Abstract
Mutations in cardiac ryanodine receptor (RYR2) and cardiac calsequestrin (CASQ2) genes are linked to catecholaminergic polymorphic ventricular tachycardia, a life-threatening genetic disease. They predispose young individuals to cardiac arrhythmia in the absence of structural abnormalities. One such mutation that changes an aspartic residue to histidine at position 307 in CASQ2 has been linked to catecholaminergic polymorphic ventricular tachycardia. In this study we made a transgenic mouse model expressing the mutant CASQ2D307H protein in a CASQ2 null background and investigated if the disease is caused by accelerated degradation of the mutant protein. Our data suggest that the mutant protein can be expressed, is relatively stable, and targets appropriately to the junctional sarcoplasmic reticulum. Moreover, it partially normalizes the ultrastructure of the sarcoplasmic reticulum, which was altered in the CASQ2 null background. In addition, overexpression of the mutant protein does not cause any pathology and/or structural changes in the myocardium. We further demonstrate, using purified protein, that the mutant protein is very stable under chemical and thermal denaturation but shows abnormal Ca2+ buffering characteristics at high calcium concentrations. In addition, trypsin digestion studies reveal that the mutant protein is more susceptible to protease activity only in the presence of high Ca2+. These studies collectively suggest that the D307H mutation can compromise the dynamic behavior of CASQ2 including supramolecular rearrangement upon Ca2+ activation.
Keywords: Calcium, Calcium/Binding Proteins, Calcium/Transport, Protein/Folding, Protein/Intracellular Trafficking, Protein/Processing, Transport/Calcium
Introduction
Calsequestrin (CASQ)2 is the major calcium buffering protein located in the lumen of the sarcoplasmic reticulum (SR) in striated muscles (1–3). The cardiac isoform CASQ2 is ∼45 kDa, binds calcium (Ca2+) with high capacity (∼60 Ca2+ ions) but with low affinity (Kd ∼ 1 mm) rendering it an excellent calcium buffer within the SR. In addition to maintaining SR calcium content through dynamic calcium binding and release, CASQ2 indirectly binds to the cardiac ryanodine receptor (RYR2) via the lumenal domains of the junctional SR (jSR) proteins, Triadin (4) and Junctin (5). This complex interaction keeps high concentrations of Ca2+, which is required for myocyte contraction, close to the site of release (6–9). Furthermore, CASQ2 along with Triadin and Junctin may also serve as sensors on the lumenal side and modulate RyR2 gating (open state) in a Ca2+-dependent manner (10). Thus, CASQ2 has been shown to play a critical role in maintaining SR Ca2+ content as well as in modulating calcium release via the RYR2 (11).
More recently, mutations in CASQ2 have been associated with ventricular arrhythmias in humans, particularly catecholaminergic polymorphic ventricular tachycardia (CPVT) (12–22). CPVT is a familial, arrhythmogenic disorder characterized by episodes of syncope and sudden death, when the individual is exposed to emotional or physical stress, in the absence of structural alterations of the heart. There are currently 7 mutations that have been identified in CASQ2 in association with CPVT (12, 14, 23, 24). Studies, both in vitro and in vivo have attempted to define the causative mechanism for the reported arrhythmia (8, 13, 15, 16, 18, 20).
The CASQ2D307H mutation, occurring in a highly conserved region of CASQ2, was suggested to cause a recessive form of CPVT (17, 23). This point mutation changes the negative aspartate residue (Asp) to a positive histidine (His) at position 307 in the C terminus, and interrupts a long stretch of negative charges. The quaternary structure of this mutant protein is such that the D307H mutation site protrudes into the inter-domain space of the CASQ2 monomer, and thus might interfere with CASQ2 calcium binding and polymerization (17, 25).
Adenoviral overexpression of CASQ2D307H in rat cardiomyocytes resulted in a net increase in protein level but significantly decreased SR Ca2+ storing capacity, Ca2+ channel-gated Ca2+ release (Ca2+ transients), and increased local Ca2+ release events (Ca2+ sparks) (15). When paced and exposed to isoproterenol treatment, these myocytes exhibited disturbances in calcium cycling (extra-systolic spontaneous calcium transients) and delayed after depolarizations. Furthermore, Houle et al. (13) showed that the mutant protein could be expressed and purified from COS cells, but had a reduced Ca2+ buffering capacity and interacted less well with Triadin and Junctin than the wild type (WT) protein. Light scattering experiments on the CASQ2D307H mutant protein also suggest that the mutant monomer cannot form a dimer that is properly oriented, subsequently affecting polymerization (18).
To understand the causal relationship between CASQ2D307H mutation and CPVT in vivo, we previously generated a mouse model that expresses this mutant protein along with wild type CASQ2 (16). 2–5-Fold expression of the mutant protein did not alter SR Ca2+ load, whereas Ca2+ transients and Ca2+ spark amplitudes were decreased. We found no compensatory alterations in the expression of other SR proteins involved in Ca2+ release. The transgenic mice developed complex ventricular arrhythmias when challenged with isoproterenol and caffeine incrementally (16). However, this model had a limitation because the mutant was expressed along with the WT CASQ2.
