues, previous results showed that LysM RLK1 is also essential for (GlcNAc)8 elicitor signaling (5, 6). It is known that the TLR4 (Toll-like receptor 4)-MD2 receptor complex recognizes lipopolysaccharides, and other proteins LBP and CD14 increase the lipopolysaccharide sensitivity of TLR4-MD2 (38). Therefore, we speculate that there might be other receptor molecules like CEBiP that can bind (GlcNAc)8 also in A. thaliana, and these receptors condense the (GlcNAc)8 molecules on the plasma membrane and allows LysM RLK1 to bind (GlcNAc)8 to activate the chitin elicitor signaling.
We also examined the kinase activity of LysM RLK1 because the kinase activity is considered to be important for signal transductions. When various concentrations of LysM RLK1-yEGFP were incubated with its substrate [γ-32P]ATP, we detected specific autophosphorylation bands corresponding to LysM RLK1-yEGFP by autoradiography (Fig. 7A). This result is consistent with the previous result, which demonstrated the kinase activity of the intracellular domain of LysM RLK1 expressed in E. coli (5). We subsequently examined whether its ligand chitin oligosaccharides or glycol chitin affected the kinase activity of LysM RLK1. Many animal receptor kinases are known to exhibit ligand-dependent autophosphorylation (24). However, neither r(GlcNAc)9 nor glycol chitin affected the autophosphorylation activity of LysM RLK1 in vitro (Fig. 7, A and B). We presume that LysM RLK1 may require other endogenous components to activate its intracellular kinase domain. A recent study reported that the brassinolide receptor BRI1 is basally phosphorylated, and its perception of brassinolide induces the phosphorylation and activation of BRI1 by a regulator component, BAK1 (39).
In legumes, it is known that other LysM RLKs, such as NFR1 and NFR5, play important roles in the perception of Nod factors, which are the lipochitin oligosaccharides secreted by symbiotic bacteria, and in the symbiotic nitrogen fixation with rhizobia (40, 41). However, it remains unclear whether these LysM RLKs directly bind to Nod factors. Interestingly, NFR1 is found to be relatively close to LysM RLK1 in the evolution of plant LysM RLKs (25). Thus, our result may be useful for examining the interaction of NFR1 and Nod factors.
In this study, we showed that LysM RLK1 directly binds to chitin. This is the first report demonstrating that LysM RLK1 directly binds to chitin, and this result may provide a new insight into chitin elicitor perception as well as the interactions of LysM RLKs and their ligands in plants.
Acknowledgments
We thank Dr. Y. Hayakawa and Dr. S. Agarie for the use of the fluorescence plate reader DTX880 and Typhoon 9200, respectively, and Dr. D. Drew for providing the plasmid pDDGFP2 and for technical suggestions. We are grateful to Dr. K. Watanabe, Dr. F. Yagi, Dr. Y. Hagihara, and Dr. S. Katayama for helpful discussions. We also thank K. Horie for technical support in the construction of the LysM RLK1 expression plasmid. The BY2777 was provided by the Yeast Genetic Resource Center.
This work was supported in part by Grant-in-aid for Scientific Research (C) from the Japan Society for the Promotion of Science 17580082 (to Y. N.) and the Foundation for Research Fellowships of Japan Society for the Promotion of Science for Young Scientists (DC1) Grant 19-8266 (to E. I.).
- PGN
- peptidoglycan
- RLK
- receptor-like kinase
- LDAO
- lauryl dimethylamine N-oxide
- r(GlcNAc)9
- reduced (GlcNAc)9
- CBB
- Coomassie Brilliant Blue
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Zipfel C. (2009) Curr. Opin. Plant Biol. 12, 414–420 [DOI] [PubMed] [Google Scholar]
- 2.Gust A. A., Biswas R., Lenz H. D., Rauhut T., Ranf S., Kemmerling B., Götz F., Glawischnig E., Lee J., Felix G., Nürnberger T. (2007) J. Biol. Chem. 282, 32338–32348 [DOI] [PubMed] [Google Scholar]
- 3.Granado J., Felix G., Boller T. (1995) Plant Physiol. 107, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miya A., Albert P., Shinya T., Desaki Y., Ichimura K., Shirasu K., Narusaka Y., Kawakami N., Kaku H., Shibuya N. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan J., Zhang X. C., Neece D., Ramonell K. M., Clough S., Kim S. Y., Stacey M. G., Stacey G. (2008) Plant Cell 20, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buist G., Steen A., Kok J., Kuipers O. P. (2008) Mol. Microbiol. 68, 838–847 [DOI] [PubMed] [Google Scholar]
- 8.Onaga S., Taira T. (2008) Glycobiology 18, 414–423 [DOI] [PubMed] [Google Scholar]
