Abstract
Yeast Zuotin and Ssz are members of the conserved Hsp40 and Hsp70 chaperone families, respectively, but compared with canonical homologs, they atypically form a stable heterodimer termed ribosome-associated complex (RAC). RAC acts as co-chaperone for another Hsp70 to assist de novo protein folding. In this study, we identified the molecular basis for the unusual Hsp70/Hsp40 pairing using amide hydrogen exchange (HX) coupled with mass spectrometry and mutational analysis. Association of Ssz with Zuotin strongly decreased the conformational dynamics mainly in the C-terminal domain of Ssz, whereas Zuotin acquired strong conformational stabilization in its N-terminal segment. Deletion of the highly flexible N terminus of Zuotin abolished stable association with Ssz in vitro and caused a phenotype resembling the loss of Ssz function in vivo. Thus, the C-terminal domain of Ssz, the N-terminal extension of Zuotin, and their mutual stabilization are the major structural determinants for RAC assembly. We furthermore found dynamic changes in the J-domain of Zuotin upon complex formation that might be crucial for RAC co-chaperone function. Taken together, we present a novel mechanism for converting Zuotin and Ssz chaperones into a functionally active dimer.
Keywords: Chaperones, Chaperones/Protein Folding, Chaperones/Heat Shock, Protein/Folding, Protein/Protein-Protein Interactions, Hsp40, Hsp70, Ribosome
Introduction
Hsp70 chaperones assist a large variety of cellular processes such as the folding of newly synthesized proteins, the refolding of stress-denatured proteins, or the transport of proteins across membranes (1–3). The diverse Hsp70 functions rely on the interaction with co-chaperones that regulate the nucleotide-controlled transient interaction of Hsp70s with their substrates. In a classical Hsp70 chaperone cycle, the Hsp40 co-chaperone stimulates the ATP hydrolysis of Hsp70 and thereby promotes tight substrate binding, whereas the nucleotide exchange factor (NEF) in turn stimulates ADP release, and rebinding of ATP triggers substrate release (4).
Members of the Hsp70 and Hsp40 family, respectively, share essential structural features, whereas NEFs are rather diverse in structures and do not belong to one conserved protein family. Hsp70 proteins typically consist of an N-terminal 44-kDa ATPase domain and a 25-kDa C-terminal domain comprising a substrate binding region and a variable C terminus (1). Hsp40 proteins contain a J-domain of ∼65 amino acids (aa), which transiently associates with the Hsp70 partner and stimulates its ATP hydrolysis. Beside this common core function, Hsp40 proteins comprise multiple accessory domains that confer them their functional specificity.
In eukaryotes, a ribosome-associated Hsp70/40 system has evolved to assist co-translational protein folding processes of nascent polypeptides (5, 6). In yeast, this system consists of Ssb1 and Ssb2, two highly similar Hsp70 isoforms (referred to hereafter as Ssb), the Hsp40 Zuotin (Zuo), and another Hsp70 termed Ssz. Ssb associates with ribosomes and nascent chains emerging from the ribosomal exit tunnel. Zuo and Ssz form a stable heterodimer termed ribosome-associated complex (RAC)5 that independently binds to ribosomes through a charged region at the C terminus of Zuo (5). The RAC complex acts as co-chaperone stimulating the ATP hydrolysis of Ssb, thereby promoting binding of Ssb to nascent polypeptides (5, 7).
RAC and Ssb form a functional entity at the ribosome. Yeast strains lacking either one or all three components of this system show similar pleiotropic phenotypes including sensitivity to high salt concentrations and low temperatures, and a hypersensitivity to cations such as aminoglycosides (5, 8–10).
Ssb/RAC reveals intriguing features that differ from classical Hsp70 chaperone systems. Unlike other J-proteins, Zuo requires stable association with Ssz to stimulate the ATPase activity of Ssb. Moreover, the Hsp70 Ssz does not require ATP binding and hydrolysis for function in vivo, nor does it seem to associate with nascent polypeptide chains (7, 9, 11). These atypical traits are not restricted to yeast cells, as it has been recently reported that the J-protein MPP11 associates with Hsp70L1 to form a stable ribosome-associated complex in mammalian cells (12, 13). In mammals, ribosome-associated RAC is suggested to target cytosolic Hsc70 to newly synthesized proteins. As yet, the molecular basis for this unusual stable Hsp70-Hsp40 pairing and the functional activation of Zuo by Ssz binding remains unclear.
In this study, we investigated the architecture and conformational dynamics of yeast RAC and its implications for co-chaperone function. We report that the complex assembly involves the C-terminal domain of Ssz and the N-terminal 62 aa of Zuo, and leads to significant stabilization of both subunits. Changes in dynamics observed in the Zuo J-domain upon complex formation might be the basis for functional activation of the co-chaperone.
EXPERIMENTAL PROCEDURES
Strains, Growth Conditions, and Complementation Analysis
Standard yeast growth and transformation procedures were used (14). For strain details, see supplemental materials. For complementation analysis, zuoΔ and sszΔ strains were transformed with a plasmid expressing either Zuo-ΔN62 (pmCUP313-ΔN62Zuo) or Zuo wt (pmCUP313-Zuo) under copper control or the empty vector pmCUP313 (HIS) (for details see supplemental materials). Strains were grown to log phase on glucose-containing -HIS medium and then spotted in a 5-fold dilution series on -HIS plates with or without hygromycin B at different concentrations and incubated at 30 °C.