To overcome this limitation and to closely relate to the human condition, we developed a mouse model that expresses only CASQ2D307H in the CASQ2 null background (26). First, we used this model to understand how this point mutation, D307H, affects protein expression, targeting, and localization of CASQ2 to the jSR; second, we determined whether expression of the mutant protein induced an ER stress response. In response to cellular stress such as expression of mutant proteins that may not be correctly folded, expression of chaperone proteins such as GRP94, GRP78, and/or calreticulin are up-regulated to help fold them. In addition, we investigated how the mutation affects calcium-dependent conformational and structural stability using bacterially expressed CASQ2D307H mutant protein. Our data collectively suggest that the mutation does not appear to affect protein expression and targeting to jSR but may compromise its ability to dynamically regulate calcium buffering.
EXPERIMENTAL PROCEDURES
Generation of CASQ2D307H Transgenic Mouse in the CASQ2 Null Background
The mouse model for this study was generated by crossing the CASQ2D307H mouse that expresses the mutant CASQ2D307H protein in the wild type background (16) with the CASQ2 null mouse (CASQ2−/−) (26). The mutant protein was expressed using the α-myosin heavy chain promoter as described previously (27–29). The CASQ2D307H mutant protein was transferred to the CASQ2 null background after three generations and confirmed by PCR as described before (16, 26). This investigation conforms to the guide for Care and Use of Laboratory Animals published by the United States National Institutes of Health (NIH Publication 85-23).
Protein Analysis
Expression of CASQ2 (antibody from Affinity Bioreagents), and ER chaperone proteins GRP78, GRP94 (Santa Cruz), and calreticulin (Affinity Bioreagents) were determined by standard Western blotting techniques (16) using whole heart homogenates and/or SR microsomes as indicated from CASQ2+/+, CASQ2D307H, and CASQ2−/− mice (n = 6). Glyceraldehyde-3-phosphate dehydrogenase (Cell Signaling) levels were used as internal controls for protein loading and quantification. Protein bands were scanned (HP Imaging) and relative intensities quantified using Image J software.
Electron Microscopy
Hearts from wild type mice and mice expressing CASQ2D307H in a null background were fixed by perfusion through the left ventricle with 3% glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.2. All mice were between the ages of 6 and 8.6 weeks. Papillary muscles from the left ventricle were dissected, postfixed in OsO4, en block stained with uranyl acetate, and embedded in epon. Thin sections were contrasted with lead citrate and examined in a Philips 410 electron microscope. Sections of hearts from three wild type, three CASQ2D307H mice, and three CASQ2 null heart tissues fixed as part of a previous set of experiments (26) were used for a quantitative analysis of dyad ultrastructure. The frequency of dyads with three appearances (“tight, intermediate, and loose”) was estimated by counting the first 10 dyads that were seen in each of 10 different grid openings in sections from each heart.
Immunofluorescent Confocal Analyses
Single ventricular cardiomyocytes were prepared as before (16) for immunofluorescence labeling and viewed by confocal microscopy as described previously (16, 30) with modifications. Cardiomyocytes from the three experimental groups were fixed with 4% paraformaldehyde and immunostained with mouse anti-α-actinin (1:500) or rabbit anti-CASQ2 (1:200) followed by Alexa 488-conjugated goat anti-mouse (1:300) and Cy3TM-conjugated donkey anti-rabbit (1:300) antibodies. The cells were viewed by excitation at 488 and 543 nm using a Zeiss laser scanning microscope (LSM510).
Ubiquitin Pulldown Assay
Whole heart homogenates from the three experimental groups were used for this assay. An ubiquitin protein enrichment kit (Calbiochem) was used according to the manufacturer's recommendations. The final protein extract was separated on a 10% SDS-polyacrylamide gel and probed for CASQ2 and ubiquitin using antibodies from Affinity Bioreagents and Cell Signaling, respectively.
Purification of Wild Type and Mutant CASQ2 Proteins
The sequence encoding the casq2 gene was cloned into pET21a between the restriction sites, NdeI and XhoI, and the D307H mutant clone was generated by site-directed mutagenesis. Bacterially expressed protein was purified using a nickel-nitrilotriacetic acid superflow column (Qiagen) using a C-terminal His tag by stepwise washing with 20 and 50 mm and eluted with 250 mm imidazole buffer. The purified protein was subjected to 10% SDS-PAGE and visualized by staining the gel with Coomassie Brilliant Blue R-250. The protein fractions were pooled and dialyzed extensively against 5 mm NaH2PO4 buffer, pH 7.4, containing 5 mm NaCl. The dialyzed protein was concentrated using a 3-kDa cut-off Centricon YM-3.
