Abstract
Nicotinic acid adenine dinucleotide phosphate (NAADP) is a potent and widespread calcium-mobilizing messenger, the properties of which have been most extensively described in sea urchin eggs. The molecular basis for calcium release by NAADP, however, is not clear and subject to controversy. Recent studies have provided evidence that members of the two-pore channel (TPC) family in mammals are the long sought after target channels for NAADP. Here, we show that the TPC3 gene, which has yet to be functionally characterized, is present throughout the deuterostome lineage but is a pseudogene in humans and other primates. We report the molecular cloning of the complete ancestral TPC gene family from the sea urchin and demonstrate that all three isoforms localize to acidic organelles to mediate NAADP-dependent calcium release. Our data highlight the functional divergence of this novel gene family during deuterostome evolution and provide further evidence that NAADP mediates calcium release from acidic stores through activation of TPCs.
Keywords: Calcium, Calcium Channels, Endosomes, Lysosomes, Sea Urchin, NAADP, Acidocalcisomes
Introduction
A multitude of extracellular stimuli mediate their cellular effects through the mobilization of intracellular calcium stores (1). Central to this ubiquitous signal transduction pathway is the production of calcium-mobilizing messengers inositol trisphosphate (IP3),3 cyclic ADP-ribose (cADPR), and NAADP (1, 2). The molecular basis of calcium release by IP3 and cADPR is established (1, 2); both release calcium from endoplasmic reticulum calcium stores through activation of IP3 and ryanodine receptors, respectively. In contrast, although NAADP has been shown to mobilize calcium in a multitude of cell types from plants to humans and to regulate a variety of calcium-dependent processes from fertilization to neuronal growth, molecular details of this pathway are lacking (3).
The calcium-mobilizing properties of NAADP were first described in eggs of the sea urchin (4), and this model organism has provided key insight into the mode of action of NAADP (5). In eggs, it is clear, based on cross desensitization studies (4), lack of feedback regulation by cytosolic calcium (6), molecular size (7), and unusual ligand binding properties (8), that NAADP activates calcium-permeable channels unrelated to IP3 and ryanodine receptors. Moreover, NAADP is highly unusual in releasing calcium not from the endoplasmic reticulum but from acidic lysosome-like calcium stores (9). Much supporting evidence has been obtained in mammalian cells (10), and recent studies in smooth muscle cells suggest that NAADP mobilizes calcium through activation of the lysosomal ion channel TRPML1 (11, 12), although these findings have yet to be independently verified. Other studies, however, indicate that NAADP does not mobilize calcium from acidic calcium stores but instead stimulates calcium influx (13), possibly through TRPM2 (14), or mobilizes endoplasmic reticulum calcium stores via direct activation of the ryanodine receptor (15, 16). The mechanism of action of NAADP is therefore controversial (17).
In three recent independent studies, direct evidence has been provided that NAADP mobilizes calcium from acidic calcium stores through activation of a previously uncharacterized family of ion channels in animals known as the two-pore channels (TPCs) (18–20). Comparative genomic analysis indicates the presence of up to three family members (TPC1–3) in the animal kingdom (18, 19). Human and mouse TPC1 and TPC2 were shown to localize to endosomes and lysosomes and to potentiate NAADP responses when overexpressed (18–20). Additionally, in a human cancer cell line, silencing of TPC1 or overexpression of TPC1 mutated within a pore region was shown to inhibit endogenous NAADP responses (18). Endogenous NAADP responses in mouse pancreatic beta cells were also abolished in mice lacking the TPC2 gene (19). Taken together, these data provide strong evidence that TPC1 and TPC2 are novel endolysosomal ion channels targeted by NAADP. Research into animal TPCs is only in its infancy, and at present, little is known concerning the properties of TPC3.
Here, we define an ancestral, three-member TPC gene family in deuterostomes and provide the first evidence of a TPC3 pseudogene in primates. We report the molecular cloning of all three TPC isoforms from the sea urchin, a model system for the study of calcium-signaling pathways, and show that they target to acidic vesicles and support calcium release by NAADP from acidic calcium stores.
MATERIALS AND METHODS
Comparative Genomics
TPC sequences were obtained from the genomic and protein databases of the National Center for Biotechnology Information and the Department of Energy Joint Genome Institute. Partial coding sequences of TPC3 pseudogenes were identified in corresponding primate genomes by significant sequence similarities with cow (GenBank™ accession number NM_001161651) and dog (GenBank accession number XM_540172) TPC3 cDNA nucleotide sequences. To allow for frameshift errors, the Wise2 form (available through the EMBL-EBI web site) was utilized for the translation of TPC3 pseudogenes by comparison with cow TPC3 protein sequence (GenBank accession number NP_001155123) followed by manual editing.