Protein Purification
Ssz, Zuo, and Zuo-ΔN62 were expressed with an Ulp1-cleavable N-terminal His6-Smt3 tag in Escherichia coli BL21/pCodonPlus by induction at 30 °C with 0.5 mm isopropyl-1-thio-d-galactopyranoside in the presence of appropriate antibiotics. Cells were resuspended in lysis buffer (40 mm HEPES-KOH pH 7.4, 500–1000 mm potassium acetate, 5 mm MgCl2, 2 mm β-mercaptoethanol (β-ME), 5% glycerol, and 1 mm ATP for Ssz), lysed by French Press, and lysates were cleared for 30 min at 4 °C at 30,000 × g. The supernatant was applied to Ni-IDA matrix (Protino; Macherey-Nagel, Düren, Germany); eluted material containing 250 mm imidazole was supplemented with His6-Ulp1 protease and dialyzed overnight. Contaminants were removed by ion exchange chromatography over a ReSourceQ 6 ml (Ssz) or a ReSourceS 6 ml (Zuo) column.
HX Experiments, Mass Spectrometry, and Data Processing
HX experiments were performed as described earlier (15, 16). 80–200 pmol of purified Zuo, Ssz, Zuo, and Ssz, or purified RAC were preincubated with or without 0.6 mm ATP for 5–10 min at 30 °C. Amide hydrogen exchange was initiated by a 20-fold dilution into D2O-based buffer (25 mm HEPES, pH 7.4, 50 mm KCl, and 5 mm MgCl2) at 30 °C. After incubation for fixed time periods (10 s to 2 h), the exchange reaction was stopped by addition of 1 volume of ice-cold quench buffer (400 mm KH2PO4/H3PO4, pH 2.2). Quenched samples were immediately injected into an HPLC setup and analyzed on an electrospray ionization-quadrupole time of flight-mass spectrometer (QSTAR Pulsar, Applied Biosystems) as described (15, 17). D2O buffer for the HX experiments was prepared using 99.85% D2O (Euriso-top), lyophilized, and redissolved five times in fresh D2O volumes. For details of the HPLC separation and data processing, see supplemental materials.
Ribosome Binding, Ssz Binding, and Tryptic Digestion
RESULTS
RAC Formation Decreases the Conformational Dynamics of Zuo and Ssz
To understand which structural elements of Zuo and Ssz contribute to RAC complex formation, we purified recombinant Zuo and Ssz individually and set out to test for their structural changes upon complex formation. We also co-expressed Ssz and Zuo in E. coli to isolate RAC as a stable complex. All purified proteins were soluble, but Ssz was recovered with low yield and required addition of 1 mm ATP during the purification procedure to improve solubility.
To define the binding interface of Ssz and Zuo, we measured the conformational and dynamics changes induced upon complex formation by amide hydrogen-deuteron exchange (HX) combined with mass spectrometry (MS) (18). In HX experiments, accessible amide protons of a protein exchange rapidly for a solvent deuteron, whereas protons buried in the protein core or involved in hydrogen bonding remain protected. Conformational changes and binding interfaces to partners are resolved by changes in solvent accessibility of amide hydrogen positions in the protein. Purified Zuo, Ssz, or RAC were diluted into a D2O-based buffer, incubated for fixed exchange periods of 10 s to 2 h, and after stopping the exchange reaction, the total number of deuterons incorporated in each protein was determined by MS (15, 16).
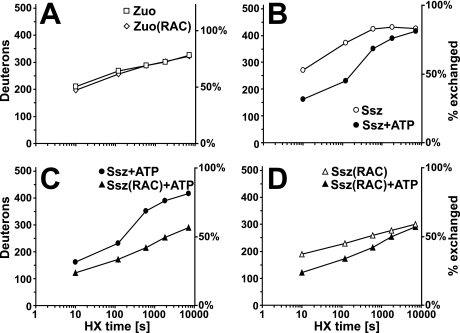
Zuo exchanged as much as 64% of all available amide hydrogens to deuterons in a 2-min HX reaction (268 deuterons, D), and thus displays high structural dynamics (Fig. 1A). Comparing the exchange properties for Zuo and Zuo assembled in RAC (designated hereafter Zuo(RAC)), we found a small but significantly decreased deuteron incorporation in Zuo(RAC) at short reaction times (Fig. 1A). Thirteen amide protons were protected from exchange at 10 s HX in RAC. This protection then decreased to 3 amide protons after 2 h. Assuming that the complex is kinetically stable (see below), this rapid decline in protection suggests that primarily fast exchanging regions within Zuo are affected by complex formation.
FIGURE 1.
Amide hydrogen exchange kinetics of Ssz, Zuo, and RAC. Time-dependent incorporation of deuterons in: Zuo versus Zuo in the complex (Zuo(RAC)) (A), in Ssz with and without ATP (B), in Ssz versus Ssz in RAC (Ssz(RAC)) in the presence of ATP (C), Ssz in RAC with and without ATP (D). The left y axis indicates the number of deuterons incorporated into the protein, and the right y axis the fraction exchanged in % of total exchangeable amides. All values in this figure are uncorrected for back-exchange. The HX reaction conditions are 0.4 μm Ssz, Zuo, or RAC with or without 0.6 mm ATP at 30 °C.