CD Spectroscopy
Circular dichroism (CD) spectra were acquired using an Aviv spectropolarimeter with a temperature-controlled cell holder. Far UV CD spectra were collected with 10 μm protein in 5 mm NaH2-PO4 buffer, pH 7.4, containing 5 mm NaCl. CD scans were recorded using a quartz cell with path length of 1 mm; response time of 2 s; scan speed of 10 nm/min; and bandwidth of 1.0 nm. Two scans were accumulated and averaged for each spectrum. For studying urea or guanidine hydrochloride (GdnHCl) induced unfolding, the protein sample was incubated with different concentrations of urea or GdnHCl, respectively, for at least 3 h before recording CD spectra. For obtaining CD spectra in the presence of Ca2+, the indicated amounts of CaCl2 was added to the protein solution, mixed by inverting, and transferred to the cuvette after a 15-min incubation period. The protein was recovered from the cuvette after recording the spectra in the presence of Ca2+ and 2 mm EGTA was added, and CD spectra were recorded. Ca2+ chelation by EGTA was repeated by addition of 2 mm EGTA until the CD spectra overlapped with the native CD spectra.
Limited Proteolysis by Trypsin
Wild type and mutant CASQ2D307H proteins were subjected to proteolysis in 50 mm Tris-HCl, pH 8.0, at 25 °C using TPCK-treated trypsin (New England Biolabs). The reactions were performed in the absence of CaCl2 at 5, 10, 20, 30, 60, and 120 min and at 1–5 mm CaCl2 for 30, 60, and 120 min (because difference in proteolytic fragments are not visible within the 5-, 10-, and 20-min digestions in the presence of CaCl2). Proteins were incubated in the reaction buffers for 20 min at 25 °C in a reaction volume of 200 μl before trypsin was added at a protease:CASQ2 ratio of 1:50 (w/w), and samples were taken at the indicated time points. The samples were quickly mixed with Laemmli SDS-PAGE sample buffer (Bio-Rad) and heated at 99 °C for 2 min before being analyzed on 12% SDS-PAGE. 5 μg of each sample were analyzed on SDS-PAGE. All experiments were repeated 4 times. The gels were stained with Coomassie Brilliant Blue R-250 staining solution (Bio-Rad).
RESULTS
Transgenic Expression of CASQ2D307H Mutant Protein in the CASQ2 Null Background
A major goal of this study was to generate a mouse model that expresses only the CASQ2D307H mutant protein. We took advantage of our earlier model, a transgenic mouse that expressed the CASQ2D307H mutant protein ∼2-fold (along with wild type CASQ2) driven by the α-myosin heavy chain promoter. By crossing this mouse model with the CASQ2−/− mouse (26) a new line that expressed only the mutant protein in a null background was developed. 2–3-Month-old male mice were used for most of the studies, unless otherwise indicated. The resulting mutant mice were compared with null and WT mice. There were no differences between the three experimental groups in survival rates (>1 year), gestation, litter sizes, and survival of litters. No incidences of sudden death were observed in the CASQ2D307H mice at any age (3–12 months) through the duration of this study. There was no significant difference between body weights or heart weight to body weight ratios at age 3 months between the 3 groups (n = 6). Histological sections from 1-year-old hearts were analyzed for gross abnormalities. No evidence of cardiac hypertrophy and/or fibrosis was found (data not shown).
CASQ2D307H Protein Is Localized in the jSR
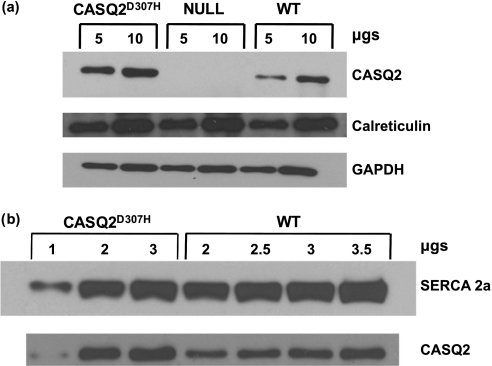
Next, expression of the mutant CASQ2D307H protein was investigated in total heart homogenates by Western blotting analyses using cardiac CASQ2-specific antibody (Fig. 1a). The expression level of mutant protein was 2-fold higher compared with wild type (n = 6) but there was no change in the level of calreticulin and SERCA2a protein. To determine whether the expressed calsequestrin is in the SR, we isolated SR microsomes from these hearts and performed Western blotting analyses. Our data indicate that the 2-fold increase in protein can be found in microsomes indicating that the expressed CASQ2D307H protein is targeted to the SR (Fig. 1b).
FIGURE 1.
Expression of CASQ2D307H in the CASQ2 null background. a, whole heart homogenates from CASQ2D307H, CASQ2 null, and WT mice were probed for CASQ2 and calreticulin by Western blotting (6 hearts per group); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control. b, SR microsomes from CASQ2D307H and WT hearts were used to stain for CASQ2 with SERCA2a as internal control.
The presence and location of the mutant CASQ2D307H were additionally verified in isolated cardiomyocytes by immunofluorescent confocal microscopy. Double immunolabeling for α-actinin, a Z-line protein, and CASQ2 showed that both WT and CASQ2D307H mutant proteins localized at the level of the Z-line at foci that were intercalated with the Z-lines of the myofibrils (Fig. 2a). The location of CASQ2-positive foci coincided with the inter-myofibrillar position of dyads, i.e. associations between jSR and T tubules (dyads), where CASQ2 is normally located. The results showed that the mutant protein was not only present but also appropriately targeted to the jSR.