Molecular Cloning of Sea Urchin TPCs and Plasmid Construction
A BLAST search of the Strongylocentrotus purpuratus genome assembly (version 2.1) with protein sequences for human TPC1 (GenBank accession number NP_060371.2) and TPC2 (GenBank accession number NP_620714.1) identified two hypothetical proteins (GenBank accession numbers XP_784701.2 and XP_796820.2) displaying significant sequence similarity. We also identified a third hypothetical protein predicted by Gnomon based on genomic contig SpuUn_WGA76937_2 (hmm 271542). A 2816-bp clone corresponding to the full-length coding sequence and 162 bp of 3′-UTR of the first homologue (designated SpTPC1) was amplified by reverse transcription-PCR using oligo(dT)-primed cDNA prepared from eggs and the following gene-specific primers (restriction sites underlined): 5′-CACCGAATTCAATGGCGGATTTGGAGGAC-3′ (forward) and 5′-AGGCTCGAGGGCTCTTTCGGAAAATGAAA-3′ (reverse). A 2391-bp clone corresponding to the second homologue (SpTPC2) was also amplified from egg cDNA using the following gene-specific primers: 5′-CACCCCATGGGAGACTACTACGAGTATGAGAG-3′ (forward) and 5′-AGTTCTAGATTAGAAGCGCAGGTTTTGTATG-3′ (reverse). A 2698-bp clone corresponding to 124 bp of 5′-UTR, full-length coding sequence, plus 51 bp of 3′-UTR of the third homologue (SpTPC3) was amplified from cDNA prepared from embryos (54 h after fertilization) using the following gene-specific primers: 5′-CAGGATCATATCCCATGCAG-3′ (forward) and 5′-ATCACTGGATGAAACGCACA-3′ (reverse). All products were subcloned into either pCR®II-TOPO® (SpTPC1 and SpTPC2) or pCR®-Blunt II-TOPO® (SpTPC3) vectors (Invitrogen) and sequenced in both directions.
Vectors harboring the SpTPC1 and SpTPC2 sequences were digested with EcoRI/XhoI and NcoI/XbaI, respectively, and the inserts were cloned into the corresponding restriction sites of pCS2 + 6MT to introduce myc tags at the N termini. We also generated constructs encoding for all three isoforms tagged with GFP at their C termini. To achieve this, the coding sequences (lacking the stop codon) were amplified using the following primers: 5′-CACCGAATTCAATGGCGGATTTGGAGGAC-3′ (forward) and 5′-AGGCTCGAGTAACTCATCTCCAAGAATATCATCATC-3′ (reverse) for SpTPC1; 5′-CACCATCGATATGGGAGACTACTACGAGTATGAGAG-3′ (forward) and 5′- AGGCTCGAGGAAGCGCAGGTTTTGTATGTG-3′ (reverse) for SpTPC2; and 5′-CACCGGATCCAAGATGGAGGGGCCAAAG-3′ (forward) and 5′-AGGCTCGAGTGCTATGTTATCCACGGAAAGA-3′ (reverse) for SpTPC3. The products were then cloned into pCS2+ GFP (18) at the EcoRI/XhoI (SpTPC1), ClaI/XhoI (SpTPC2), and BamHI/XhoI (SpTPC3) restriction sites. Plasmids encoding for GFP-tagged HsTPC1 and TPC2 have been described previously (18).
Subcellular Localization and Calcium Imaging
TPCs were heterologously expressed in Xenopus laevis oocytes, SKBR3 cells, and HEK cells for subcellular localization and calcium imaging as described previously (18). Briefly, oocytes were subject to nuclear plasmid microinjection and maintained for 48 h prior to confocal microscopy of live oocytes. SKBR3 and HEK cells were transiently transfected with plasmids using Lipofectamine™ 2000 transfection according to the manufacturer's instructions. After 24–48 h, cells were fixed for confocal microscopy. In some experiments, SKBR3 cells were loaded with LysoTracker Red (1 μm, Invitrogen) for 30 min prior to fixation. Cytosolic calcium concentration in SKBR3 cells was measured by imaging of fura-2 fluorescence. NAADP and cyclic ADP-ribose were delivered by microinjection.