The Ssz subunit showed even faster exchange of amide hydrogens over time than Zuo did (Fig. 1B). Ssz exchanged 73% of all available amide hydrogens in a 2-min reaction and 84% in 30 min (incorporation of 373 and 431 deuterons, respectively, Fig. 1B). This unusually fast exchange kinetics is indicative of a highly dynamic and loosely folded protein conformation. Because nucleotide binding affects the conformational dynamics of other Hsp70 family members (15, 19, 20), we added 0.6 mm ATP to the reaction. The presence of ATP resulted in strongly decreased deuteron incorporation to 45% after a 2-min HX reaction (Fig. 1B), indicating a substantial stabilization of Ssz by ATP. Under these conditions, the exchange rate corresponds to that observed for canonical Hsp70 chaperones like DnaK (15, 20). The nucleotide-induced stabilization, however, vanished with longer incubation times in D2O, implying that the nucleotide is released within minutes. We could not conduct experiments using nucleotide-free Ssz because the protein was too unstable under this condition, and purified Ssz used in this study contained residual amounts of ATP and ADP (data not shown).
Next, we compared Ssz to Ssz assembled in RAC (designated hereafter Ssz(RAC)) in the presence of ATP to specifically report about the effect of Zuo binding (Fig. 1C). The binding of Zuo to Ssz resulted in a pronounced exchange protection of Ssz(RAC); Ssz(RAC) incorporated 59 deuterons less than Ssz alone in a 2-min reaction. This protection persisted up to HX reaction times of 2 h, indicating that the complex is kinetically very stable. Interestingly, comparison of HX kinetics of Ssz(RAC) in the presence and absence of ATP showed that Ssz(RAC) was also stabilized by the addition of nucleotides in its complexed form (Fig. 1D), whereas Zuo in RAC was not affected by ATP (supplemental Fig. S1B). We conclude that the Ssz stabilization effect of ATP binding is not transmitted further to Zuo within the complex.
The C-terminal Domain of Ssz Constitutes the Major Binding Site for Zuo
Next, we combined our HX setup with on-line pepsination prior to MS analysis to map the specific regions within Ssz and Zuo that are affected by complex formation and thus are potentially involved in the interaction surface.
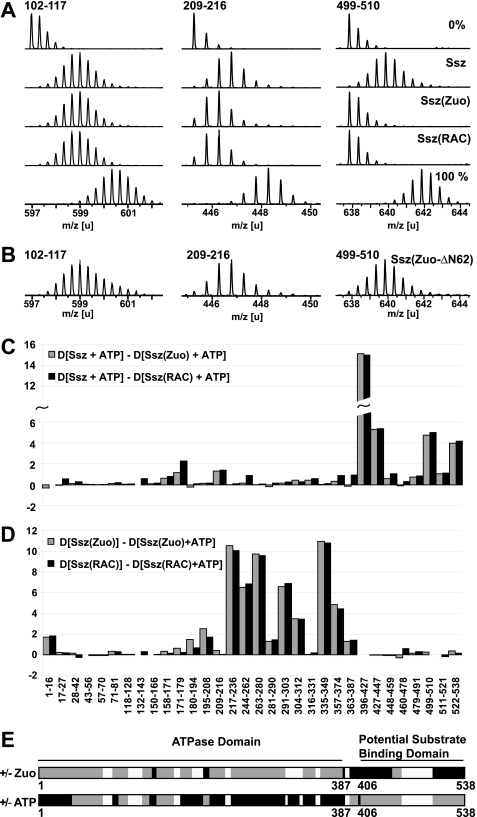
We first compared the HX peptide profiles of Ssz and Ssz(RAC) in the presence of ATP. We detected two categories of peptides: (i) peptides that incorporated less deuterons in the complex than in Ssz alone (HX protection), and (ii) peptides that exhibited no changes between the two conditions. Fig. 2A shows representative mass spectra for Ssz peptides harboring HX protection (peptides 209–216 and 499–510), or no changes in HX properties (peptide 102–117). Overall, we found particularly strong HX protection effects in regions 396–447 and 479–538 of the C-terminal domain of Ssz (Fig. 2, C and E and supplemental Fig. S2). These regions localized to the predicted β-sheet subdomain and α-helical lid region of the C-terminal domain, respectively and are likely to include the major Zuo interaction surface. Additional protection was observed for peptides encompassing aa 171–179 and aa 209–216 (Fig. 2, A, C, and E) of the ATPase domain of Ssz(RAC). Importantly, purified Ssz co-incubated in vitro with Zuo, designated hereafter Ssz(Zuo), yielded very similar results compared with purified RAC (Fig. 2, A and C). This confirms that purified Zuo and Ssz properly assembled into RAC, and that uncomplexed Ssz and Zuo can be taken as bona fide reference components.
FIGURE 2.