- 9.Ohnuma T., Onaga S., Murata K., Taira T., Katoh E. (2008) J. Biol. Chem. 283, 5178–5187 [DOI] [PubMed] [Google Scholar]
- 10.Newstead S., Kim H., von Heijne G., Iwata S., Drew D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13936–13941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeuniaux C. (1958) Arch. Int. Physiol. Biochim. 66, 408–427 [DOI] [PubMed] [Google Scholar]
- 12.Yabuki M., Hirano M., Ando A., Fujii T., Amemiya Y. (1987) Tech. Bull. Fac. Hort. Chiba Univ. 39, 23–27 [Google Scholar]
- 13.Yamada H., Imoto T. (1981) Carbohydr. Res. 92, 160–162 [DOI] [PubMed] [Google Scholar]
- 14.Mitsutomi M., Takahara S., Kobayashi S., Ueda M., Arai M., Miyatake K. (2002) Chitin Chitosan. Res. 8, 228–229 [Google Scholar]
- 15.Barker S. A., Foster A. B., Stacey M., Webber J. M. (1958) J. Chem. Soc. 1958, 2218–2227 [Google Scholar]
- 16.Mitsutomi M., Ohtakara A., Fukamizo T., Goto S. (1990) Agric. Biol. Chem. 54, 871–877 [PubMed] [Google Scholar]
- 17.Iizasa E., Nagano Y. (2006) BioTechniques 40, 79–83 [DOI] [PubMed] [Google Scholar]
- 18.Kohara J., Tsuneyoshi N., Gauchat J. F., Kimoto M., Fukudome K. (2006) Protein Expr. Purif. 49, 276–283 [DOI] [PubMed] [Google Scholar]
- 19.Carninci P., Nishiyama Y., Westover A., Itoh M., Nagaoka S., Sasaki N., Okazaki Y., Muramatsu M., Hayashizaki Y. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjoelker L. W., Gosting L., Frey S., Hunter C. L., Trong H. L., Steiner B., Brammer H., Gray P. W. (2000) J. Biol. Chem. 275, 514–520 [DOI] [PubMed] [Google Scholar]
- 21.Hardt M., Laine R. A. (2004) Arch. Biochem. Biophys. 426, 286–297 [DOI] [PubMed] [Google Scholar]
- 22.Nam K. H., Li J. (2002) Cell 110, 203–212 [DOI] [PubMed] [Google Scholar]
- 23.Reese T. A., Liang H. E., Tager A. M., Luster A. D., Van Rooijen N., Voehringer D., Locksley R. M. (2007) Nature 447, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlessinger J. (2000) Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 25.Zhang X. C., Wu X., Findley S., Wan J., Libault M., Nguyen H. T., Cannon S. B., Stacey G. (2007) Plant Physiol. 144, 623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaaje-Kolstad G., Houston D. R., Riemen A. H., Eijsink V. G., van Aalten D. M. (2005) J. Biol. Chem. 280, 11313–11319 [DOI] [PubMed] [Google Scholar]
- 27.Joshi M. C., Sharma A., Kant S., Birah A., Gupta G. P., Khan S. R., Bhatnagar R., Banerjee N. (2008) J. Biol. Chem. 283, 28287–28296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheer J. M., Ryan C. A. (1999) Plant Cell 11, 1525–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones D. G., Boller T., Felix G. (2006) Cell 125, 749–760 [DOI] [PubMed] [Google Scholar]
- 30.Shimosato H., Yokota N., Shiba H., Iwano M., Entani T., Che F. S., Watanabe M., Isogai A., Takayama S. (2007) Plant Cell 19, 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim C. Y., Gal S. W., Choe M. S., Jeong S. Y., Lee S. I., Cheong Y. H., Lee S. H., Choi Y. J., Han C. D., Kang K. Y., Cho M. J. (1998) Plant Mol. Biol. 37, 523–534 [DOI] [PubMed] [Google Scholar]
- 32.Park H. C., Kim M. L., Kang Y. H., Jeon J. M., Yoo J. H., Kim M. C., Park C. Y., Jeong J. C., Moon B. C., Lee J. H., Yoon H. W., Lee S. H., Chung W. S., Lim C. O., Lee S. Y., Hong J. C., Cho M. J. (2004) Plant Physiol. 135, 2150–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B., Ramonell K., Somerville S., Stacey G. (2002) Mol. Plant Microbe Interact 15, 963–970 [DOI] [PubMed] [Google Scholar]
- 34.Mulder L., Lefebvre B., Cullimore J., Imberty A. (2006) Glycobiology 16, 801–809 [DOI] [PubMed] [Google Scholar]
- 35.Shibuya N., Ebisu N., Kamada Y., Kaku H., Cohn J., Ito Y. (1996) Plant Cell Physiol. 37, 894–898 [DOI] [PubMed] [Google Scholar]
- 36.Day R. B., Okada M., Ito Y., Tsukada K., Zaghouani H., Shibuya N., Stacey G. (2001) Plant Physiol. 126, 1162–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albert P., Miya A., Hiratsuka K., Kawakami N., Shibuya N. (2006) Plant Biotechnol. 23, 459–466 [Google Scholar]
- 38.Jerala R. (2007) Int. J. Med. Microbiol. 297, 353–363 [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., Huber S. C., Clouse S. D. (2008) Dev. Cell 15, 220–235 [DOI] [PubMed] [Google Scholar]
- 40.Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003) Nature 425, 585–592 [DOI] [PubMed] [Google Scholar]
- 41.Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003) Nature 425, 637–640 [DOI] [PubMed] [Google Scholar]