RESULTS AND DISCUSSION
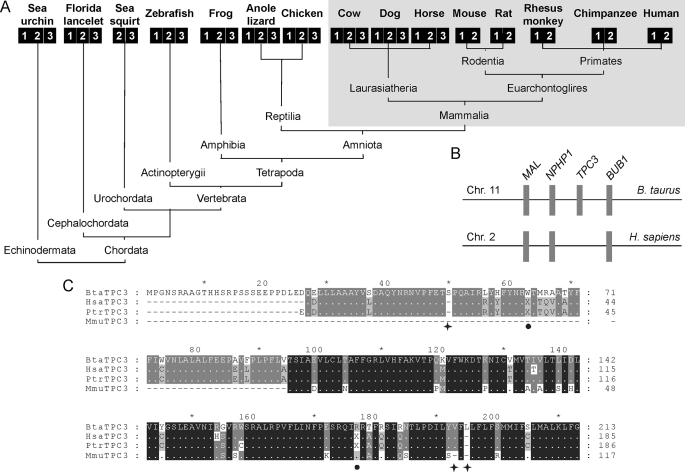
As reported recently, the TPC gene underwent expansion early in animal evolution (18). Fig. 1A shows the TPC gene complement of several deuterostomes for which complete genomes are available. Deuterostomes are animals in which the blastopore of the embryo forms the anus. Three TPC genes are found in sea urchins (a basal deuterostome) and in most other animals representative of the major clades within the deuterostome lineage. In mammals, however, the TPC3 gene is apparently lacking in humans and other primates and also in rats and mice (Fig. 1A). Strikingly, other closely related placental mammals such as cows, dogs, and horses possess the full TPC gene complement (Fig. 1A). These data indicate that loss of TPC3 in mammals is an unusually rare, relatively recent event likely beginning after the split of Euarchontoglires from Laurasiatheria, its diverse sister group. In cows, the TPC3 gene is flanked by NPHP1 and BUB1 (Fig. 1B). This arrangement is conserved in dogs, horses, chickens, and frogs (data not shown). Based on this synteny, we inspected the corresponding genomic sequence in humans for TPC3-related sequences. We found sequences that showed significant sequence identity to the N terminus of TPC3 (supplemental Fig. S1). Similar results were obtained in monkeys and chimpanzees (supplemental Fig. S1). Intriguingly, we noted the presence of conserved nucleotide deletions in primate sequences, the first of which introduces a downstream stop codon that would result in the generation of a truncated protein that is unlikely to function as a channel (supplemental Fig. S1, circle). Fig. 1C shows an alignment of the cow TPC3 amino acid sequence and those deduced from primate TPC3 genomic sequences following manual editing to preserve the reading frame. This analysis reveals marked sequence similarity at the protein level. These data provide syntenic and genomic evidence that the TPC3 gene has degenerated into a pseudogene in humans and other primates as proposed for the monkey (19), indicating loss of functions associated with this isoform in these animals. TPC3, together with TRPC2 (21), represent the only known calcium-permeable channels in mammals for which pseudogenes have been described.
FIGURE 1.
TPC3 is a pseudogene in primates. A and B, deuterostome phylogeny of TPC genes (unscaled). Each isoform is represented by a numbered square. The TPC3 gene complement of mammals is shaded. B, chromosomal synteny between cow and human genomes in the vicinity of the TPC3 gene. Chr. 11, chromosome 11; Chr. 2, chromosome 2; B. taurus, Bos taurus; H. sapiens, Homo sapiens. C, multiple sequence alignment of translated amino acid sequences of cow TPC3 (BtaTPC3) and TPC3 pseudogenes from human (Hsa), monkey (Mmu), and chimpanzee (Ptr). Missing amino acids resulting from nucleotide deletion in primate genomes are marked with asterisks. The reading frame was manually adjusted at these positions to maximize subsequent sequence alignment with the cow sequence. In-frame stop codons (after adjustment) are indicated with filled circles.
