Abstract
Clustering or overexpression of the transmembrane form of the extracellular matrix proteoglycan agrin in neurons results in the formation of numerous highly motile filopodia-like processes extending from axons and dendrites. Here we show that similar processes can be induced by overexpression of transmembrane-agrin in several non-neuronal cell lines. Mapping of the process-inducing activity in neurons and non-neuronal cells demonstrates that the cytoplasmic part of transmembrane agrin is dispensable and that the extracellular region is necessary for process formation. Site-directed mutagenesis reveals an essential role for the loop between β-sheets 3 and 4 within the Kazal subdomain of the seventh follistatin-like domain of TM-agrin. An aspartic acid residue within this loop is critical for process formation. The seventh follistatin-like domain could be functionally replaced by the first and sixth but not by the eighth follistatin-like domain, demonstrating a functional redundancy among some follistatin-like domains of agrin. Moreover, a critical distance of the seventh follistatin-like domain to the plasma membrane appears to be required for process formation. These results demonstrate that different regions within the agrin protein are responsible for synapse formation at the neuromuscular junction and for process formation in central nervous system neurons and suggest a role for agrin's follistatin-like domains in the developing central nervous system.
Keywords: Extracellular Matrix/Heparan Sulfate, Glycoproteins/Structure, Neurobiology/Neuroscience, Organisms/Bird, Protein/Motifs, Protein/Structure, Tissue/Organ Systems/Nerve, Synapse Formation
Introduction
Agrin is a proteoglycan with a molecular mass of >500 kDa that is expressed in many tissues (1, 2). The function of agrin is best characterized in skeletal muscle where it is a key organizer during formation, maintenance, and regeneration of the neuromuscular junction (2–4). Accordingly, mice with an inactivation of the agrn gene die at birth due to non-functional neuromuscular junctions and consequent respiratory failure (5).
Little is known about the role of agrin in tissues other than skeletal muscle, in particular in the central nervous system (for review see Refs. 1, 2, 6). Although neurons from mice with a targeted deletion of the agrn gene form synaptic specializations in vitro and in vivo (7, 8), the acute suppression of agrin expression or function by antisense oligonucleotides or antibodies influences the formation and function of interneuronal synapses (9, 10). Likewise, brains of agrin-deficient mice, whose perinatal death was prevented by the re-expression of agrin in motor neurons, have a severely reduced number of pre- and postsynaptic specializations as well as functional deficits at excitatory synapses in the CNS3 (11). Although these data are consistent with a role of agrin during CNS synaptogenesis, the precise function of agrin during CNS development remains unclear.
Agrin has been cloned from several species, and the sequences are highly homologous. The agrin cDNAs predict a number of domains with similarity to other extracellular matrix proteins, including four EGF-like repeats and three domains with similarity to globular domain of the laminin α-chain (LG domain) within the C-terminal half of the protein. In skeletal muscle fibers the C-terminal LG domains of agrin bind to α-dystroglycan (12–14) and to the low density lipoprotein receptor-like protein 4 (LRP4 (15, 16)). The binding of agrin to LRP4 activates the tyrosine kinase MuSK and initiates a complex intracellular signaling cascade resulting in the formation of most if not all pre- and postsynaptic specializations (1, 2). The functions of the other domains, particularly of the 9 follistatin-like domains within the N-terminal half of the agrin protein are unknown.
Alternative first exon usage generates either a secreted soluble agrin molecule (NtA-agrin) or a transmembrane form of agrin (TM-agrin (17, 18)). The secreted form of agrin binds to the laminin γ1-chain via its NtA domain resulting in stable association of this agrin isoform with basal laminae (19). In contrast, in TM-agrin the NtA domain is replaced by a non-cleaved signal anchor that converts agrin into a type II transmembrane protein and localizes the agrin protein in Ncyto/Cexo orientation in the plasma membrane (17, 18).
TM-agrin is primarily expressed on axons and dendrites of CNS neurons (20–22), and it has been hypothesized that neurite-associated TM-agrin might serve as a receptor or co-receptor (21). In agreement with this hypothesis, it was recently shown that clustering or overexpression of TM-agrin in neurons during the phase of active neurite growth reorganizes the actin cytoskeleton and induces the rapid formation of numerous filopodia-like processes extending from the primary neurite (21, 22). The formation of these processes is caused by the initiation of a complex signaling cascade, which involves lipid rafts, as well as the activation of mitogen-activated protein kinase, Fyn, and Cdc42 (22, 23). The function of the filopodia-like processes in the developing CNS is unknown, but they might represent initial stages during the formation of excitatory spine synapses (24). Consistent with this idea, mice that lack CNS agrin develop 30% fewer glutamatergic synapses (11).
In this study we demonstrate that overexpression of TM-agrin in non-neuronal cell lines can induce processes similar to those induced in CNS neurites, suggesting that the process-inducing activity of TM-agrin is not limited to nerve cells. We also investigated the structural requirements for the process-inducing activity of TM-agrin by mapping the region necessary for this activity. Our results show that an aspartic acid residue within the seventh follistatin-like domain is essential for process induction. This particular amino acid is highly conserved in other follistatin-like domains of TM-agrin. Accordingly, the seventh follistatin-like domain could functionally be replaced by the first or sixth follistatin-like domain. These data show that follistatin-like domains are critical for process formation and support a role for TM-agrin as a receptor or co-receptor in the developing CNS.
EXPERIMENTAL PROCEDURES
Animals
Fertile White Leghorn (Gallus gallus domesticus) chicken eggs were purchased from a local hatchery and incubated at 38 °C in a humid atmosphere. The age of the embryos was expressed as embryonic day (E). All experiments were conducted in accordance with the guidelines for the welfare of experimental animals issued by the Federal Government of Germany.
Antibodies
The following antibodies against agrin were used: rabbit anti-agrin antiserum #46 (25–27). This antiserum was generated against the C-terminal half of agrin and, thus, specifically reacts with the extracellular part of all TM-agrin and NtA-agrin isoforms in Western blotting and immunohistochemistry. In contrast, sheep anti-TM-agrin (generated against a peptide of the intracellular domain of TM-agrin) reacts only with TM-agrin but not with NtA-agrin (21, 23). Actin filaments were stained with Alexa594-conjugated phalloidin (Molecular Probes, Eugene, OR (21)). Tectal neurons were stained with the monoclonal antibody against β-tubulin (Clone TUB 2.1, Sigma). GFP was stained with a rabbit anti-GFP antiserum (Molecular Probes, Leiden, Netherlands).
Expression Constructs
The cDNA sequence for chick TM-agrin has been described elsewhere (18). Deletion constructs from these sequences were generated using the PCR and Phusion high fidelity DNA-polymerase (BioCat, Heidelberg, Germany). The constructs were cloned via EcoRI and XbaI or via HindIII and EcoRI restriction sites into the pcDNA4TO expression vector (Invitrogen). The following primers were used: TM-agrin Δ IC: 5′-GTGCGCGAATTCATGATCCCGTGCAACATT-3′ (sense) and 5′-CGGGCCCTCTAGAAGGGAAGGAGAAGGAA-3′ (antisense); TMFD9: 5′-GGTGGAATTCACAGCATGACGGCTTGCCAGTA-3′ (sense) and 5′-CGTGTGAGTTCTAGACTCGTGGCATTGCCCC-3′ (antisense); TMFD8: 5′-GTGTGGTGGAATTCACAGCATGACGGC-3′ (sense) and 5′-GCTCTAGACTGACTGCAGTGCACAACTGG-3′ (antisense); TMFD6: 5′-GTGTGGTGGAATTCACAGCATGACGGC-3′ (sense) and 5′-GCTCTAGAGTCATAGGTGAGGCCATCGGTGC-3′ (antisense); TMFD7–572: 5′- ATAAAGCTTAGCATGACGGCTTGCCAGTACC-3′ (sense) and 5′-AATGAATTCTTAGCCACATTCGTCCTCACACG-3′ (antisense); TMFD7–568: 5′-ATAAAGCTTAGCATGACGGCTTGCCAGTACC-3′ (sense) and 5′-AATGAATTCTTACTCACACGGCCCCATCTTGG-3′ (antisense); and TMFD7–566: 5′-ATAAAGCTTAGCATGACGGCTTGCCAGTACC-3′ (sense) and 5′-AATGAATTCTTACGGCCCCATCTTGGCCACC-3′ (antisense).