FIGURE 2.
a, CASQ2D307H localizes to the jSR. Isolated cardiac myocytes from CASQ2D307H and WT hearts were immunostained to show CASQ2 localization to the jSR as indicated by the merge with α-actinin (at the Z-line). b, electron micrographs from thin sections of age-matched WT, CASQ2−/− and CASQ2D307H myocardium from 3 hearts per group. T-tubules are shaded yellow and the SR vesicles are green. The electron-dense “clumps” are indicated by arrows.
Expression of Mutant CASQ2D307H Protein Restores the Ultrastructural Alterations of the CASQ2 Null Background
Electron microscopy illustrates the disposition of CASQ2 within the jSR cisternae. In cardiac myocytes from wild type mice (Fig. 2b, top row), the jSR lumen has a fairly uniform, narrow width and is occupied by a tight arrangement of small electron-dense “clumps,” presumably composed of CASQ2 associated with Triadin1 and Junctin (31). We call this a “tight” configuration. In CASQ2 null hearts (Fig. 2b, middle row), on the other hand, the jSR cisternae are apparently empty and have a “loose” configuration and a variable width (26). In contrast, most jSR cisternae in the mutant hearts expressing CASQ2D307H on a null background (Fig. 2b, bottom row) were identical to those in wild type myocytes: they have a narrow width and show a dark, tight staining pattern within the SR lumen similar to WT CASQ2.
Differences in the appearance of jSR from WT, CASQ2 null, and CASQ2D307H hearts are quite obvious to the eye and this allowed us to obtain a fairly precise estimate of the relative frequency of jSR cisternae with the appearances described above. Table 1 combines counts from three wild type hearts from the current set of experiments (first row) with counts from three CASQ2 null hearts, from previous experiments (middle row), and three CASQ2D307H hearts (bottom row). In wild type hearts the great majority (93%) of jSR cisternae has a clear tight configuration; a few cisternae are intermediate and none are totally loose. In null hearts practically all jSR cisternae (93%) are loose; in CASQ2D307H rescued hearts the majority (73%) are tight and the remaining have an intermediate or loose configuration. The rescue, although excellent, is not complete, because the mean values for the tight configuration are lower in rescued than in wild type hearts.
TABLE 1.
CASQ2D307H restores tight jSR configuration in CASQ2 null myocytes
jSR profiles were visually classified as tight if the CASQ2 content of the jSR appeared clustered into small electron dense spots or loose if CASQ2 was either absent or not clustered. The majority of jSR profiles are tight in WT cells and loose in CASQ2 null cells. Values are shown as mean ± S.D. calculated for 100 jSR cisternae from 3 mice for each point.
| Type | Tight jSR | Intermediate jSR | Loose jSR |
|---|---|---|---|
| % of total | |||
| WT | 93 ± 7a | 7 ± 7 | 0 |
| CASQ2 null | 0 | 11 ± 12 | 89 ± 12b |
| CASQ2D307H | 73 ± 16a | 17 ± 11 | 10 ± 10b |
a Values are significantly different (Student's t test, p < 0.4.10−6).
b Values are highly significantly different (Student's t test, p < 0.9.10−34).
This confirms the correct localization of the mutant CASQ2D307H protein to the jSR and its interaction with the proteins (Triadin1 and Junctin) that are responsible for clustering of CASQ2. More importantly, it indicates that the expression of CASQ2D307H partially restores the jSR modifications reported in the CASQ2 null mouse hearts.
The Mutant CASQ2D307H Protein Gets Ubiquitinated at the Same Rate as the Wild Type CASQ2
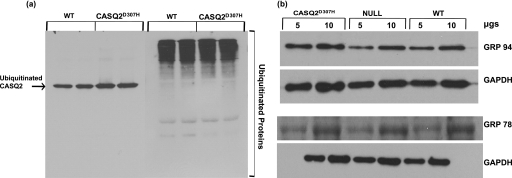
A recent study, using a D307H knock-in mouse model, showed that the CASQ2D307H protein is almost undetectable in these hearts and therefore suggested that the mutation could render the protein highly unstable, which could get degraded (20). We therefore investigated whether the mutant protein is degraded faster than WT protein through the proteasomal pathway. Ubiquitination is an early step for proteins destined for degradation via the proteasome. Using a ubiquitin pulldown assay, the relative abundance of ubiquitinated protein was estimated for mutant CASQ2D307H in comparison to wild type CASQ2 (n = 4; protein quantified by Image J software). The summarized data from the 4 separate experiments show that the mutant and WT proteins are ubiquitinated at similar rates (Fig. 3a).
FIGURE 3.
a, the mutant CASQ2D307H gets ubiquitinated at the same rate as the WT CASQ2. Whole heart homogenates from WT and CASQ2D307H (transgenic) (n = 4) were enriched for ubiquitinated proteins, separated on a 10% SDS gel, and probed for CASQ2 (left) and ubiquitin (right). The experiment was repeated 4 times. b, expression of CASQ2D307H protein does not affect the expression level of ER chaperone proteins GRP94 and GRP78 relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as a standard.