TPC1 and TPC2 have recently emerged as credible candidates for the elusive NAADP-sensitive calcium channel in mammals (18–20). To gain insight in to the ancestral TPC family, we determined the nucleotide sequences of TPCs from the purple sea urchin, which possesses the full TPC complement and in which the effects of NAADP have been extensively characterized (5). We have previously shown that transcripts for sea urchin TPC1 (SpTPC1) and TPC2 (SpTPC2) are readily detectable in eggs, whereas those for TPC3 (SpTPC3) are more abundant in embryos (18). The coding sequences were therefore amplified by reverse transcription-PCR using either egg or embryo cDNA as appropriate. The predicted molecular masses of SpTPC1 and SpTPC2 were 101 and 92 kDa, respectively. The corresponding value for SpTPC3 (98 kDa) was similar. A multiple sequence alignment of sea urchin and human TPCs is presented in supplemental Fig. S2. Overall similarity was 30–40%, which is substantially lower than that between members of the IP3 and ryanodine receptor family of intracellular calcium channels (∼70%). Notably, sequence similarity between sea urchin TPCs was particularly high in the two putative pores (supplemental Fig. S2, red). These data provide a comprehensive molecular description of the TPC family in this model organism.
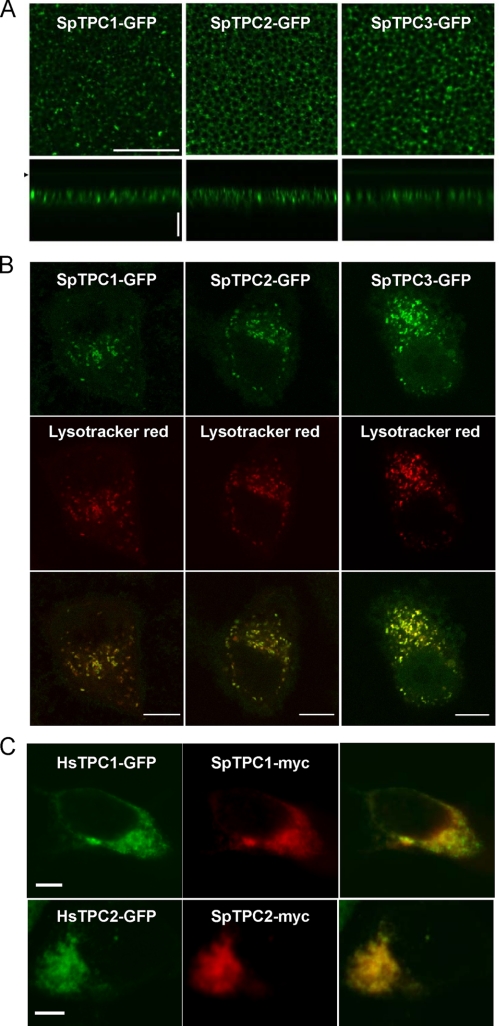
To define the properties of sea urchin TPCs, we determined their subcellular location. Fig. 2A shows the distribution of GFP-tagged SpTPC1 and SpTPC2 heterologously expressed in Xenopus oocytes. Both isoforms showed a punctate intracellular distribution in the oocyte periphery, localizing to a heterogeneous population of vesicular structures. A punctuate distribution was also observed in SKBR3 cells and HEK cells (Fig. 2, B and C). Co-labeling of cells with the fluorescent weak base LysoTracker Red revealed marked co-localization (Fig. 2B), indicating that SpTPC1 and SpTPC2 target to acidic vesicles, probably endosomes and lysosomes, as reported for mammalian TPCs (18–20) and consistent with functional data showing release of calcium by NAADP from these organelles (9, 22). Indeed, both urchin isoforms co-localized with human TPC1 and TPC2 when co-expressed (Fig. 2C). Furthermore, SpTPC2 co-localized with the late endosome/lysosome marker, LAMP-1 (data not shown). We also determined the subcellular location of SpTPC3. As with SpTPC1 and SpTPC2, SpTPC3 showed a punctate intracellular distribution, which overlapped with that of LysoTracker Red (Fig. 2B). SpTPCs therefore appear to localize to acidic organelles.
FIGURE 2.
Sea urchin TPCs localize to acidic organelles. A–C, confocal fluorescence images of X. laevis oocytes (A), SKBR3 cells (B), and HEK cells (C) expressing the indicated sea urchin TPC fusion protein. A, images of the animal hemisphere taken in lateral (xy, scale = 20 μm) and axial (xz, scale = 10 μm) sections. The arrow indicates the position the of coverslip in z-scan (depth). In B, cells were loaded with LysoTracker Red, whereas in C, the cells were cotransfected with human TPCs.