Association with Zuo affects the C-terminal domain of Ssz, whereas ATP binding stabilizes the ATPase domain of Ssz. A, HX properties of Ssz uncomplexed and bound to Zuo. Mass spectra of representative peptic peptides of Ssz (aa 102–117, m/z = 596.98, z = +3; aa 209–216, m/z = 445.27, z = +2; aa 499–510, m/z = 637.83, z = +2) incubated in H20 (0%) or for 10 s in D2O buffer in the uncomplexed state (Ssz), in the complexed state (Ssz(RAC)), and in the reconstituted complex (Ssz(Zuo)). 100% designates a control spectrum for the same peptides obtained from fully deuterated RAC. The samples during HX contained 0.5 μm Ssz, or 0.8 μm RAC, or 0.3 μm Ssz and 0.6 μm Zuo, and 0.6 mm ATP. B, truncated Zuo-ΔN62 mutant protein does not induce HX protection in Ssz. Mass spectra of the same peptides from Ssz incubated in the presence of the Zuo-ΔN62 for 10 s in D2O buffer. The samples during HX contained 0.4 μm Ssz and 0.8 μm Zuo-ΔN62, and 0.6 mm ATP. C, difference in deuteron incorporation between uncomplexed Ssz and Ssz(Zuo) (gray bars) or Ssz(RAC) (black bars) after 10 s of incubation in D2O buffer. The data were resolved to individual peptic peptides as indicated by the start and end residue numbers of the corresponding segments. D, difference in deuteron incorporation for Ssz(Zuo) (gray bars) and Ssz(RAC) (black bars) in the presence or absence of 0.6 mm ATP after 10 s of incubation in D2O buffer. For the effect of ATP on Ssz alone, see supplemental Fig. S2. E, schematic model of the domain architecture of Ssz colored according to the observed HX protection patterns induced by Zuo and ATP binding, respectively. Black, HX protection induced by Zuo, resp. ATP binding. Gray, no changes in HX properties upon Zuo or ATP binding and white, no data available. Protection effects were considered that were ≥ 1D and ≥ 10% of the total exchangeable sites for all peptides covering a particular region.
Taken together, these findings suggest that the C-terminal domain of Ssz is the main site of association with Zuo, and that there is a possible second point of contact or docking on the ATPase domain. This assumption fits well with a recent deletion analysis of Ssz showing that a fragment encompassing the ATPase domain but lacking the C-terminal substrate binding domain is strongly impaired in Zuo binding (11).
ATP and Zuo Binding Have Distinct Stabilizing Effects on Ssz
Using the same methodology, we furthermore localized the regions of Ssz, Ssz(RAC), and Ssz(Zuo) affected by ATP binding. Strong HX protection upon addition of ATP was observed for segments 217–280, 291–312, 335–349, and 358–374 in the ATPase domain, whereas no effects were observed in the C-terminal domain of Ssz (Fig. 2, D and E and supplemental Fig. S3). The protected segments constitute most of lobe II of the Ssz ATPase domain according to the Hsp70 standard domain nomenclature (21) and do not coincide with any regions affected by Zuo binding (Fig. 2, C, D, and E). We conclude that ATP binding predominantly affects the ATPase domain of Ssz, involving regions that are distinct from the segments stabilized by Zuo binding.
The Flexible N Terminus of Zuo Is Stabilized upon Binding to Ssz
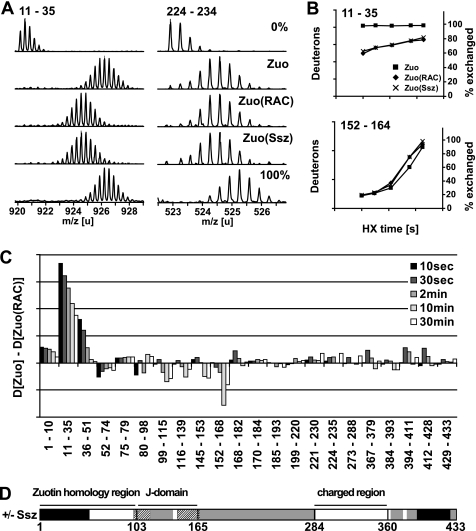
We next sought to identify regions of Zuo involved in Ssz binding and compared Zuo with Zuo(RAC) or Zuo in the in vitro assembled complex (Zuo(Ssz)) in HX pepsination experiments. We found extensive HX protection upon complex formation that localized to a single region at the N terminus of Zuo covering aa 1–51 (Fig. 3, A–D and supplemental Fig. S4). Interestingly, this N-terminal region revealed very fast HX exchange kinetics with almost complete exchange of its amide hydrogens within 10 s in uncomplexed Zuo (Fig. 3B). It appears highly flexible and loosely folded in the absence of Ssz, whereas it becomes less dynamic in the presence of Ssz. This region thus constitutes the potential binding site for Ssz. Moreover, additional smaller protection effects were observed in the C terminus of Zuo (peptides covering aa 394–428, Fig. 3C and supplemental Fig. S4). However, the functional importance of these changes in conformational dynamics in the C terminus of Zuo is unclear.
FIGURE 3.