To obtain a GFP tag at the C terminus the TMFD8 and TMFD6 constructs were cloned via EcoRI and BamHI restriction sites into the pEGFP-N1 expression vector (Clontech, Mountain View, CA) using the following primers: TMFD8-GFP: 5′-GTGTGGTGGAATTCACAGCATGACGGC-3′ (sense) and 5′-CGCGGATCCGTCTGACTGCAGTGC-3′ (antisense); and TMFD6-GFP: 5′-GTGTGGTGGAATTCACAGCATGACGGC-3′ (sense) and 5′-CGCGGATCCTCATAGGTGAGGCCATCG-3′ (antisense).
To generate the constructs with an altered order of the follistatin-like domains, an intermediate called TMFD6–503 was created and cloned via HindIII and EcoRI restriction sites into the pEGFP-N1 vector. Other follistatin-like domains were added at the 3′-end via EcoRI and BamHI restriction sites. For this, the following primers were used: TMFD6–503: 5′-ATAAAGCTTAGCATGACGGCTTGCCAGTACC-3′ (sense) and 5′-TAGAATTCTTGCAGTCGCCTTGGGCAGC-3′ (antisense); TMFD6+FD1-GFP: 5′-ATGAATTCAGATGCCTGCCGAGGGATGC-3′ (sense) and 5′-ATGGATCCGTGCTGCCAAAGCTGC-3′ (antisense); TMFD6+FD6-GFP: 5′-ATGAATTCCGACCGCTGTGGCAAGTGC-3′ (sense) and 5′-ATGGATCCAAGGAGCACACCGTCGTGC-3′ (antisense); and TMFD6+FD8-GFP: 5′-ATGAATTCTGAGGACGAATGTGGCTCAGG-3′ (sense) and 5′-CGCGGATCCGTCTGACTGCAGTGC-3′ (antisense).
To generate the constructs containing successively less N-terminal follistatin-like domains an intermediate called intTM-Linker was created and cloned via NheI and XhoI restriction sites into the pEGFP-N1 vector, using the endogenous XhoI restriction site in the TM-agrin cDNA sequence at amino acid position 209. Other follistatin-like domains were then added at the 3′-end via XhoI and BamHI restriction sites using the following primers: intTM-Linker: 5′-ATAGCTAGCAGCATGACGGCTTGCCAGTACC-3′ (sense) and 5′- ATTGAATTCATCTCGAGGGGTCGGCGG-3′ (antisense); TMFD2–8-GFP: 5′-ATACTCGAGGACCATGTGGCTCCAAGGACC-3′ (sense) and 5′-CGCGGATCCGTCTGACTGCAGTGC-3′ (antisense); and TMFD3–8-GFP: 5′-ATACTCGAGTCTTCAAGAAGTTTGATGGAGCC-3′ (sense) and 5′-CGCGGATCCGTCTGACTGCAGTGC-3′ (antisense).
Site-directed mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the TMFD8-GFP construct as template. The point mutations were introduced successively using the following oligonucleotides (mutated bases are shown in lowercase letters). Only the sense primers are shown as the antisense primers had the reverse complementary sequence TMFD8 C535G: 5′-CCTGGCCCAGGTCgGTGGCACCGATGGC-3′; TMFD8C567G: 5′-CCAAGATGGGGCCGgGTGAGGACGAATGTGG-3′; TMFD8 N544D: 5′-GGCCTCACCTATGACgACCGCTGCGAGCTCCG-3′; TMFD8 C470G: 5′-CCTCCCAGCCTGTCgGTGGCACAGATGGCAACACC-3′; TMFD8 QQ555,556EE: 5′-GCAGCCTCCTGCCAAGAGgaGAAGAGCATCG-3′; TMFD8 C553A: 5′-CTCCGAGCAGCCTCCgcCCAACAGCAGAAGAG-3′; TMFD8 C546A: 5′-CCTATGACAACCGCgcCGAGCTCCGAGCAG-3′; TMFD8 P530A: 5′-GTGAGCAGCAGgCCCTGGCCCAG-3′; TMFD8 G539P: 5′-GGTCTGTGGCACCGATccCCTCACCTATGACA-3′; TMFD8 V561N: 5′-GCAGAAGAGCATCGAGaacGCCAAGATGGGGCCT-3′; TMFD8 D538N: 5′-CAGGTCTGTGGCACCaacGGCCTCACCTATGAC-3′; and TMFD8 D538G: 5′-AGGTCTGTGGCACCGgcGGCCTCACCTATGAC-3′. The exact sequences of all mutated and non-mutated expression constructs were verified by sequencing.
Cell Culture, Transfections, and Immunohistochemistry
Human embryonic kidney cells (HEK293), COS cells and PC12 cells were grown as described previously (18) and transfected using the SuperFect transfection reagent (Qiagen). Expression and correct insertion of the protein into the plasma membrane were verified by live staining prior to fixation with antibodies against the extracellular part of agrin (antiserum #46), or with antibodies against the extracellular GFP tag. When necessary, antibodies against the intracellular part of TM-agrin (sheep anti-TM-agrin (18)) were used to verify TM-agrin expression and membrane orientation. Processes were analyzed by manual counting them after fixation and staining. Neurons were identified by staining with antibodies against β-tubulin. For quantification, only isolated neurons where the filopodia-like processes clearly emanated from identified neurites were evaluated and scored according to the criteria defined previously (21–24, 28, 29). Briefly, non-neuronal cells were scored “process-containing” if they had at least 10 processes or alternatively 2 or more processes each with a length of more than twice the cell's diameter. Neurons were scored process-containing if their neurites had at least 2 segments of at least 40 μm in length with >2 filopodia-like processes per 10-μm neurite. Only processes with a length of >3 μm were scored. Although these criteria allowed the unambiguous identification of neurons being either process-containing or “not process-containing,” they do not reveal minor differences between neurons transfected with the various constructs. One-tailed t-tests were conducted to evaluate the significance of differences in the mean values of process density using GraphPad Prism version 4.03 for Windows (GraphPad Software, San Diego, CA).
Chick tectal neurons from E7 embryos were prepared as described (30) with minor modifications. The tecta were dissected free of meningeal tissue, cut into small pieces, and incubated in cell dissociation buffer (Sigma) containing 0.002% Trypsin for 15 min at 37 °C. Cells were dissociated using fire-polished Pasteur pipettes and separated from debris by centrifugation. Constructs were transfected into chick neurons using the Nucleofector System according to the protocol provided by the manufacturer (Lonza, Cologne, Germany). Briefly, 3 × 106 cells were resuspended in 100 μl of Chicken Neuron Nucleofector Solution (Lonza) with 8 μg of plasmid DNA. The cell suspension was electroporated using the G-13 program (Lonza). After transfection, the cell suspension was diluted with RPMI 1640 supplemented with 10% fetal calf serum, penicillin (100 units/ml), streptomycin (100 μg/ml), and Glutamax (2 mm, Invitrogen). Approximately 5–6 × 105 cells were plated on a 1-cm2 poly-l-lysine/laminin-coated coverslips placed in a 35-mm cell culture dish. Four hours after electroporation, the medium was removed and replaced with Neurobasal A medium containing 2% B27 supplement (Invitrogen), 0.5 mm l-glutamine, penicillin, and streptomycin. Cultures were maintained at 37 °C with 5% CO2. After 1 or 3 days in vitro cells were fixed, and the transfected cells were selectively stained with rabbit-anti-GFP antiserum (Molecular Probes) and co-stained with the neuron-specific monoclonal anti-β-tubulin antibody (Sigma) or with Alexa594-conjugated phalloidin to reveal the actin cytoskeleton (21). The cultures were subsequently incubated with the appropriate Alexa594- or Alexa488-conjugated secondary antibodies (donkey anti-sheep, goat anti-mouse, or goat anti-rabbit; Molecular Probes; 24 μg/ml final concentration). All secondary antibodies were pre-absorbed against IgG of other species, eliminating cross-reactivity. After 1.5 h the cultures were again washed in phosphate-buffered saline containing 0.1% Triton X-100, and embedded in Citifluor mounting medium (Plano, Wetzlar, Germany). Specimens were analyzed with a photomicroscope (DMRA, Leica, Solms, Germany) equipped with epifluorescence optics using fluorescence filters of the appropriate wavelength for the complete separation of the different chromophores. Pictures were acquired with a digital camera (DC200, Leica) using the Leica data acquisition software. Contrast and brightness of entire images were adjusted using Photoshop (version 7.0, Adobe, Mountain View, CA).