Expression of Mutant CASQ2D307H Protein Does Not Induce ER Stress Response in Mouse Hearts
In response to cellular stress such as expression of mutant proteins that may not be correctly folded, expression of chaperone proteins such as GRP94, GRP78, and/or calreticulin are up-regulated to help fold them. This unfolded protein response helps proper folding of the damaged proteins (32, 33). Therefore we determined if overexpression of the CASQ2D307H mutant protein in the heart can induce an ER stress response. We analyzed expression levels of ER stress markers GRP94, GRP78, and calreticulin by Western blotting (n = 6) in WT, CASQ2 null, and CASQ2D307H hearts. There were no significant differences in the expression of these proteins between the three groups (Figs. 1a and 3b).
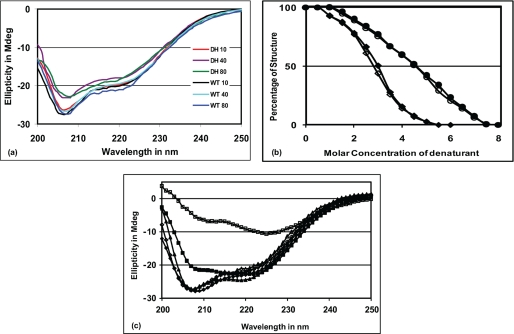
CASQ2D307H Protein Shows Similar CD Spectra as Wild Type CASQ2, when Exposed to High Temperature and Denaturing Conditions
The expression profile of the mutant CASQ2D307H in vivo prompted us to further verify the stability of the mutant protein in vitro using bacterially expressed CASQ2D307H. Our studies show that both WT and D307H mutants have similar CD spectra (Fig. 4a). The CD spectra did not show any significant change for both WT and mutant protein even at 80 °C (Fig. 4a). We next studied protein unfolding following chemical denaturation using urea and GdnHCl and by plotting ellipticity at 222 nm as a function of denaturant concentration (Fig. 4b). The denaturation profile of the WT and mutant proteins nearly overlapped with each other. They are structurally intact at lower concentrations of urea and start to denature only when the urea concentration reaches about 2 m. Complete unfolding was achieved with 7 m urea. Concentration of urea corresponding to 50% unfolding (Cm) was 4.5 m in both cases. In the presence of GdnHCl the proteins underwent sharper denaturation. They started to denature at 0.5 m GdnHCl concentration and complete denaturation was achieved with 4.0 m GdnHCl and Cm for GdnHCl was 2.7 and 3 m for D307H mutant and wild type proteins, respectively. Therefore these studies indicate that the WT and mutant protein have similar conformations in the absence of calcium and are comparably stable at higher temperatures and under chemical denaturation conditions.
FIGURE 4.
CASQ2D307H mutant is relatively stable when treated with (a) temperature, (b) denaturants, but shows (c) altered behavior to calcium. Solid symbols are used for WT and open symbols are used for D307H protein. a, far-UV CD spectra of both proteins at 10, 40, and 80 °C shows that both proteins are equally stable at higher temperatures. b, unfolding curves of the two proteins in the presence of urea (●/○) and GdnHCl (♦/◇) are very similar. c, when [Ca2+] was increased from 0 to 5 mm, the CD spectrum of the mutant protein became drastically different from that of WT (■/□). Upon chelation of Ca2+ with EGTA (♦/◇), both proteins could regain CD ellipticity comparable with their native conformation at 25 °C (▴/Δ), although the WT protein needed only ∼3 mm EGTA, the mutant protein required ∼5.5 mm EGTA for the same.
D307H Mutant Protein Shows Altered Aggregation in the Presence of Ca2+
We studied the conformational changes of the mutant protein under increasing Ca2+ concentration from 0 to 5 mm (Fig. 4c). In the presence of 5 mm Ca2+ the mutant protein lost its CD ellipticity significantly more than the WT protein and the CD spectra were completely different. Additionally, while the WT protein solution visually remained in a crystalline form, the D307H protein solution became more turbid and the calcium-induced aggregations were irregular in shape; upon addition of EGTA (a Ca2+ chelator), both proteins regained ellipticity similar to their native CD spectral pattern. Interestingly, the wild type CASQ2 regained its secondary structure comparable with native conformation with ∼3 mm EGTA, while the mutant protein required ∼5.5 mm EGTA to attain its native conformation. This study indicates that the mutant protein behaves quite differently only at high calcium concentrations, indicating a defect in its ability to dynamically handle calcium.