Zuo N terminus is strongly stabilized by Ssz binding. A, HX properties of Zuo uncomplexed and bound to Ssz. Mass spectra of representative peptic peptides of Zuo (aa 11–35, m/z = 920,1776, z = +3; aa 224–235, m/z = 571,9333, z = +3) incubated in H2O (0%) or for 2 min in D2O in the uncomplexed state (Zuo), in the complexed state (Zuo(RAC)), and in the reconstituted complex (Zuo(Ssz)). 100% designates a control spectrum for the same peptides obtained from fully deuterated RAC. The samples during HX contained 0.5 μm Zuo, or 0.5 μm RAC, or 0.3 μm Zuo and 0.6 μm Ssz, and 0.6 mm ATP. B, kinetics of deuteron incorporation into selected segments of Zuo in the absence (■) and the presence of Ssz (Zuo(RAC), ♦; Zuo(Ssz), X). The axis on the left indicates the number of deuterons incorporated into the particular peptide, and the axis on the right the corresponding fraction exchanged in % of total exchangeable amides. C, difference of deuteron incorporation between Zuo and Zuo(RAC) after different incubation times (10 s to 30 min) in D2O. The data were resolved to individual peptic peptides as indicated by the start and end residue numbers of the corresponding segments (see also supplemental Fig. S3). D, schematic model of the domain structure of Zuo colored according to the observed HX protection patterns induced by Ssz binding. Black, HX protection upon Ssz binding; dashed, HX deprotection upon Ssz binding, gray, no changes in HX properties upon Ssz binding, and white, no data available. Protection and deprotection effects were considered that were ≥ 1 D and ≥ 10% of the total exchangeable sites for at least one HX time point measured in all peptides covering a particular region.
Interestingly, the comparison of peptide HX kinetics also revealed peptides that incorporated more deuterons in Zuo(RAC) than in uncomplexed Zuo. This HX deprotection localized in the J-domain of Zuo. Specifically, peptides covering the region of residues 99–168 showed a bimodal HX behavior (Fig. 3, B and C), indicating that opening of the structure in this region is slower than the hydrogen exchange reaction itself. In the presence of Ssz, the rate of opening of the Zuo J-domain is increased.
We next tested whether the protection of the N terminus in Zuo(RAC) could be detected with a different method probing protein backbone accessibility. Purified Zuo or RAC was subjected to limited trypsination, and the resulting fragments were analyzed by SDS-PAGE with Coomassie Blue staining or by mass spectrometry to identify products. In the absence of Ssz, Zuo was rapidly cleaved by trypsin into three distinct bands, which remained stable with respect to further digestion under our assay conditions (Fig. 4A). The three cleavage products could be assigned to the Zuo fragments 63–433, 46–433, and 43–433, respectively. This rapid cleavage confirms that the N terminus of Zuo is loosely folded and accessible to degradation by the protease, whereas the residual portion of Zuo is more stable. In contrast, when RAC was incubated with trypsin, cleavage of Zuo within the N terminus was prevented, and only a minor cleavage still occurred after position 62 of Zuo. This result confirms that the N terminus of Zuo is protected by the association with Ssz (Fig. 4A). Tryptic proteolysis of Ssz led to formation of small fragments under our assay conditions, which were invisible or visible only as faint bands in our gel.
FIGURE 4.
N-terminal 62 amino acids of Zuo are required for RAC formation. A, time-dependent trypsin (0.4 ng/μl) digest of Zuo, RAC, and Ssz (each 0.3 μg/μl). Samples were analyzed by ESI-MS and on SDS-PAGE with Coomassie Blue staining. B, Zuo-ΔN62 does not associate with Ssz. Zuo wt and Zuo-ΔN62 (1.6 μm) were incubated with N-terminally His6-tagged Ssz (1.6 μm) in the presence and absence of 2 mm ATP. The mixture was applied to a Ni-NTA matrix. After washing, the fractions eluted by imidazole were applied on SDS-PAGE and were Coomassie Blue-stained. C, Zuo-ΔN62 binds to yeast ribosomes but fails to target Ssz to the ribosome. 4 μm Zuo wt and Zuo-ΔN62 were incubated with 2 μm purified 80 S yeast ribosomes with or without Ssz, followed by ultracentrifugation through a sucrose cushion to separate unbound chaperones in the supernatant (S) from ribosome-associated chaperones in the pellet (P) fraction. The fractions were separated by SDS-PAGE and were Coomassie Blue-stained.
The N-terminal 62 Amino Acids of Zuo Are Essential for Complex Formation with Ssz in Vitro
To investigate whether the N-terminal Zuo segment is directly involved in binding of Ssz, we created a truncated Zuo mutant lacking the first N-terminal 62 aa (Zuo-ΔN62). The HX peptide profiles of this mutant protein were comparable to that of wt Zuo, with the exception of the absent N-terminal region, confirming the structural integrity of the mutant protein (data not shown). We tested whether Zuo-ΔN62 was able to associate with Ssz and performed binding assays with purified components (Fig. 4B). Zuo-ΔN62 and wt Zuo were incubated at 30 °C with Ssz carrying an N-terminal His6-Smt3 tag, and the protein mixtures were subsequently applied to a Ni-NTA matrix. After washing, bound protein fractions were eluted with imidazole and analyzed by SDS-PAGE. Whereas wt Zuo formed a stable complex with tagged Ssz and co-eluted with a 1:1 stoichiometry, Zuo-ΔN62 did not form any detectable complex irrespective of the presence or absence of 1 mm ATP (Fig. 4B). In agreement with these findings, Zuo-ΔN62 did not induce any detectable HX protection in Ssz (compare Fig. 2, B to A).
