Abstract
The pancreatic beta cell is sensitive to even small changes in PDX1 protein levels; consequently, Pdx1 haploinsufficiency can inhibit beta cell growth and decrease insulin biosynthesis and gene expression, leading to compromised glucose-stimulated insulin secretion. Using metabolic labeling of primary islets and a cultured β cell line, we show that glucose levels modulate PDX1 protein phosphorylation at a novel C-terminal GSK3 consensus that maps to serines 268 and 272. A decrease in glucose levels triggers increased turnover of the PDX1 protein in a GSK3-dependent manner, such that PDX1 phosphomutants are refractory to the destabilizing effect of low glucose. Glucose-stimulated activation of AKT and inhibition of GSK3 decrease PDX1 phosphorylation and delay degradation. Furthermore, direct pharmacologic inhibition of AKT destabilizes, and inhibition of GSK3 increases PDX1 protein stability. These studies define a novel functional role for the PDX1 C terminus in mediating the effects of glucose and demonstrate that glucose modulates PDX1 stability via the AKT-GSK3 axis.
Keywords: Hormones/Insulin, Metabolism/Glucose, Phosphorylation/Kinases/Serine-Threonine, Phosphorylation/Transcription Factors, Protein/Degradation, Tissue/Organ Systems/Pancreatic Islet
Introduction
The homeodomain protein PDX1 has been shown to be a critical regulator of pancreatic development, in both humans and in mice (1, 2). Whereas genetic ablation or the total functional inhibition of PDX1 results in pancreatic agenesis, haploinsufficiency of PDX1 in humans leads to diabetes (3) and is associated with diminished glycemic control in mice (4, 5). Loss of a single allele of Pdx1 results in increased beta cell death (6) and a diminished capacity to mount a compensatory response in some models of insulin resistance (7). Thus, the beta cell is highly sensitive to total cellular levels of PDX1 protein.
The mechanisms by which PDX1 may regulate glucose homeostasis have been widely examined. In addition to regulating the insulin gene (8, 9), PDX1 has been shown to activate Glut2 and glucokinase genes, two key players required for glucose sensing (10, 11). PDX1 can also impact glucose sensing by influencing expression of mitochondrial metabolic pathways (12, 13). Accordingly, proteins important in glucose sensing are down-regulated in Pdx1+/− mice (5, 7).
In the beta cell, PDX1 is thought to be a direct mediator of glucose. Several studies have suggested that glucose-stimulated phosphorylation of PDX1 impacts DNA binding (14, 15) and the nucleocytoplasmic shuttling of PDX1 (16). Glucose-dependent phosphorylation has also been suggested to increase the transactivation potential of PDX1 by increasing recruitment of chromatin-modifying proteins such as p300 (17, 18) or by decreasing binding with histone deacetylases (19). Other studies suggest direct, glucose-dependent phosphorylation of PDX1 by specific kinases, including ERK2 1 and 2 (20), and perhaps p38 MAPK (14, 16). However, to date, the specific amino acid residues at which glucose-induces PDX1 phosphorylation, within the context of the cell, are not known.
The N terminus of PDX1 harbors a strong transactivation domain, flanked by the DNA-binding homeodomain (8, 21, 22). The C terminus of PDX1 has been shown to be a weak transactivation domain (21) and can also recruit inhibitor proteins (19, 23). Here, we show that glucose regulates steady-state levels of PDX1 protein via a novel phosphorylation site that maps to its C terminus. Although published studies suggest a stimulatory effect of glucose on PDX1 phosphorylation (15, 16, 20), our results show that glucose stimulation of primary islets and cultured Min6 beta cells decreases PDX1 phosphorylation. A decrease in C-terminal phosphorylation of PDX1 was associated with an increase in protein stability and was regulated by the reciprocal action of AKT and GSK3. Because glucose-stimulated insulin secretion results in autocrine stimulation of the insulin receptor in the beta cell (24), our results imply that glucose-dependent insulin secretion regulates steady-state levels of PDX1 protein in the beta cell.
EXPERIMENTAL PROCEDURES
Reagents
Commercial antibodies used in the study include mouse anti-hemagglutinin (HA), rabbit anti-AKT, anti-phospho-473-AKT, anti-GSK-3β, anti-phospho-21/9-GSK-3a/β (Cell Signaling, Beverly, MA), and rabbit anti-HA antibodies (Santa Cruz Biotechnology, Santa Cruz, CA). Mouse anti-HA-agarose beads, and M2-anti-FLAG-agarose beads were from Sigma. Anti-PDX1 antibodies were a gift from Dr. Marc Montminy (25). Protein A-Sepharose-conjugated anti-PDX1 antibodies were generated as described previously (26), and glutathione-Sepharose beads were from Amersham Biosciences. Horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were detected using Supersignal West-Pico chemiluminescence reagents (Pierce). [32P]Orthophosphate, [γ-32P]ATP, and [35S]methionine, [35S]cysteine (35S-Met/35S-Cys) were purchased from MP Biomedicals (Solon, OH). Recombinant GSK-3β was from New England Biolabs (Ipswich, MA). GSK3β inhibitor II, AKT inhibitor VIII, and cycloheximide were purchased from EMD Biosciences, San Diego(full description of inhibitor mechanism available on EMD website), and sequencing grade trypsin was from Hoffmann-La Roche.