SDS-PAGE and Immunoblotting
Transiently transfected HEK293 cells expressing various TM-agrin deletion and mutation constructs were scraped into-cold lysis buffer containing 1% Nonidet P-40, 1% Triton X-100, 50 mm Tris (pH 7,4), 500 mm NaCl, 2 mm EDTA, and complete-EDTA free-protease inhibitor mixture (Roche Diagnostics, Penzberg, Germany). After 30-min incubation on ice, the cell lysate was passed through a 22- and a 26-gauge needle (6 times both). Insoluble material was removed by centrifugation at 20,000 × g for 3 min at 4 °C, and the supernatant was dissolved in Tris-HCl buffer containing 4% SDS, 10% β-mercaptoethanol, and 20% glycerol. Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Schwalbach, Germany) by electroblotting. The membranes were blocked with 4% nonfat milk in Tris-buffered saline containing 0.1% Tween 20 for 1 h at room temperature, blots were probed either with rabbit anti-GFP antiserum (Molecular Probes) or sheep anti-TM-agrin antiserum and co-probed with a monoclonal antibody against β-actin (MAB1501, Chemicon) overnight at 4 °C. Horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch (West Grove, PA). Peroxidase-associated signals were detected using the enhanced chemiluminescence method (SuperSignal West Pico, Pierce, Bonn, Germany), followed by exposure to BioMax HyperFilm ECL (Amersham Biosciences).
Protein bands in Western blots were quantified, and densitometric analysis was performed using the AIDA Image Analyzer Software (version 3.28, Raytest, Straubenhardt, Germany) according to the manufacturer's instructions and as described previously (23). Protein expression of the constructs with point mutations was monitored and quantified using the Li-Cor Odyssey infrared system (Li-Cor Bioscience, Bad Homburg, Germany). To this end, Western blot membranes were incubated with the primary rabbit anti-GFP antibody (Molecular Probes) and subsequently with Alexa680-conjugated anti-rabbit IgG secondary antibodies (Molecular Probes). The signal was visualized and quantified after washing the blot by scanning with the Li-Cor Odyssey infrared imager and the Odyssey application software (version 3.0). Mean normalized values from the indicated number of experiments were plotted and compared statistically as previously described (31). Two-tailed Student t-tests were performed to evaluate the significance of differences in mean values using GraphPad Prism version 4.03 for Windows (GraphPad Software).
The molecular model of follistatin-like domains is based on the data specified in Refs. 32 and 33. For supplemental Fig. S4 the crystal structure of the follistatin-like domain 1, the heparin-binding domain of follistatin, was used (PDB identifier code: 1LR9). The display style, the viewing angle, and the color code of specific amino acid residues were modified using the ViewerPro program (version 4.2, Accelrys).
RESULTS
Expression of TM-agrin in Non-neuronal Cells Induces Filopodia-like Processes
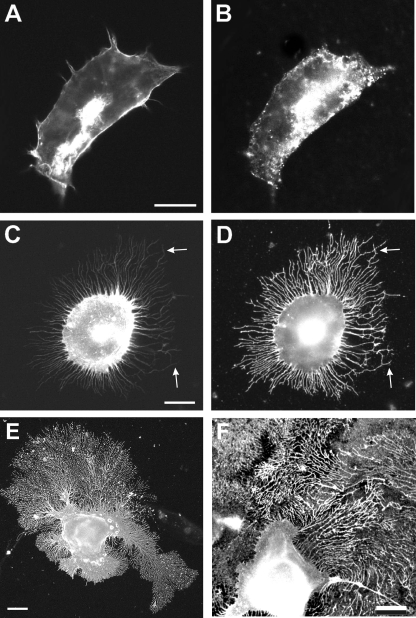
Overexpression of TM-agrin in skeletal muscle cells, hippocampal neurons, and in SY5Y neuroblastoma cells results in the formation of numerous filopodia-like processes (21, 22, 34). To determine if these processes can also be observed in human cells and in other non-neuronal cell lines, we transfected HEK293 cells with full-length cDNA coding for either chick NtA-agrin or chick TM-agrin. As shown in Fig. 1, expression of NtA-agrin had no obvious effect on the overall cell shape and on the cytoskeleton of the transfected cell, as revealed by staining with antibodies against agrin (Fig. 1B) or with phalloidin (Fig. 1A). In contrast, expression of TM-agrin changed the morphology of the transfected cell. TM-agrin-transfected cells extended numerous processes, which contained actin filaments (Fig. 1C) and also displayed agrin immunoreactivity on their surface (Fig. 1D). Quantification showed that >85% of the cells transfected with the TM-agrin cDNA had extended processes, whereas <2% of the NtA-agrin-expressing cells showed processes (Fig. 2). In contrast to the transiently forming filopodia-like processes induced on neurons (21, 22), the processes extended by the transfected non-neuronal cells had no visible growth cone at their tips, were much less dynamic and did not collapse and retract, but instead continued to grow for several days (Fig. 1, E and F) and eventually covered the entire surface of the culture dish. However, similar to the processes extended by axons and dendrites, the processes formed by HEK293 cells contained a complex cytoskeleton, including actin indicated by their staining with phalloidin (Fig. 1C and data not shown). The actin filaments were present within the entire length of the processes, extending almost to their tips (arrows in Fig. 1 (C and D)). Similar actin-containing processes were observed after expression of full-length chick TM-agrin (but not of NtA-agrin) cDNA in monkey COS-7 cells, in primary chick myoblasts, in rat PC12 cells or in Chinese hamster ovary cells (data not shown). These results demonstrate that overexpression of TM-agrin induces the formation of filopodia-like process not only in neurons but also in non-neuronal cells and that this effect is independent of the type and species of the transfected cell.
FIGURE 1.
Expression of TM-agrin induces processes in non-neuronal cells. Human embryonic kidney cells (A–D) or COS-7 cells (E and F) were transiently transfected with a cDNA coding for either NtA-agrin (A and B) or for TM-agrin (C–F). Cells were stained with Alexa594-conjugated phalloidin to reveal the actin filaments (A and C) and with anti-agrin antibodies (B, D, E, and F). Although the expression of NtA-agrin did not apparently affect the morphology of the transfected HEK293 cells (A and B), expression of TM-agrin induced the formation of numerous actin filament-containing processes by the transfected cells (C). The actin filaments extended until close to the tip of the processes (arrows in C and D). The processes did not collapse and retract but instead continued to grow in culture (A–D show cells 24 h after transfection; E and F show cells 48 h post transfection). Scale bars in A, C, E, and F: 10 μm.
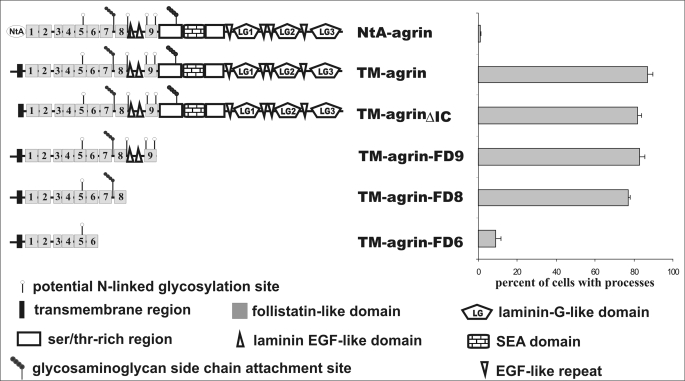
FIGURE 2.
Follistatin-like domains are required for process-inducing activity. Schematic representation of the different TM-agrin constructs used to localize the region responsible for the formation of the filopodia-like processes (left). The different domains within the TM-agrin protein are specified on the bottom. The nine follistatin-like domains are numbered according to their position starting from the N terminus. Quantification of the transfected cells extending processes is shown on the right. Note that deletion of most of the extracellular part of TM-agrin has no influence on process formation. However, deletion of follistatin-like domains 7 and 8 (TM-agrin-FD6 construct) abolished process-inducing activity. The bars represent the mean ± S.E. with n = 3.