Because Zuo targets Ssz to the ribosome via complex formation, we tested whether Zuo-ΔN62 has lost this capability by performing in vitro ribosome binding experiments. Purified Zuo or Zuo-ΔN62 with or without Ssz was incubated with purified 80 S ribosomes from Saccharomyces cerevisiae in a 3:1 molar ratio. The mixtures were loaded onto a sucrose cushion, and unbound protein (supernatant, S) was separated from ribosome-associated protein (pellet, P) by ultracentrifugation and subsequently analyzed by SDS-PAGE and Coomassie Blue staining. Wt Zuo and Zuo-ΔN62 were detected only in the pellet fraction together with 80 S ribosomes in a 1:1 stoichiometry, demonstrating the intact ribosomal binding capacity of the Zuo-ΔN62 mutant (Fig. 4C, compare lane 12 with lanes 4 and 8). However, Ssz targeting to ribosomes was only possible in the presence of wt Zuo, and not in the presence of Zuo-ΔN62 (Fig. 4C, lanes 6 and 10). Thus, Zuo-ΔN62 does not stably associate with Ssz and is no longer able to target Ssz to ribosomes. We conclude that the N-terminal 62 aa of Zuo are essential for binding to its Hsp70 partner Ssz and targeting it to the translation apparatus.
The N-terminal 62 Amino Acids of Zuo Are Essential and Sufficient to Mediate Complex Formation with Ssz in Vivo
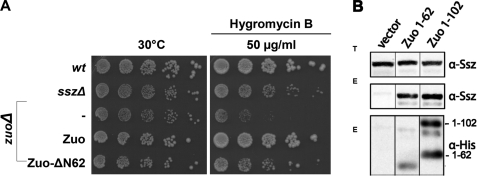
To elucidate the role of the N-terminal domain of Zuo in vivo, we analyzed the ability of Zuo-ΔN62 to complement the hygromycin B sensitivity phenotype of yeast cells deleted for Zuo. Expression of wt Zuo from a plasmid at wt level fully restored growth of zuoΔ cells (Fig. 5A). Whereas zuoΔ cells expressing the Zuo-ΔN62 mutant displayed ameliorated growth on hygromycin B compared with zuoΔ cells, they were still impaired in comparison to zuoΔ cells complemented with Zuo or wt cells (Fig. 5A). Similar results were obtained in complementation analysis of the NaCl-sensitive phenotype of zuoΔ cells (supplemental Fig. S5). Importantly, the hygromycin B sensitivity of zuoΔ cells expressing Zuo-ΔN62 corresponded to the growth properties of sszΔ cells (Fig. 5A). Hygromycin B sensitivity of cells lacking Ssz is less pronounced than that of cells deleted for Zuo, most likely due to residual activity of Zuo in the absence of its complex partner (11). Zuo-ΔN62 thus also displays residual function, but its inability to associate with Ssz results in a phenotype similar to that of complete absence of Ssz.
FIGURE 5.
The N-terminal 62 aa of Zuo are sufficient for RAC assembly in vivo. A, complementation analysis of the hygromycin B sensitivity of zuoΔ cells. Haploid yeast strains (wt and different mutants expressing indicated constructs from plasmid) were grown in minimal medium to early log phase and after serial 5-fold dilutions spotted on −HIS plates or −HIS plates containing hygromycin B as indicated. Plates were incubated for 3 days at 30 °C. The symbol (−) indicates cells carrying empty vector for control. B, association with Ssz of chimeric fusion proteins carrying the N-terminal zuotin region. The N-terminal aa 1–62 and 1–102 of Zuo fusion proteins carrying a C-terminal His6 tag were expressed in zuoΔ cells and subsequently affinity purified using a Ni-NTA matrix and tested for co-purification of Ssz. Totals (T) show the levels of Ssz in the lysates used for the affinity pull-downs. Eluates (E): Western blotting against Ssz and the His6 tag of the Zuo mutants 1–62 and 1–102 showing co-purification of Zuo mutants with Ssz. Zuo-(1–102) Western blot detection revealed two additional signals for unknown reasons.
Finally, we fused the first 62 aa of Zuo, or the first 102 aa representing the entire N-terminal segment preceding the J-domain, to the unrelated J-domain of Ydj1 together with a C-terminal His6 tag for affinity isolation. Fusion to the J-domain of Ydj1, a Hsp40 homolog in the yeast cytosol that does not react with Ssz or Ssb (7), was important to stabilize the unfolded N terminus of Zuo. These two constructs, designated Zuo-(1–62) and Zuo-(1–102), were expressed in yeast cells deleted for endogenous Zuo, and their association with Ssz was probed by pull-down experiments using the His tag of the recombinant Zuo fusion proteins (Fig. 5B). Whereas no Ssz protein was present in the control reaction, Ssz specifically co-purified with both the Zuo fusion proteins in the eluates (Fig. 5B). As control, we also expressed the His-tagged J-domain of Ydj1 lacking the Zuo moiety in zuoΔ cells and confirmed that this domain did not interact on its own with Ssz in vivo (data not shown). We conclude that the N-terminal 62-aa Zuo segment is portable to other proteins and sufficient to mediate interaction with Ssz in vivo. Taken together, our data demonstrate that the N terminus of Zuo comprises the major structural element for binding the partner chaperone Ssz both, in vitro and in vivo.