The Mutant Protein Is More Susceptible to Proteolysis under High Ca2+
Trypsin digestion has been used to test conformational changes of CASQ in the presence of Ca2+ (21). Therefore we treated both WT and mutant CASQ2D307H to trypsin digestion in the absence and presence of increasing CaCl2. We found that in the absence of CaCl2, both proteins were highly sensitive to trypsin digestion and the proteolytic patterns of the two proteins were very similar within 5–120 min digestion (Fig. 5a). In contrast, in the presence of CaCl2 the speed of the enzymatic digestion of both proteins became slower (Fig. 5, b and c) with distinct proteolytic fragments becoming apparent only after 20-30 min. The WT CASQ2 became increasingly resistant to trypsin digestion as the Ca2+ concentration increased, whereas the mutant CASQ2D307H remained susceptible to trypsin digestion (Fig. 5, b and c). At 1 mm CaCl2, WT and mutant proteins show proteolytic fragments that are different between 20–25 and 37 kDa, and between 2 and 5 mm CaCl2, proteolytic fragments around 37 kDa are different (Fig. 5, b and c). These data suggest that conformational differences between these two proteins become apparent only when the concentration of CaCl2 increases, supporting the CD data.
FIGURE 5.
The mutant CASQ2D307H is more susceptible to proteolysis. WT and D307H mutant CASQ2 proteins were subjected to proteolysis by TPCK-treated trypsin. a, in the absence of CaCl2 (b and c) and presence of CaCl2 at 1, 2, and 5 mm. 5–120 m refers to duration of digestion. The tryptic fragments that are different between WT and D307H mutant are indicated by black arrows. The molecular weight of the protein marker bands are indicated on the right side of the gels. Experiments were repeated 4 times.
DISCUSSION
Cardiac SR Ca2+ release is orchestrated by a macromolecular complex involving multiple proteins both at the cytoplasmic and the lumenal side of the jSR membrane. It is believed that close interactions involving RYR2, Triadin1 and Junctin, and CASQ2 at the luminal side serve to regulate the opening, closing, and the refractory state (gating behavior) of the channel (4, 5, 10, 11). In this regard CASQ2 is believed to act as a Ca2+ sensor and regulator of Ca2+ release, both by controlling local Ca2+ reservoir and through its dynamic interaction with RYR2 along with Triadin1 and Junctin. The role of CASQ2 as a regulator of Ca2+ release has gained much attention by the recent discovery of several point mutations leading to CPVT.
The goal of this study was to understand how a point mutation D307H affects CASQ2 structure and function and how it contributes to the CPVT phenotype. To critically examine this question, the mutant CASQ2D307H protein was expressed 2-fold under the control of α-myosin heavy chain promoter, in the CASQ2 null background. A key question was if the expressed protein was properly targeted to the SR. Confocal immunofluorescent images, and electron microscopy indicate that the mutant protein is targeted to the jSR. Furthermore, histological analyses of the mutant hearts did not reveal evidence of cardiac hypertrophy and/or structural disease (data not shown). Interestingly, the expression of this mutant protein in the null background partially restores the ultrastructural modifications that were reported when CASQ2 is totally absent (26), showing that the presence of CASQ2, even if mutated, is critical for maintaining the normal jSR architecture. Previous studies showed that 10-fold overexpression of canine CASQ2 (1) showed a large number of membrane-limited vesicles of CASQ2 indicating a localization that was not limited to the jSR when CASQ2 is overexpressed. However, in the present mouse model we did not observe any abnormal distribution of CASQ2 and 2-fold expression of this mutant is well tolerated. In contrast to the 10-fold CASQ2 overexpression mouse model (1) and adenoviral gene transfer studies (15), which suggested that increased expression of CASQ2D307H up to 3-fold produced a gain of function and increased the Ca2+ load, we did not find a corresponding increase in the SR calcium load in our mouse model (data not shown).
Song et al. (20) recently reported a knock-in mouse model for the D307H mutation wherein the expression of the mutant CASQ2D307H protein was below the detection level despite normal mRNA expression. This finding raised the possibility that the mutation could compromise the stability of the protein. Because there is no protein data available from CPVT patients with the D307H mutation, it has not been possible to confirm if this mutant protein is expressed or not. We and others have previously shown that the mutant protein can be expressed in cardiac myocytes and mouse models (13, 15, 16). In our studies we have employed the α-myosin heavy chain promoter that drives higher protein expression and makes it possible to examine protein stability. Data shown in this