The Extracellular Part of TM-agrin Is Necessary for Process Formation
To identify the region in TM-agrin responsible for process formation HEK293 cells were transfected with TM-agrin cDNA deletion and mutation constructs (see Fig. 2 for a schematic representation of the constructs). In agreement with previous results (22), deletion of the intracellular domain of TM-agrin (TM-agrinΔIC in Fig. 2) did not abolish the process-inducing activity. Because replacement of the entire extracellular part of TM-agrin by GFP resulted in the loss of the process-inducing activity (data not shown but see Ref. 22) our results confirm that the intracellular 34 amino acids are not required for this effect and suggest an essential role for the extracellular part of TM-agrin. Because the sequences of the extracellular parts of TM-agrin and NtA-agrin are identical, our results also show that membrane association of TM-agrin is required for the process-inducing activity.
We next mapped the region within the extracellular part of TM-agrin responsible for the induction of the filopodia-like processes. To this end, we successively deleted parts of the TM-agrin cDNA and expressed the corresponding constructs in HEK293 cells. Deletion of the three C-terminal LG domains together with the four epidermal growth factor-like repeats (EGF-like repeats) did not influence the process-inducing activity (data not shown, but see Ref. 22), demonstrating that the region involved in binding to the agrin receptor LRP4 and responsible for the formation of the postsynaptic specializations at the neuromuscular junction is dispensable for process formation. Likewise, additional deletion of the two serine/threonine-rich regions together with the SEA domain (a module first identified in sea urchin sperm protein, enterokinase, and agrin) did not alter the process-inducing activity of TM-agrin (construct TM-agrin-FD9 in Fig. 2). Similarly, overexpression of a construct in which the two laminin-EGF-like domains were deleted together with the C-terminal ninth follistatin-like domain (construct TM-agrin-FD8 in Fig. 2) in HEK cells resulted in >80% of the transfected cells extending processes. In contrast, subsequent deletion of follistatin domains 7 and 8 (construct TM-agrin-FD6) completely abolished the process-inducing activity of the corresponding TM-agrin protein, despite comparable expression levels and appropriate insertion and orientation of the transfected protein into the membrane (data not shown; see supplemental Figs. S1 and S2). In summary, these results demonstrate that the C-terminal half of TM-agrin is not required for process induction, and, therefore, the region involved in process induction and the domains responsible for the synaptogenic role of agrin at the neuromuscular junction are localized within different parts of the agrin protein.
The Seventh Follistatin-like Domain Is Essential for Process Formation
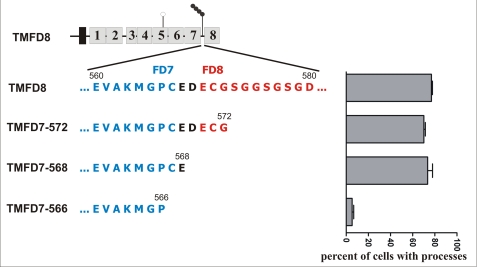
The results suggested that either follistatin-like domains 7 or 8 or the glycosaminoglycan (GAG) side chains, which are attached to the core protein between the two follistatin-like domains 7 and 8 (35, 36), are responsible for the induction of the processes. To determine, if GAG side chains are involved in the formation of the filopodia-like processes, all three serine residues (serines 573, 576, and 578, respectively), which serve as acceptors for the GAG side chains (35), were mutated to alanine. These mutations resulted in a decrease in the number of process-forming cells to ∼30% (data not shown). However, deletion of the entire GAG side-chain attachment region restored process-inducing activity of the corresponding agrin protein (construct TMFD7–572 in Fig. 3). This result indicates that the GAG side chains are dispensable for process formation. Likewise, transfection of HEK293 cells with a construct in which additional four amino acids were deleted from the C terminus (construct TMFD7–568) resulted in ∼80% of the cells extending processes. Deletion of the next two amino acids (constructs TMFD7–566 in Fig. 3), however, resulted in a protein that completely lacked process-inducing activity. This suggests that cysteine 567 or glutamic acid 568 is crucial for process induction. We next mutated cysteine 567 to glycine by site-directed mutagenesis (construct TMFD8 C567G in Fig. 4). In addition, we tagged the protein with a C-terminal GFP tag (indicated by an asterisk in Figs. 4A and 6) to follow expression and membrane insertion of the fusion protein. Fig. 4B shows that TMFD8-GFP encodes a protein with similar process-inducing activity as full-length TM-agrin, demonstrating that the GFP tag does not significantly influence process-inducing activity. In contrast, the Cys-567 → Gly point mutation within the TMFD8-GFP protein completely abolished the process-inducing activity demonstrating an essential role for cysteine 567 in TM-agrin-mediated process formation.
FIGURE 3.
The seventh follistatin-like domain is required for process formation. To determine the role of the heparan sulfate side chains in the formation of the filopodia-like processes, the construct TMFD8 was truncated in the region between follistatin-like domain 7 (FD7; shown in blue) and 8 (FD8, shown in red). The exact amino acid sequence containing the three serine residues (serine 573, 576, and 578, respectively) used as GAG chain attachment sites of chick TM-agrin is shown on the left. The percentage of transfected cells having filopodia-like processes is shown on the right (mean ± S.E. with n = 3). Complete deletion of the GAG side-chain attachment sites did not result in a loss of the process-inducing activity (constructs TMFD7–572 and TMFD7–568), demonstrating that the GAG side chains are dispensable for process induction. In contrast, deletion of two additional amino acids (cysteine at position 567 and glutamic acid at position 568) completely abolished process-inducing activity (construct TMFD7–566).
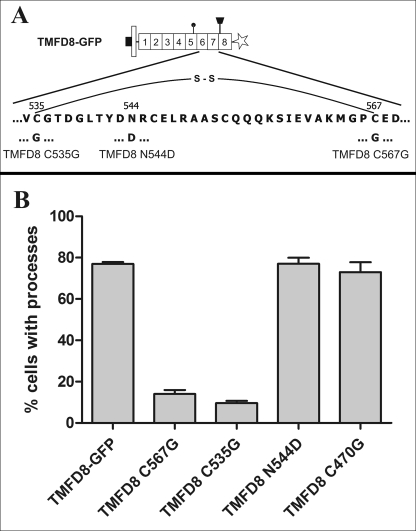
FIGURE 4.
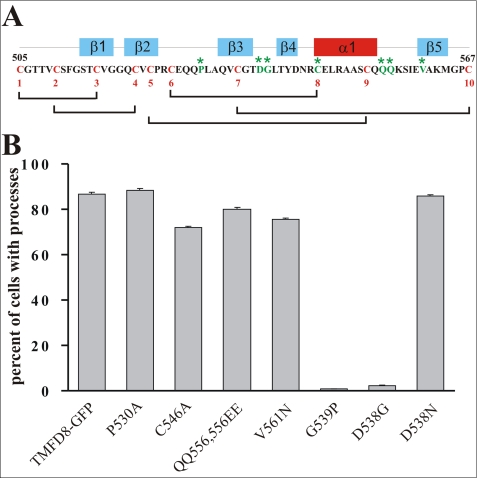
The disulfide bond between cysteines 535 and 567 within the seventh follistatin-like domain is critical for process-inducing activity. To interrupt the disulfide bond between cysteine 535 and 567, both residues were individually mutated to glycine giving rise to the TMFD8 C535G and TMFD8 C567G constructs, respectively. Each mutation resulted in a loss of process-inducing activity. In contrast, mutation of the cysteine corresponding to the cysteine 535 in the neighboring sixth follistatin-like domain (construct TMFD8 C470G) or mutation of the asparagine residue to aspartic acid within the seventh follistatin-like domain (TMFC8 N544D) did not result in a loss of process-inducing activity. The bars in B show the mean ± S.E. with n = 3.
FIGURE 6.
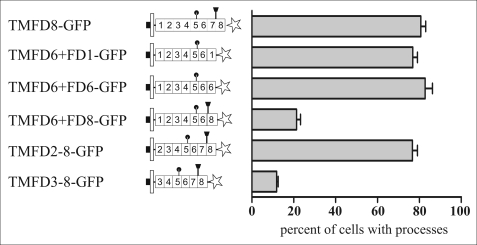
Follistatin-like domain 7 can be replaced by domains 1 and 6 but not by domain 8. To determine if other follistatin-like domains of TM-agrin also have process-inducing activity when placed at the appropriate position, constructs were generated where the seventh follistatin-like domain was replaced by either the first follistatin-like domain (construct TMFD6+FD1-GFP), by the sixth follistatin-like domain (construct TMFD6+FD6-GFP), or by the eighth follistatin-like domain (construct TMFD6+FD8-GFP). Although the first and sixth follistatin-like domains can functionally substitute for the seventh follistatin-like domain, replacing follistatin-like domain 7 with the eighth follistatin-like domain resulted in a severe reduction of the number of processes extended by the transfected cells. Moreover, a critical distance of the seventh follistatin-like domain to the plasma membrane appears to be necessary for process formation, because deletion of the first and second follistatin-like domain (construct TMFD3–8-GFP) resulted in a loss of the process-formation activity, whereas deletion of only the first follistatin-like domain (construct TMFD2–8-GFP) did not influence process formation.