DISCUSSION
Zuo and Ssz are members of the Hsp40 and Hsp70 chaperone families, respectively, but display a number of atypical features compared with their canonical homologs. One of the most remarkable of them is the formation of a stable complex that is required for function of this system as co-chaperone for another Hsp70, Ssb. How Zuo and Ssz assemble into RAC and how their association enables Zuo to act catalytically onto Ssb was as yet not understood in the absence of structural information.
In this study, we provide information on the complex architecture and implications for its function. We combined amide hydrogen exchange experiments and traditional biochemistry to define the complex interface and its conformational dynamics, and found that (i) the subunits Ssz and Zuo individually are highly dynamic and strongly stabilized by their association into RAC, (ii) the Zuo-specific N-terminal extension is an essential structural element of RAC, (iii) the complex interface involves the N-terminal region of Zuo and the C-terminal domain of Ssz, and (iv) association into RAC results in increased dynamics of the Zuo J-domain.
The intrinsically dynamic conformation of Ssz is stabilized both by ATP binding and by Zuo binding. The regions affected by the respective ligands are distinct: ATP binding specifically causes the compaction of the ATPase domain, whereas Zuo binding affects most of the C-terminal domain of Ssz (Fig. 6). We note that ATP stabilizes the ATPase domain of Ssz also in the complex with Zuo, but Zuo does not sense this effect. From our data, we thus expect the binding of ATP and of Zuo to serve distinct functions.
FIGURE 6.
Model for the RAC complex assembly and architecture. ATP (yellow) binding strongly stabilizes the ATPase domain of Ssz (blue). However, ATP binding is only transient, and complex formation with Zuo (pink) is thought to be independent of the ATP status of Ssz. RAC assembly involves the C-terminal domain of Ssz and the N terminus of Zuo, and leads to a pronounced stabilization of the engaged moieties in both proteins and to the formation of a kinetically very stable heterodimer. Domains with significant changes in conformational dynamics upon ligand/partner binding are indicated in dark blue (Ssz) or in purple (Zuo). Moreover, association of Zuo with Ssz results in increased dynamics of the Zuo J-domain region carrying the HPD motif, which is likely to contribute to the activation of RAC as co-chaperone. ATP binding to RAC has no effect on the conformational dynamics of Zuo but additionally stabilizes the ATPase domain of Ssz in the complex.
The pattern of nucleotide-induced HX protection in Ssz does not correlate with those observed for the classical Hsp70 DnaK (15). Specifically, the ATP-induced stabilization of the ATPase domain is more extensive in Ssz, and no deprotection of segments in the predicted β-sheet region of the C-terminal potential substrate binding domain is observed upon ATP binding. Ssz thus does not display conformational changes characteristic of canonical Hsp70 chaperones in response to nucleotide binding. Other findings also indicate that Ssz cannot be considered a classical Hsp70 chaperone: its C-terminal domain is shorter than that of other family members (5) and it appears to not bind to substrates (7). Moreover, Ssz does not efficiently hydrolyze ATP (11).
Following these considerations, the importance of the nucleotide-induced stabilization remains unclear. We could not specifically test whether ATP binding is required for complex formation or RAC activity because no nucleotide-free Ssz in soluble state could be obtained. However, a recent study showed that a Ssz mutant deficient in ATP binding can complement the phenotype of sszΔ cells in vivo, suggesting that the assembly of a functional RAC can occur even in the absence of bound nucleotide (11).
We found that binding of Zuo to Ssz induces major structural or dynamic changes in most of the C-terminal domain of Ssz. The observed stabilization effects can result from direct involvement of the affected regions in the complex interface or from reduced conformational flexibility indirectly induced by complex formation. In both cases, we conclude that this domain plays an important role in complex formation. In agreement with these findings, a study by Rospert and co-workers (11) demonstrated that a Ssz mutant comprising the ATPase domain and lacking the C-terminal domain could not stably interact with Zuo. In classical Hsp70 chaperones, the C-terminal domain is dedicated to substrate binding. In Ssz however, this domain seems to assume a different role and respond to interaction with the N terminus of Zuo (Fig. 6). We find here again evidence for a divergence in function of Ssz as compared with classical members of the Hsp70 family. Another example of such divergence was reported for yeast Sse. Although Sse shares structural elements with Hsp70 chaperones, it functions as a nucleotide exchange factor and uses its C-terminal domain to embrace its partner Hsp70 (22, 23).
Our analysis of the Ssz HX properties shows that a second region comprising segments 171–179 and 209–216 of the ATPase domain is affected by binding to Zuo. Making use of sequence alignments and known Hsp70 structures, we find that these segments localize more precisely to lobe Ia, in the region thought to be involved in docking of the C-terminal domain onto the ATPase domain in Hsp70 chaperones. We thus hypothesize that the binding of Zuo to the C-terminal domain of Ssz affects interdomain docking. Alternatively, Zuo may contact both domains directly. Further investigations will be necessary to clarify the details of Ssz mode of association with Zuo.