article suggest that the point mutation D307H does not affect protein stability: expression of the CASQ2D307H mutant protein (2-fold) does not result in an increase in ER stress response as evidenced by unchanged levels of ER chaperones. Second, the ubiquitin pulldown assay showed that the mutant CASQ2D307H protein is ubiquitinated at the same level as that of the WT CASQ2. We found a 2-fold difference in the ubiquitinated fraction of CASQ2 in CASQ2D307H samples indicative of the inherent difference in the CASQ2 expression in these hearts (if the mutant protein was prone to misfolding and rapid degradation, we expected to see an increase in the proportion of ubiquitinated CASQ2) and last, in vivo treatment with the proteasomal inhibitor MG 132 did not show an increase in the mutant protein level, which suggested that the mutant is not preferentially degraded.3
Thermal and chemical denaturation studies with purified CASQ2 protein also suggest that both wild type and D307H mutant proteins are equally stable and their folding patterns are very similar. This is additionally supported by the finding that in the absence of Ca2+, both WT and mutant CASQ2 protein show increased sensitivity to trypsin within 5 min exposure to the enzyme. Both proteins show identical digestion patterns and result in similar proteolytic fragments suggesting similar conformational or folding states in the absence of Ca2+ when they are mostly in a monomeric state. Exposure to increased [Ca], a condition that promotes higher order polymers (8), protects both proteins from rapid enzymatic digestion to 30 min (data not shown). However, conformational differences between WT and mutant CASQ2 become apparent under longer periods of trypsin digestion (30–120 min). With increasing [Ca] concentration, WT protein becomes increasingly resistant to trypsin indicative of higher order packing that masks sites that were previously accessible to trypsin (34), whereas the mutant CASQ2D307H continues to be susceptible to trypsin digestion. An increased susceptibility to trypsin was also observed with other CASQ2 mutations, R33Q and L167H suggesting that these mutations affect conformational flexibility of CASQ2 (21, 22). Our finding indicates that the mutant protein exhibits an altered conformation only under high [Ca], and could potentially affect the dynamic Ca2+ buffering and release properties. Interestingly, the mutant protein required twice the amount of EGTA to regain its original CD spectra (similar to the native conformation) following Ca2+-induced aggregation. It is known that polymerization of CASQ2 depends on the concentration of positively charged ions and is strongly affected by divalent cations like calcium (35). With increasing [Ca], CASQ2 undergoes aggregation and in solution can be seen as needle-shaped crystalline structures (6). During our high [Ca] experiments we could observe similar needle-shaped crystalline shapes only with the WT protein, whereas aggregation formed by the mutant protein was predominantly irregular in shape. These studies suggest that the mutation affects the supramolecular organization and architecture of CASQ2 in the presence of Ca2+.
In summary, by generating a mouse model expressing the mutant D307H protein in a CASQ2 null background, we demonstrate that the mutant CASQ2D307H protein is expressed and targeted to the jSR similar to the WT CASQ2 and is capable of restoring jSR ultrastructure. Our data collectively suggest that the mutation does not affect protein stability and targeting to jSR but may compromise its dynamic regulation of calcium buffering.
This work was supported, in whole or in part, by National Institutes of Health Grant R0-1 HL64014 (to M. P.) and an American Heart Association pre-doctoral fellowship (to A. K.).
A. Kalyanasundaram, N. C. Bal, C. Franzini Armstrong, B. C. Knollmann, and M. Periasamy, unpublished data.
- CASQ
- calsequestrin
- CPVT
- catecholaminergic polymorphic ventricular tachycardia
- GdnHCl
- guanidine hydrochloride
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- CD
- circular dichroism
- SR
- sarcoplasmic reticulum
- jSR
- junctional sarcoplasmic reticulum
- WT
- wild type
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Jones L. R., Suzuki Y. J., Wang W., Kobayashi Y. M., Ramesh V., Franzini-Armstrong C., Cleemann L., Morad M. (1998) J. Clin. Invest. 101, 1385–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terentyev D., Viatchenko-Karpinski S., Györke I., Volpe P., Williams S. C., Györke S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11759–11764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard N. A., Laver D. R., Dulhunty A. F. (2004) Prog. Biophys. Mol. Biol. 85, 33–69 [DOI] [PubMed] [Google Scholar]
- 4.Guo W., Campbell K. P. (1995) J. Biol. Chem. 270, 9027–9030 [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Kelley J., Schmeisser G., Kobayashi Y. M., Jones L. R. (1997) J. Biol. Chem. 272, 23389–23397 [DOI] [PubMed] [Google Scholar]