The precise structures of follistatin-like domains from BM-40 and from follistatin have been determined using x-ray crystallography (32, 33). The crystal structures predict that cysteine 567 forms an intradomain disulfide bond with cysteine 535 (Fig. 4A; see supplemental Fig. S4 for a schematic three-dimensional structure of a follistatin domain and the location of the disulfide bond). To determine if this disulfide bond is required for the process-inducing activity, we mutated cysteine 535 to glycine by site-directed mutagenesis (construct TMFD8 C535G in Fig. 4). As shown in Fig. 4B this construct encodes a protein without process-inducing activity, indicating that this disulfide bond is critical for the structural integrity of the follistatin-like domain and, thus, for process formation. In contrast, mutation of the corresponding cysteine in the neighboring sixth follistatin-like domain (construct TMFD8 C470G in Fig. 4B) had no influence on process formation. Likewise, mutation of the asparagine residue within the seventh follistatin-like domain to aspartic acid (construct TMFD8 N544D in Fig. 4, A and B) did not influence process-inducing activity, demonstrating that not all mutations within follistatin-like domain 7 influence process formation. In summary, these results demonstrate that the disulfide bond between cysteine 535 and cysteine 567 within the seventh follistatin-like domain is required for the process-inducing activity of TM-agrin.
The Loop between β-Sheets 3 and 4 Is Involved in Process Formation
Mutations of cysteine residues most likely have severe repercussions on the structure of the entire follistatin-like domain. To more precisely map the region responsible for process formation, several other amino acids within the seventh follistatin-like domain were mutated. Follistatin-like domains consist of an N-terminal EGF-like subdomain, formed by the two antiparallel β-sheets 1 and 2 and a C-terminal Kazal-like subdomain, formed by three short antiparallel β-sheets and an α-helix (33) (see supplemental Fig. S4). To investigate which of the two subdomains is responsible for the process-inducing activity, we mutated several amino acids within the Kazal part of the seventh follistatin-like domain (Fig. 5A). Neither mutation of proline 530 to alanine nor cysteine 546 to alanine or the double mutation of the glutamine residues 555 and 556 to glutamic acid had an influence on the process-inducing activity of the corresponding protein (Fig. 5B; see supplemental Fig. S4 for the localization of the mutated amino acids within the follistatin-like domain). In contrast, mutating aspartic acid 538 within the Kazal-like subdomain to glycine completely abolished the process-inducing activity of the encoded protein (construct TMFD8 D538G in Fig. 5B). To determine if the length or the charge of the side chain was responsible for the process-inducing activity of TM-agrin, aspartic acid 538 was mutated to asparagine. In contrast to the mutation of this amino acid to glycine, the mutation to asparagine did not influence process-inducing activity (Fig. 5B) suggesting that length of the side chain rather than its charge is crucial for process formation. Because Asp-538 is part of a reverse type I β-turn between β-sheets 3 and 4 (32), we mutated the neighboring glycine 539 to proline to disrupt this turn. As shown in Fig. 5B, the TMFD8 G539P protein had no process-inducing activity. In contrast, mutation of glycine 539 to alanine did not abolish process-inducing activity (data not shown). These results demonstrate that the structural integrity of the Kazal-like subdomain within the seventh follistatin-like domain, particularly the loop between β-sheets 3 and 4, is essential for the process-inducing activity of TM-agrin.
FIGURE 5.
The loop between β-sheet 3 and 4 within the seventh follistatin-like domain is critical for process formation. The amino acid sequence of follistatin-like domain 7 is shown in A. The 10 cysteine residues are shown in red, and their disulfide bonds are schematically indicated. The location of the α-helix and the five β-sheets within the follistatin-like domain are schematically indicated on top of the amino acid sequence. The mutated amino acids are shown in green and are marked by an asterisk. B shows the quantification of the process formation activity of the various mutated constructs. The values represent the mean ± S.E. with n = 3.
Follistatin-like Domains 1 and 6 Can Functionally Substitute for the Seventh Follistatin-like Domain
Because Asp-538 is conserved in the corresponding loops between β sheets 3 and 4 of all follistatin-like domains of TM-agrin (see supplemental Fig. S3 for an amino acid alignment of the eight follistatin-like domains of agrin), we replaced the seventh follistatin-like domain by other follistatin-like domains. Exchange of the seventh follistatin-like domain by the first follistatin-like domain (construct TMFD6+FD1-GFP in Fig. 6) resulted in a protein that had process-inducing activity similar to full-length TM-agrin (Fig. 6). Likewise, replacing the seventh follistatin-like domain with an additional copy of the sixth follistatin-like domain (construct TMFD6+FD6-GFP in Fig. 6) resulted in the synthesis of a protein with process-inducing activity (Fig. 6). In contrast, replacing the seventh follistatin-like domain by the eighth follistatin-like domain (construct TMFD6-FD8-GFP in Fig. 6) resulted in the synthesis of a protein with little if any process-inducing activity, although the aspartic acid residue between β-sheet 3 and 4 is also conserved in the eighth follistatin-like domain (see supplemental Fig. S3). These data indicate that some follistatin-like domains, including follistatin-like domains 1 and 6, can functionally substitute for the seventh domain, whereas follistatin-like domain 8 cannot, suggesting a functional redundancy of the follistatin-like domains. However, because mutation of the cysteine in the sixth follistatin-like domain that is homologous to the critical cysteine 535 in the seventh follistatin-like domain had no influence on process formation (construct TMFD8 C470G in Fig. 4B) the functional redundancy might require an appropriate localization of the domain. The presence of a high molecular mass smear characteristic of glycosylated proteins in Western blots of proteins that have process-inducing activity (i.e. TMFD8-GFP) and in those without process-inducing activity (i.e. for example TMFD6-FD8-GFP; see supplemental Fig. S2A) indicates that the eighth follistatin-like domain is glycosylated independent of its position within the FD8 protein and further supports the conclusion that the process-inducing activity of TM-agrin is independent of GAG side-chain attachment.
A Minimal Distance to the Membrane Is Required
Because the seventh follistatin-like domain is identical in NtA and TM-agrin, but only TM-agrin has process-inducing activity, our results indicated that membrane anchoring is critical for process formation. We therefore addressed the question of whether the distance to the plasma membrane of the follistatin-like domains is important for TM-agrin process-inducing activity. To this end, we deleted the juxtamembrane first follistatin-like domain (construct TMFD2–8-GFP in Fig. 6). This did not influence the process-inducing activity of the encoded protein (Fig. 6). In contrast, deletion of the first follistatin-like domain together with the adjacent second follistatin-like domain (construct TMFD3–8-GFP in Fig. 6) completely abolished the process-inducing activity of the encoded protein, consistent with the hypothesis that a critical distance between the plasma membrane and the seventh follistatin-like domain is necessary for process formation. Analysis of the membrane orientation and of the expression levels showed that all constructs were expressed at approximately similar levels and that the mutated proteins were targeted to the plasma membrane of the transfected cells (see supplemental Figs. S1 and S2 for details).