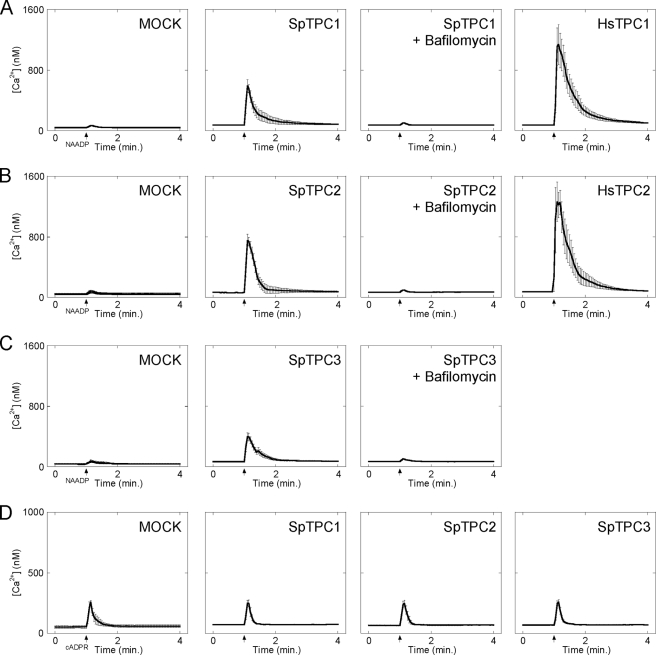
To determine whether SpTPCs are sensitive to NAADP, we examined the effect of microinjecting NAADP on cytosolic calcium levels in SKBR3 cells overexpressing SpTPCs. As reported previously (18), microinjection of 10 nm NAADP (pipette concentration) had little effect on cytosolic calcium levels in mock-transfected cells but induced robust responses in cells overexpressing human TPC1 (Fig. 3A). Similar results were obtained with human TPC2 (Fig. 3B). Importantly, enhanced responses to NAADP were also observed in cells overexpressing SpTPC1 and SpTPC2 (Fig. 3, A and B). These data are entirely consistent with the notion that these proteins are NAADP-sensitive calcium-permeable channels. The effects of NAADP in cells overexpressing sea urchin TPCs were abolished by pretreating cells with the V-type ATPase inhibitor bafilomycin A-1 (Fig. 3, A and B). This compound collapses the pH gradient across acidic organelles and is thereby thought to compromise calcium uptake (9). These results, together with our localization data (Fig. 2), are consistent with SpTPC1 and SpTPC2 mediating calcium release from an acidic calcium store. Finally, we examined the effects of overexpressing SpTPC3, the functional properties of which are completely unexplored. Again, as with SpTPC1 and SpTPC2, cytosolic calcium responses to NAADP were markedly potentiated in cells overexpressing SpTPC3 in the absence but not the presence of bafilomycin (Fig. 3C). The magnitudes of the NAADP-mediated calcium signals in cells expressing SpTPC3, however, were smaller than those in cells overexpressing SpTPC1 and SpTPC2 (Fig. 3, A and B). In stark contrast to the effects of NAADP, responses to a submaximal cADPR concentration were unaffected by overexpressing any of the sea urchin TPCs (Fig. 3D), thereby highlighting specificity. All members of the TPC family are thus likely NAADP-sensitive calcium-permeable channels.
FIGURE 3.
Sea urchin TPCs mediate NAADP-dependent calcium release from acidic calcium stores. A–D, cytosolic calcium responses of individual fura-2-loaded SKBR3 cells microinjected with either 10 nm NAADP (A–C) or 50 nm cADPR (D). Cells were from mock-transfected cultures (MOCK) or cultures expressing the indicated GFP-tagged TPC construct from sea urchin (SpTPC1–3) or human (HsTPC1–2). In some experiments, cells were pretreated with bafilomycin A1 (1 μm, 60 min) prior to injection. Data are expressed as mean fluorescence ratios (±S.E.) from 4–12 cells.
To conclude, we provide new molecular insight into the evolution, location, and function of animal TPCs, a poorly characterized family of ion channels, which have recently emerged as targets for NAADP. We find evidence that the TPC3 gene, which although widely present in deuterostomes, is a pseudogene in humans and other primates, indicative of functional divergence within closely related mammals. We define the molecular nature of the ancestral TPC family in a key model organism and demonstrate that all isoforms localize to acidic organelles and that their overexpression enhances NAADP-mediated calcium release in a bafilomycin-sensitive manner. Our data provide further mechanistic evidence supporting the concept that NAADP releases calcium from acidic organelles via TPCs.