We identified the N-terminal 62 aa of Zuo as an essential and sufficient structural element for stable interaction with Ssz, both in vitro and in vivo (Fig. 6). This N-terminal extension is a feature specific to Zuo-like proteins, and the interaction reported here is therefore characteristic for RAC complexes. Because Zuo only exhibits marginal stimulation of Ssb ATPase activity in the absence of its partner Ssz (7), we reason that this particular association mode must influence Zuo function. Specifically, it must affect the Zuo J-domain, responsible for interaction with Ssb and ATPase stimulation. Our HX data revealed increased dynamics in the J-domain of Zuo in the RAC complex (Fig. 6). The affected segments (aa 99–168) comprise the HPD-motif, which is known to be the site of interaction with Ssb (7). We thus speculate that this change in dynamics of the J-domain upon association with Ssz results in activation of Zuo.
In sum, these data allow us to derive a model suggesting a novel mechanism for converting Zuotin and Ssz chaperones into a functional dimer (Fig. 6). The C-terminal domain of Ssz is engaged into the stable interaction with the J-protein Zuo and thus displays an altered function compared with canonical Hsp70 homologs, which use this domain for the transient ATP-controlled substrate binding. Although ATP binding significantly stabilizes Ssz in its monomeric form as well as in the heterodimer, nucleotide binding is rather transient and does not seem essential to drive complex formation. However, complex formation essentially relies on the interaction of Ssz with the N-terminal segment of Zuo. Additionally, binding of Ssz to the N terminus of Zuo transmits a signal into the adjacent J-domain thereby increasing the accessibility of its HPD motif and activating RAC as co-chaperone. Further experiments are required to unravel the details of this mechanism of activation. The results of this study provide the essential groundwork to understand the architecture of this atypical complex and enable the design of appropriate strategies for further investigations.
Supplementary Material
Acknowledgments
We thank Renate Dippel for excellent technical assistance and the members of the Deuerling and Bukau laboratories for helpful discussions and comments on the manuscript.
This work was supported in part by the Konstanz Graduate School Chemical Biology and the Interdisciplinary Research Center Proteostasis, a fellowship of the Zukunftskolleg (to S. P.), an HFSP fellowship (to J. F.), and DFG Grants DE-783/3-1 (to E. D.) and SFB638 (to B. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental materials and Figs. S1–S5.
- RAC
- ribosome-associated complex
- aa
- amino acids
- wt
- wild type
- MS
- mass spectrometry
- HX
- hydrogen exchange
- NTA
- nitrilotriacetic acid.
REFERENCES
- 1.Mayer M. P., Bukau B. (2005) Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartl F. U., Hayer-Hartl M. (2002) Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 3.Wegrzyn R. D., Deuerling E. (2005) Cell. Mol. Life Sci. 62, 2727–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukau B., Weissman J., Horwich A. (2006) Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 5.Gautschi M., Lilie H., Fünfschilling U., Mun A., Ross S., Lithgow T., Rücknagel P., Rospert S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3762–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig E. A., Eisenman H. C., Hundley H. A. (2003) Curr. Opin. Microbiol. 6, 157–162 [DOI] [PubMed] [Google Scholar]
- 7.Huang P., Gautschi M., Walter W., Rospert S., Craig E. A. (2005) Nat. Struct. Mol. Biol. 12, 497–504 [DOI] [PubMed] [Google Scholar]
- 8.Yan W., Schilke B., Pfund C., Walter W., Kim S., Craig E. A. (1998) EMBO J. 17, 4809–4817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hundley H., Eisenman H., Walter W., Evans T., Hotokezaka Y., Wiedmann M., Craig E. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4203–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S. Y., Craig E. A. (2005) Eukaryot. Cell 4, 82–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conz C., Otto H., Peisker K., Gautschi M., Wölfle T., Mayer M. P., Rospert S. (2007) J. Biol. Chem. 282, 33977–33984 [DOI] [PubMed] [Google Scholar]
- 12.Otto H., Conz C., Maier P., Wölfle T., Suzuki C. K., Jenö P., Rücknagel P., Stahl J., Rospert S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10064–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hundley H. A., Walter W., Bairstow S., Craig E. A. (2005) Science 308, 1032–1034 [DOI] [PubMed] [Google Scholar]
- 14.Guthrie C., Fink G. R. (2004) Guide to Yeast Genetics and Molecular Biology, Elsevier Academic Press, San Diego [Google Scholar]
- 15.Rist W., Graf C., Bukau B., Mayer M. P. (2006) J. Biol. Chem. 281, 16493–16501 [DOI] [PubMed] [Google Scholar]
- 16.Rist W., Jørgensen T. J., Roepstorff P., Bukau B., Mayer M. P. (2003) J. Biol. Chem. 278, 51415–51421 [DOI] [PubMed] [Google Scholar]
- 17.Rist W., Mayer M. P., Andersen J. S., Roepstorff P., Jørgensen T. J. (2005) Anal. Biochem. 342, 160–162 [DOI] [PubMed] [Google Scholar]
- 18.Wales T. E., Engen J. R. (2006) Mass Spectrom. Rev. 25, 158–170 [DOI] [PubMed] [Google Scholar]
- 19.Andréasson C., Fiaux J., Rampelt H., Druffel-Augustin S., Bukau B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16519–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 21.Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 22.Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.