Similar Structural Requirements for Process Formation in Neurons and Non-neuronal Cells
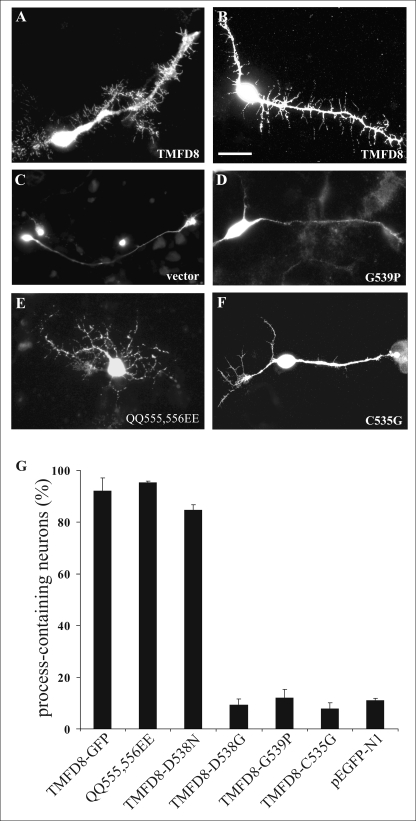
To determine if the seventh follistatin-like domain also regulates process induction in nerve cells, chick tectal neurons, which have previously been shown to form filopodia-like processes in response to TM-agrin clustering (21), were transfected with the various constructs. Fig. 7 shows representative neurons and their processes 72 h post-transfection. Although transfection of the TMFD8-GFP or of the TMFD8 QQ555,556EE construct into tectal neurons resulted in the formation of numerous filopodia-like processes by the axon and dendrite of the transfected neuron (Fig. 7, A, B, and E), expression of the empty vector (Fig. 7C), of the TMFD8 G539P-GFP (Fig. 7D), or of the TMFD8 C535G-GFP (Fig. 7F) construct had no influence on the number of processes extended by the neurites. Fig. 7G shows the quantification of the process-inducing activity of the various constructs after overexpression in tectal neurons. All tested constructs that induced process formation in transfected non-neuronal cells also induced processes in transfected CNS neurons, whereas those mutated proteins inactive in process formation in non-neuronal cells also did not induce processes in transfected tectal neurons. We observed no apparent difference in the total neurite length of neurons transfected with process-inducing constructs compared with neurons transfected with non process-inducing constructs (data not shown). However, our results do not exclude subtle differences between the different constructs with respect to the morphology or stability of the induced processes. In any case, our results demonstrate a critical role for the loop between β-sheets 3 and 4 within the seventh follistatin-like domain also for process formation in CNS neurons.
FIGURE 7.
Overexpression of TM-agrin mutation constructs in tectal neurons. Representative images of tectal neurons after transfection with TMFD8-GFP (A and B), the empty EGFP-N1 vector (C), TMFD8-G539P-GFP (D), TMFD8-QQ555,556EE-GFP (E), or TMFD8-C535G-GFP (F) are shown. Tectal neurons were stained 3 days after transfection with Alexa594-conjugated phalloidin (A, C, D, and E) or with anti-GFP antibodies (B and F). When TMFD8 or TMFD8 QQ555,556EE were overexpressed, many filopodia-like processes extending from the main neurites were observed (A, B, and E). In contrast, neurons transfected with the TMFD8-G539P construct (D), with the TMFD8-C535G construct (F), or with the empty vector (C) showed only few filopodia-like processes. Quantification of the percent of process-containing tectal neurons (identified according to the criteria detailed under “Experimental Procedures”) after transfection with the TM-agrin mutation constructs is shown in G. All tested constructs that had process-inducing activity in HEK293 cells also induced the formation of processes in tectal neurons. Moreover, mutations that affected the process-inducing activity in HEK cells also resulted in a loss of the process formation activity in tectal neurons. Bars in G represent mean ± S.E. with n = 3. Scale bar = 20 μm.
DISCUSSION
The aim of this study was to identify the structural basis for the TM-agrin-mediated formation of filopodia-like processes. Our results demonstrate three requirements for TM-agrin process-inducing activity: An association of TM-agrin with the plasma membrane, the integrity of the loop between β-sheet 3 and 4 within the seventh follistatin-like domain, and a critical distance of the follistatin-like domain to the plasma membrane. Follistatin-like domains are common structural components of extracellular matrix proteins, including for example SPARC/osteonectin/BM-40 (32, 37), complement proteins C6 and C7 (38), testican-2 (39), SMOC-1 and -2 (40, 41), and tomoregulin (42). Although follistatin-like domains are present in a number of extracellular matrix proteins, little is known about their function. It is, however, generally assumed that follistatin-like modules acquire different functions depending on their structural setting (43). We show here that follistatin-like domain 7 of TM- agrin is essential for the formation of filopodia-like processes when expressed in neurons and non-neuronal cells. Moreover, because the expression of chick TM-agrin cDNA can induce processes for example in human HEK293 cells or monkey COS-7 cells, the effect appears to be species-independent, suggesting a conserved molecular mechanism for process induction. In agreement with this result is the recent demonstration that the formation of the processes is the result of the activation of a signaling cascade that involves partitioning of TM-agrin into sphingolipid- and cholesterol-rich membrane subdomains called lipid rafts (23). Lipid rafts are almost ubiquitously present signaling platforms with important functions during CNS development and synapse formation (for review see Refs. 44, 45). The widespread presence of lipid rafts in almost all eukaryotic cells might explain the formation of processes after transfection of TM-agrin into a wide variety of cell types from different species. Likewise, the involvement of the ubiquitous lipid rafts might also explain the formation of filopodia-like processes in response to overexpression of several other proteins, including for example the transmembrane proteoglycan syndecan (46), Tenascin-R (47), or thrombospondin (48).
X-ray crystallographic analyses of the heparan sulfate-binding domain of follistatin (33) or of a dimer of the first two follistatin-like domains from SPARC/osteonectin/BM-40 (32) have revealed a stereotyped elongated structure determined primarily by the 10 cysteine residues forming 5 specific intradomain disulfide bonds. These disulfide bonds subdivide follistatin-like domains into two separate substructures, an N-terminal EGF-like subdomain characterized by two β-sheet structure (β1 und β2) connected via a β-hairpin, and a C-terminal part with similarity to the Kazal-type family of protease inhibitors consisting of three short antiparallel β-sheets (β3, β4, and β5) packed against an α-helix (32, 33; see supplemental Fig. S4 for the structure of both subdomains). Our results show that the reverse type I β-turn between β-sheets 3 and 4 is critical for the formation of the filopodia-like processes in neurons and non-neuronal cells. This region has so far no function assigned in any follistatin-like domain. For example, the interaction between the EF-hand calcium-binding site-containing domain and the follistatin-like domain within BM-40 is mediated by the strand β5 (32). Likewise, the binding of activin to follistatin, although not yet precisely mapped, is likely to involve the N-terminal hairpin structure between β-sheets 1 and 2 of the first follistatin domain of follistatin (49, 50). The same area also harbors the heparin-binding region, consistent with heparin being able to block the activin-follistatin interaction (33, 51). On the other hand, in the follistatin-like domain of BM-40 an asparagine residue located between β-sheet 3 and 4 (Asn-99 in human BM-40) has been shown to be covalently linked to two N-acetylglucosamine sugars (32). However, this asparagine is not conserved in any of the follistatin-like domains of agrin, suggesting that glycosylation at this position is not likely to influence process formation.
A number of functions of agrin have been assigned to the heparan sulfate and chondroitin sulfate side chains (35), including their interaction with low molecular weight growth factors, like fibroblast-like growth factors (52) or their inhibition of neurite growth (36). Our results show that GAG side chains are dispensable for the formation of filopodia-like processes in neurons and non-neuronal cells, eliminating an effect of growth factor- or GAG side chain-mediated signaling pathways and their modulation by agrin on process formation. Interestingly, when all three serines that serve as glycosaminoglycan side-chain attachment sites between follistatin-like domains 7 and 8 (35) were mutated to alanine, the protein had a reduced process-inducing activity, whereas complete deletion of the eighth follistatin-like domain together with the GAG side-chain attachment sites restored process-forming activity. One possible explanation might be that the GAG side chains have a role in maintaining the structural integrity of the eighth follistatin-like domain. In the absence of GAG side chains, the eighth follistatin-like domain might adopt an altered conformation and might interfere with the function of follistatin-like domain 7. This effect would not be detectable in the absence of both the GAG side chains and follistatin-like domain 8 explaining the process-inducing activity of the TMFD7–572 construct.