Supplementary Material
Acknowledgments
We thank Dev Churamani, George Dickinson, Chi Li, Latha Ramakrishnan, and Victor Vacquier for useful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM088790 (to J. S. M.), HL 090804 (to E. B.), and HL 090804-01A2S109 (to E. B.). This work was also supported by Biotechnology and Biological Sciences Research Council Grants BB/D018110/1 and BB/G013721/1 (to S. P.) and American Heart Association Fellowship Award 0625403U (to X. C.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FN598566, FN598567, and FN598568.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- IP3
- inositol trisphosphate
- cADPR
- cyclic ADP-ribose
- NAADP
- nicotinic acid adenine dinucleotide phosphate
- TPC
- two-pore channel
- GFP
- green fluorescent protein
- HEK
- human embryonic kidney
- UTR
- untranslated region
- Sp
- S. purpuratus.
REFERENCES
- 1.Berridge M. J., Lipp P., Bootman M. D. (2000) Nat. Rev. Mol. Cell Biol. 1, 11–21 [DOI] [PubMed] [Google Scholar]
- 2.Lee H. C. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 317–345 [DOI] [PubMed] [Google Scholar]
- 3.Guse A. H., Lee H. C. (2008) Sci. Signal. 1, re10. [DOI] [PubMed] [Google Scholar]
- 4.Lee H. C., Aarhus R. (1995) J. Biol. Chem. 270, 2152–2157 [DOI] [PubMed] [Google Scholar]
- 5.Galione A., Patel S., Churchill G. C. (2000) Biol. Cell 92, 197–204 [DOI] [PubMed] [Google Scholar]
- 6.Chini E. N., Dousa T. P. (1996) Biochem. J. 316, 709–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berridge G., Dickinson G., Parrington J., Galione A., Patel S. (2002) J. Biol. Chem. 277, 43717–43723 [DOI] [PubMed] [Google Scholar]
- 8.Churamani D., Dickinson G. D., Ziegler M., Patel S. (2006) Biochem. J. 397, 313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchill G. C., Okada Y., Thomas J. M., Genazzani A. A., Patel S., Galione A. (2002) Cell 111, 703–708 [DOI] [PubMed] [Google Scholar]
- 10.Lee H. C. (2005) J. Biol. Chem. 280, 33693–33696 [DOI] [PubMed] [Google Scholar]
- 11.Zhang F., Li P. L. (2007) J. Biol. Chem. 282, 25259–25269 [DOI] [PubMed] [Google Scholar]
- 12.Zhang F., Jin S., Yi F., Li P. L. (2008) J. Cell Mol. Med., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim D., Kyozuka K., Gragnaniello G., Carafoli E., Santella L. (2001) FASEB J. 15, 2257–2267 [DOI] [PubMed] [Google Scholar]
- 14.Beck A., Kolisek M., Bagley L. A., Fleig A., Penner R. (2006) FASEB J. 20, 962–964 [DOI] [PubMed] [Google Scholar]
- 15.Gerasimenko J. V., Sherwood M., Tepikin A. V., Petersen O. H., Gerasimenko O. V. (2006) J. Cell Sci. 119, 226–238 [DOI] [PubMed] [Google Scholar]
- 16.Dammermann W., Zhang B., Nebel M., Cordiglieri C., Odoardi F., Kirchberger T., Kawakami N., Dowden J., Schmid F., Dornmair K., Hohenegger M., Flügel A., Guse A. H., Potter B. V. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10678–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guse A. H. (2009) Curr. Biol. 19, R521–R523 [DOI] [PubMed] [Google Scholar]
- 18.Brailoiu E., Churamani D., Cai X., Schrlau M. G., Brailoiu G. C., Gao X., Hooper R., Boulware M. J., Dun N. J., Marchant J. S., Patel S. (2009) J. Cell Biol. 186, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calcraft P. J., Ruas M., Pan Z., Cheng X., Arredouani A., Hao X., Tang J., Rietdorf K., Teboul L., Chuang K. T., Lin P., Xiao R., Wang C., Zhu Y., Lin Y., Wyatt C. N., Parrington J., Ma J., Evans A. M., Galione A., Zhu M. X. (2009) Nature 459, 596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong X., Schieder M., Cuny H., Fenske S., Gruner C., Rötzer K., Griesbeck O., Harz H., Biel M., Wahl-Schott C. (2009) Pflugers Arch. 458, 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vannier B., Peyton M., Boulay G., Brown D., Qin N., Jiang M., Zhu X., Birnbaumer L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2060–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menteyne A., Burdakov A., Charpentier G., Petersen O. H., Cancela J. M. (2006) Curr. Biol. 16, 1931–1937 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.