- 6.Wang S., Trumble W. R., Liao H., Wesson C. R., Dunker A. K., Kang C. H. (1998) Nat. Struct. Biol. 5, 476–483 [DOI] [PubMed] [Google Scholar]
- 7.Glover L., Culligan K., Cala S., Mulvey C., Ohlendieck K. (2001) Biochim. Biophys. Acta 1515, 120–132 [DOI] [PubMed] [Google Scholar]
- 8.Kang C., Trumble W. R., Dunker A. K. (2002) Methods Mol. Biol. 172, 281–294 [DOI] [PubMed] [Google Scholar]
- 9.MacLennan D. H., Abu-Abed M., Kang C. (2002) J. Mol. Cell. Cardiol. 34, 897–918 [DOI] [PubMed] [Google Scholar]
- 10.Györke I., Hester N., Jones L. R., Györke S. (2004) Biophys. J. 86, 2121–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Györke S., Györke I., Terentyev D., Viatchenko-Karpinski S., Williams S. C. (2004) Biol. Res. 37, 603–607 [DOI] [PubMed] [Google Scholar]
- 12.di Barletta M. R., Viatchenko-Karpinski S., Nori A., Memmi M., Terentyev D., Turcato F., Valle G., Rizzi N., Napolitano C., Gyorke S., Volpe P., Priori S. G. (2006) Circulation 114, 1012–1019 [DOI] [PubMed] [Google Scholar]
- 13.Houle T. D., Ram M. L., Cala S. E. (2004) Cardiovasc. Res. 64, 227–233 [DOI] [PubMed] [Google Scholar]
- 14.Postma A. V., Denjoy I., Hoorntje T. M., Lupoglazoff J. M., Da Costa A., Sebillon P., Mannens M. M., Wilde A. A., Guicheney P. (2002) Circ. Res. 91, e21–26 [DOI] [PubMed] [Google Scholar]
- 15.Viatchenko-Karpinski S., Terentyev D., Györke I., Terentyeva R., Volpe P., Priori S. G., Napolitano C., Nori A., Williams S. C., Györke S. (2004) Circ. Res. 94, 471–477 [DOI] [PubMed] [Google Scholar]
- 16.Dirksen W. P., Lacombe V. A., Chi M., Kalyanasundaram A., Viatchenko-Karpinski S., Terentyev D., Zhou Z., Vedamoorthyrao S., Li N., Chiamvimonvat N., Carnes C. A., Franzini-Armstrong C., Györke S., Periasamy M. (2007) Cardiovasc. Res. 75, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eldar M., Pras E., Lahat H. (2003) Trends Cardiovasc. Med. 13, 148–151 [DOI] [PubMed] [Google Scholar]
- 18.Kim E., Youn B., Kemper L., Campbell C., Milting H., Varsanyi M., Kang C. (2007) J. Mol. Biol. 373, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 19.Lahat H., Pras E., Eldar M. (2004) Ann. Med. 36, Suppl. 1, 87–91 [DOI] [PubMed] [Google Scholar]
- 20.Song L., Alcalai R., Arad M., Wolf C. M., Toka O., Conner D. A., Berul C. I., Eldar M., Seidman C. E., Seidman J. G. (2007) J. Clin. Invest. 117, 1814–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzi N., Liu N., Napolitano C., Nori A., Turcato F., Colombi B., Bicciato S., Arcelli D., Spedito A., Scelsi M., Villani L., Esposito G., Boncompagni S., Protasi F., Volpe P., Priori S. G. (2008) Circ. Res. 103, 298–306 [DOI] [PubMed] [Google Scholar]
- 22.Valle G., Galla D., Nori A., Priori S. G., Gyorke S., de Filippis V., Volpe P. (2008) Biochem. J. 413, 291–303 [DOI] [PubMed] [Google Scholar]
- 23.Lahat H., Pras E., Olender T., Avidan N., Ben-Asher E., Man O., Levy-Nissenbaum E., Khoury A., Lorber A., Goldman B., Lancet D., Eldar M. (2001) Am. J. Hum. Genet. 69, 1378–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terentyev D., Nori A., Santoro M., Viatchenko-Karpinski S., Kubalova Z., Gyorke I., Terentyeva R., Vedamoorthyrao S., Blom N. A., Valle G., Napolitano C., Williams S. C., Volpe P., Priori S. G., Gyorke S. (2006) Circ. Res. 98, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 25.Lahat H., Eldar M., Levy-Nissenbaum E., Bahan T., Friedman E., Khoury A., Lorber A., Kastner D. L., Goldman B., Pras E. (2001) Circulation 103, 2822–2827 [DOI] [PubMed] [Google Scholar]
- 26.Knollmann B. C., Chopra N., Hlaing T., Akin B., Yang T., Ettensohn K., Knollmann B. E., Horton K. D., Weissman N. J., Holinstat I., Zhang W., Roden D. M., Jones L. R., Franzini-Armstrong C., Pfeifer K. (2006) J. Clin. Invest. 116, 2510–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulick J., Subramaniam A., Neumann J., Robbins J. (1991) J. Biol. Chem. 266, 9180–9185 [PubMed] [Google Scholar]
- 28.Subramaniam A., Gulick J., Neumann J., Knotts S., Robbins J. (1993) J. Biol. Chem. 268, 4331–4336 [PubMed] [Google Scholar]
- 29.Subramaniam A., Jones W. K., Gulick J., Wert S., Neumann J., Robbins J. (1991) J. Biol. Chem. 266, 24613–24620 [PubMed] [Google Scholar]
- 30.Babu G. J., Zheng Z., Natarajan P., Wheeler D., Janssen P. M., Periasamy M. (2005) Cardiovasc. Res. 65, 177–186 [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Franzini-Armstrong C., Ramesh V., Jones L. R. (2001) J. Mol. Cell. Cardiol. 33, 233–247 [DOI] [PubMed] [Google Scholar]
- 32.Schröder M., Kaufman R. J. (2005) Mutat. Res. 569, 29–63 [DOI] [PubMed] [Google Scholar]
- 33.Shen X., Zhang K., Kaufman R. J. (2004) J. Chem. Neuroanat. 28, 79–92 [DOI] [PubMed] [Google Scholar]
- 34.Mitchell R. D., Simmerman H. K., Jones L. R. (1988) J. Biol. Chem. 263, 1376–1381 [PubMed] [Google Scholar]
- 35.Ikemoto N., Nagy B., Bhatnagar G. M., Gergely J. (1974) J. Biol. Chem. 249, 2357–2365 [PubMed] [Google Scholar]