Although follistatin-like domain 7 could be functionally replaced by domains 1 and 6, the eighth follistatin-like domain could not substitute for the seventh domain. While the eighth follistatin-like domain of TM-agrin also contains the conserved aspartic acid residue between β-sheet 3 and 4, it differs from follistatin-like domains 1 and 6 in at least two aspects: an ∼8-amino acid-long stretch between β-sheet 1 and 2 and the GAG side-chain attachment sites at its N terminus (see supplemental Fig. S3 for a sequence alignment of the 8 different follistatin-like domains of TM-agrin). Because the amino acid insertion between β-sheets 1 and 2 is in the EGF-like subdomain, which is apparently not involved in process formation, the more likely explanation for the failure of the eighth follistatin-like domain to functionally substitute for follistatin-like domain 7 is the presence of the GAG side chains. Although we have not specifically addressed the glycosylation of the proteins encoded by the various constructs, Western blotting of proteins containing the eighth follistatin-like domain showed a high molecular mass smear characteristic of highly glycosylated proteins in our study (see supplemental Fig. S2A for details) as well as in previous studies (35, 36). The absence of this smear in constructs lacking the eighth follistatin-like domain suggests that proteins containing the eighth follistatin-like domain have GAG side chains attached. Therefore, in the TMFD6+FD8-GFP construct, the GAG side chains might block the interaction of the follistatin-like domain with other proteins or might interfere with the protein's ability to partition into lipid rafts.
Agrin contains nine follistatin-like domains, eight of which are arranged as an uninterrupted tandem repeat. Rotary shadowing followed by electron microscopy demonstrates a rather rigid rod-like arrangement of the follistatin-like domains in agrin (53). A model has been proposed for the structure of the tandem of follistatin-like domains (32). This model predicts that the β1–β2 hairpin of the N-terminal part of follistatin-like domain interacts with the small three-stranded β-sheet of the preceding follistatin-like domain (32). Because the loop between β-sheet 3 and 4 is part of the three-stranded C-terminal β-sheet, it is possible that mutations in this region influence the interaction between two follistatin-like domains. However, the process-inducing activity of follistatin-like domain 7 is not influenced by the deletion of the eighth follistatin-like domain. Therefore, the presence of a C-terminal interaction partner appears not to be necessary for process formation.
Dendritic filopodia-like processes are known to serve as precursors for new synapses during development as well as during activity-dependent synapse remodeling (for review see Refs. 54, 55). Once formed, the filopodia can mature into spine synapses upon contact with a presynaptic bouton (56–58). However, the number of successfully established functional synapses is low; >90% of the new filopodia dissolve within days, and only a small minority matures into synapse-bearing spines, consistent with a massive overproduction and a constant turnover of these processes (59). Interestingly, dendritic spine precursor processes and the filopodia-like processes induced by TM-agrin are very similar with respect to length, time course of appearance, and disappearance and to their dependence on specific intracellular signaling proteins (56, 57, 60–62). This similarity suggests that TM-agrin might be involved in the formation of spine synapses in the developing CNS by regulating the formation of filopodia-like processes on dendrites. Consistent with this hypothesis, inhibition of TM-agrin synthesis markedly reduced the number of synapses in hippocampal neurons (29). In addition, mice that lack agrin in the CNS have a 30% reduction in the number of excitatory synapses and spines in the cerebral cortex (11).
At mature interneuronal synapses neurotrypsin, a serine protease that is concentrated in the presynaptic active zone and in the synaptic cleft, has been shown to cleave TM-agrin (63). Cleavage of agrin by neurotrypsin occurs at two conserved sites within the agrin protein, generating a truncated plasma membrane-associated TM-agrin protein as well as two soluble fragments from the C terminus with molecular masses of 90 and 22 kDa, respectively (64). Recruitment to synapses as well as exocytosis of neurotrypsin depends on presynaptic activity, whereas agrin cleavage requires the additional and coincident postsynaptic activity and involves N-methyl-d-aspartic acid receptor activation as well as calcium influx (24, 65). Interestingly, neurotrypsin-deficient mice form significantly fewer long term potentiation-associated dendritic filopodia-like processes (24), demonstrating that neurotrypsin-dependent agrin cleavage is required for process formation. The C-terminal 22-kDa agrin fragment generated by neurotrypsin is able to induce filopodia-like processes when added to mature hippocampal slices, and the same fragment is able to rescue the long term potentiation-induced decrease in filopodia-like processes in neurotrypsin-deficient mice (24). Thus, overexpression of TM-agrin as well as direct addition of the soluble 22-kDa C-terminal agrin fragment generated by neurotrypsin can induce the formation of processes. Therefore, TM-agrin and neurotrypsin might act synergistically on process formation and interneuronal synapse formation. Another possibility is that the two proteins directly interact. This hypothesis would predict that binding of the 22-kDa C-terminal agrin fragment to TM-agrin or proteolytic cleavage of TM-agrin would lead to the formation of filopodia-like processes for example by dimerization of TM-agrin and subsequent partitioning into lipid rafts. Thus, TM-agrin would be a receptor or coreceptor for the binding of the 22-kDa agrin fragment generated by neurotrypsin. This hypothesis does not, however, exclude additional involvement of the tyrosine kinase MuSK (11), of the agrin receptor LRP4 (15, 16), or of the Na/K ATPase (66) all of which are expressed at synapses in the CNS. In any case, the results presented in this study demonstrate a critical involvement of the seventh follistatin-like domain within TM-agrin in the formation of filopodia-like processes.
Supplementary Material
Acknowledgments
We thank A. Maelicke and M. Götz for support and encouragement and M. Rüegg and J. Bixby for critical reading of the manuscript. The technical assistance of A. Rohrbacher, S. Lochbrunner, and K. Peters is gratefully acknowledged.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- CNS
- central nervous system
- E
- embryonic day
- LRP4
- low density lipoprotein-like receptor 4
- TM-agrin
- transmembrane form of agrin
- HEK
- human embryonic kidney cell
- GAG
- glycosaminoglycan
- EGF
- epidermal growth factor
- GFP
- green fluorescent protein
- NtA-agrin
- NtA domain-containing agrin isoform.
REFERENCES
- 1.Bezakova G., Ruegg M. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 295–308 [DOI] [PubMed] [Google Scholar]
- 2.Kröger S., Pfister H. (2009) Fut. Neurol. 4, 67–86 [Google Scholar]
- 3.Sanes J. R., Lichtman J. W. (2001) Nat. Rev. Neurosci. 2, 791–805 [DOI] [PubMed] [Google Scholar]
- 4.Lin S., Landmann L., Ruegg M. A., Brenner H. R. (2008) J. Neurosci. 28, 3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautam M., Noakes P. G., Moscoso L., Rupp F., Scheller R. H., Merlie J. P., Sanes J. R. (1996) Cell 85, 525–535 [DOI] [PubMed] [Google Scholar]
- 6.Kröger S., Schröder J. E. (2002) News Physiol. Sci. 17, 207–212 [DOI] [PubMed] [Google Scholar]
- 7.Li Z., Hilgenberg L. G., O'Dowd D. K., Smith M. A. (1999) J. Neurobiol. 39, 547–557 [DOI] [PubMed] [Google Scholar]
- 8.Serpinskaya A. S., Feng G., Sanes J. R., Craig A. M. (1999) Dev. Biol. 205, 65–78 [DOI] [PubMed] [Google Scholar]
- 9.Ferreira A. (1999) J. Cell Sci. 112, 4729–4738 [DOI] [PubMed] [Google Scholar]
- 10.Böse C. M., Qiu D., Bergamaschi A., Gravante B., Bossi M., Villa A., Rupp F., Malgaroli A. (2000) J. Neurosci. 20, 9086–9095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ksiazek I., Burkhardt C., Lin S., Seddik R., Maj M., Bezakova G., Jucker M., Arber S., Caroni P., Sanes J. R., Bettler B., Ruegg M. A. (2007) J. Neurosci. 27, 7183–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowe M. A., Deyst K. A., Leszyk J. D., Fallon J. R. (1994) Neuron 12, 1173–1180 [DOI] [PubMed] [Google Scholar]
- 13.Hopf C., Hoch W. (1996) J. Biol. Chem. 271, 5231–5236 [DOI] [PubMed] [Google Scholar]
- 14.Gesemann M., Brancaccio A., Schumacher B., Ruegg M. A. (1998) J. Biol. Chem. 273, 600–605 [DOI] [PubMed] [Google Scholar]
- 15.Zhang B., Luo S., Wang Q., Suzuki T., Xiong W. C., Mei L. (2008) Neuron 60, 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim N., Stiegler A. L., Cameron T. O., Hallock P. T., Gomez A. M., Huang J. H., Hubbard S. R., Dustin M. L., Burden S. J. (2008) Cell 135, 334–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess R. W., Skarnes W. C., Sanes J. R. (2000) J. Cell Biol. 151, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neumann F. R., Bittcher G., Annies M., Schumacher B., Kröger S., Ruegg M. A. (2001) Mol. Cell. Neurosci. 17, 208–225 [DOI] [PubMed] [Google Scholar]
- 19.Denzer A. J., Brandenberger R., Gesemann M., Chiquet M., Ruegg M. A. (1997) J. Cell Biol. 137, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annies M., Kröger S. (2002) Mol. Cell. Neurosci. 20, 525–535 [DOI] [PubMed] [Google Scholar]
- 21.Annies M., Bittcher G., Ramseger R., Löschinger J., Wöll S., Porten E., Abraham C., Rüegg M. A., Kröger S. (2006) Mol. Cell. Neurosci. 31, 515–524 [DOI] [PubMed] [Google Scholar]
- 22.McCroskery S., Chaudhry A., Lin L., Daniels M. P. (2006) Mol. Cell. Neurosci. 33, 15–28 [DOI] [PubMed] [Google Scholar]
- 23.Ramseger R., White R., Kröger S. (2009) J. Biol. Chem. 284, 7697–7705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto-Miyai K., Sokolowska E., Zurlinden A., Gee C. E., Lüscher D., Hettwer S., Wölfel J., Ladner A. P., Ster J., Gerber U., Rülicke T., Kunz B., Sonderegger P. (2009) Cell 136, 1161–1171 [DOI] [PubMed] [Google Scholar]
- 25.Tsen G., Halfter W., Kröger S., Cole G. J. (1995) J. Biol. Chem. 270, 3392–3399 [DOI] [PubMed] [Google Scholar]
- 26.Kröger S., Horton S. E., Honig L. S. (1996) J. Neurobiol. 29, 165–182 [DOI] [PubMed] [Google Scholar]
- 27.Kröger S. (1997) Mol. Cell. Neurosci. 10, 149–161 [DOI] [PubMed] [Google Scholar]
- 28.Grutzendler J., Kasthuri N., Gan W. B. (2002) Nature 420, 812–816 [DOI] [PubMed] [Google Scholar]
- 29.McCroskery S., Bailey A., Lin L., Daniels M. P. (2009) Neuroscience 163, 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dityateva G., Hammond M., Thiel C., Ruonala M. O., Delling M., Siebenkotten G., Nix M., Dityatev A. (2003) J. Neurosci. Methods 130, 65–73 [DOI] [PubMed] [Google Scholar]
- 31.White R., Gonsior C., Krämer-Albers E. M., Stöhr N., Hüttelmaier S., Trotter J. (2008) J. Cell Biol. 181, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hohenester E., Maurer P., Timpl R. (1997) EMBO J. 16, 3778–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Innis C. A., Hyvönen M. (2003) J. Biol. Chem. 278, 39969–39977 [DOI] [PubMed] [Google Scholar]
- 34.Uhm C. S., Neuhuber B., Lowe B., Crocker V., Daniels M. P. (2001) J. Neurosci. 21, 9678–9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winzen U., Cole G. J., Halfter W. (2003) J. Biol. Chem. 278, 30106–30114 [DOI] [PubMed] [Google Scholar]
- 36.Baerwald-De la Torre K., Winzen U., Halfter W., Bixby J. L. (2004) J. Neurochem. 90, 50–61 [DOI] [PubMed] [Google Scholar]
- 37.Murphy-Ullrich J. E., Lane T. F., Pallero M. A., Sage E. H. (1995) J. Cell. Biochem. 57, 341–350 [DOI] [PubMed] [Google Scholar]
- 38.Ullman C. G., Perkins S. J. (1997) Biochem. J. 326, 939–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vannahme C., Schübel S., Herud M., Gösling S., Hülsmann H., Paulsson M., Hartmann U., Maurer P. (1999) J. Neurochem. 73, 12–20 [DOI] [PubMed] [Google Scholar]
- 40.Vannahme C., Smyth N., Miosge N., Gösling S., Frie C., Paulsson M., Maurer P., Hartmann U. (2002) J. Biol. Chem. 277, 37977–37986 [DOI] [PubMed] [Google Scholar]
- 41.Vannahme C., Gösling S., Paulsson M., Maurer P., Hartmann U. (2003) Biochem. J. 373, 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uchida T., Wada K., Akamatsu T., Yonezawa M., Noguchi H., Mizoguchi A., Kasuga M., Sakamoto C. (1999) Biochem. Biophys. Res. Commun. 266, 593–602 [DOI] [PubMed] [Google Scholar]
- 43.Bork P., Downing A. K., Kieffer B., Campbell I. D. (1996) Q. Rev. Biophys. 29, 119–167 [DOI] [PubMed] [Google Scholar]
- 44.Simons K., Toomre D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 45.Allen J. A., Halverson-Tamboli R. A., Rasenick M. M. (2007) Nat. Rev. Neurosci. 8, 128–140 [DOI] [PubMed] [Google Scholar]
- 46.Yoneda A., Couchman J. R. (2003) Matrix Biol. 22, 25–33 [DOI] [PubMed] [Google Scholar]
- 47.Zacharias U., Leuschner R., Nörenberg U., Rathjen F. G. (2002) Mol. Cell. Neurosci. 21, 626–633 [DOI] [PubMed] [Google Scholar]
- 48.Adams J. C., Schwartz M. A. (2000) J. Cell Biol. 150, 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Inouye S., Guo Y., Ling N., Shimasaki S. (1991) Biochem. Biophys. Res. Commun. 179, 352–358 [DOI] [PubMed] [Google Scholar]
- 50.Sumitomo S., Inouye S., Liu X. J., Ling N., Shimasaki S. (1995) Biochem. Biophys. Res. Commun. 208, 1–9 [DOI] [PubMed] [Google Scholar]
- 51.Wang Q., Keutmann H. T., Schneyer A. L., Sluss P. M. (2000) Endocrinology 141, 3183–3193 [DOI] [PubMed] [Google Scholar]
- 52.Kim M. J., Cotman S. L., Halfter W., Cole G. J. (2003) J. Neurobiol. 55, 261–277 [DOI] [PubMed] [Google Scholar]
- 53.Denzer A. J., Schulthess T., Fauser C., Schumacher B., Kammerer R. A., Engel J., Ruegg M. A. (1998) EMBO J. 17, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuste R., Bonhoeffer T. (2004) Nat. Rev. Neurosci. 5, 24–34 [DOI] [PubMed] [Google Scholar]
- 55.Holtmaat A. J., Trachtenberg J. T., Wilbrecht L., Shepherd G. M., Zhang X. Q., Knott G. W., Svoboda K. (2005) Neuron 45, 279–291 [DOI] [PubMed] [Google Scholar]
- 56.Ziv N. E., Smith S. J. (1996) Neuron 17, 91–102 [DOI] [PubMed] [Google Scholar]
- 57.Fiala J. C., Feinberg M., Popov V., Harris K. M. (1998) J. Neurosci. 18, 8900–8911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knott G., Holtmaat A. (2008) Brain Res. Rev. 58, 282–289 [DOI] [PubMed] [Google Scholar]
- 59.Lohmann C., Bonhoeffer T. (2008) Neuron 59, 253–260 [DOI] [PubMed] [Google Scholar]
- 60.Dailey M. E., Smith S. J. (1996) J. Neurosci. 16, 2983–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parnass Z., Tashiro A., Yuste R. (2000) Hippocampus 10, 561–568 [DOI] [PubMed] [Google Scholar]
- 62.Knott G. W., Holtmaat A., Wilbrecht L., Welker E., Svoboda K. (2006) Nat. Neurosci. 9, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 63.Stephan A., Mateos J. M., Kozlov S. V., Cinelli P., Kistler A. D., Hettwer S., Rülicke T., Streit P., Kunz B., Sonderegger P. (2008) FASEB J. 22, 1861–1873 [DOI] [PubMed] [Google Scholar]
- 64.Reif R., Sales S., Hettwer S., Dreier B., Gisler C., Wölfel J., Lüscher D., Zurlinden A., Stephan A., Ahmed S., Baici A., Ledermann B., Kunz B., Sonderegger P. (2007) FASEB J. 21, 3468–3478 [DOI] [PubMed] [Google Scholar]
- 65.Frischknecht R., Fejtova A., Viesti M., Stephan A., Sonderegger P. (2008) J. Neurosci. 28, 1568–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hilgenberg L. G., Su H., Gu H., O'Dowd D. K., Smith M. A. (2006) Cell 125, 359–369 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.