Plasmids
Full-length rat PDX1 was subcloned into pGEX4T using BamHI/NotI sites and subcloned into both pcDNA3-HA and pcDNA3-FLAG using XhoI/Xbal sites. PDX1 deletion mutants were generated by PCR, and point mutations were generated using QuikChange site-directed mutagenesis kit as per the manufacturer's instructions (Stratagene, La Jolla, CA). All constructs were verified by sequencing. GSK3β and AKT constructs were purchased from Addgene (Cambridge, MA).
Adult Human Islets
Human adult islets were provided by Islet Cell Resource Center Basic Science Human Islet Distribution Program and the Islet Transplant Program, University of Illinois, Chicago. Islets were purified by handpicking and cultured overnight in CRML supplemented with 10% FBS. Prior to incubation with inhibitors, islets were cultured for at least 8 h in RPMI 1640 medium supplemented with 10% FBS.
Islet Isolation
Islet isolation was performed as described previously (27). Briefly, pancreata from 3-month-old Sprague-Dawley rats or C57Bl/6J mice (Jackson Laboratory, Bar Harbor, ME) were distended with collagenase P (1.5 mg/ml; Roche Applied Science) or liberase (0.2 mg/ml; Roche Applied Science), respectively, and digested at 37 °C for 15 min. Preparations were washed with Hanks' buffered salt solution, and the dissociated islets were purified on Ficoll gradients and cultured in RPMI 1640 medium supplemented with 10% FBS. For metabolic labeling studies, islets from 80 rats were isolated in two batches, and the pooled islets were cultured overnight in RPMI medium prior to metabolic labeling.
Cell Culture and Transfections
Min6 cells (passages 15–18) were grown in DMEM containing 24 mm glucose supplemented with 4% batch-tested, heat-inactivated FBS and 50 μm β-mercaptoethanol. Transfections were performed using Lipofectamine 2000 (Invitrogen) as per manufacturer's instructions. For cycloheximide chase and static treatment, 6-well dishes were transfected with 0.5 μg of HA-PDX1 plasmid and where indicated with 0.4 μg of kinase plasmid. For phosphorylation studies, Min6 cells in 10-cm dishes were transfected with 5 μg of PDX1 plasmid, and where indicated 3 μg of kinase plasmid was included.
Metabolic Labeling and Phosphopeptide Mapping and Phosphoamino Acid Analysis
Two groups of 6000 rat islets each, handpicked and counted, were incubated in low glucose (4 mm) medium containing phosphate-free DMEM, 5% dialyzed serum, and 0.5 mCi/ml [32P]orthophosphate for 5 h, followed by a either a 45-min stimulus with glucose to a final concentration of 24 mm or nonmetabolizable sugar Xylitol (used as a control to maintain osmotic balance). In case of transfected Min6 cells, metabolic labeling was conducted in 10-cm dishes 40 h post-transfection. The cells were washed with cold phosphate-buffered saline and harvested in SDS-lysis buffer (0.5% SDS, 50 mm Tris, pH 8.0, 1 mm EDTA) containing phosphatase inhibitors. Lysates were boiled, cooled on ice, and precleared with pansorbin, and supernatants were used for immunoprecipitation with protein A-Sepharose-conjugated PDX1 antibodies or agarose-conjugated anti-FLAG antibodies. Following SDS-PAGE and autoradiography of the gel, PDX1 protein was extracted directly from the gel (28) or transferred to nitrocellulose, exposed to film, and then used for PDX1 protein extraction. To ensure equal loading, the membrane was used for Western blot analysis for PDX1 protein or for FLAG tag using alkaline phosphatase color detection system. The gel band bearing labeled PDX1 was excised, and the protein was extracted by boiling followed by two overnight extractions in 50 mm NH4HCO3, 1% SDS, and 0.05% β-mercaptoethanol, precipitated using trichloroacetic acid, and digested using trypsin as described previously (29). PDX1 bands excised from the membrane were blocked in 1% PVP-40 in 0.1 m acetic acid at 37 °C, washed extensively, and digested using trypsin. Tryptic peptides generated by either technique were oxidized in performic acid, washed extensively, and resolved by two-dimensional analysis using electrophoresis (pH 8.9) in the first dimension, followed by thin layer chromatography (n-butyl alcohol and pyridine), and visualized by autoradiography as described previously (29). After an initial comparison of two-dimensional maps derived from the two methods of labeled PDX1 extraction, the membrane-based method was used for the rest of the study. For phosphoamino acid analysis, labeled phosphopeptides identified from the two-dimensional maps were scraped off the silica plates and hydrolyzed in 6 n HCl at 110 °C; unlabeled phosphoamino acids were added to the hydrolyzed protein, and the mixture was separated by two-dimensional high voltage thin layer electrophoresis at pH 1.9 in the first dimension and pH 3.5 in the second dimension. Phosphoamino acids were detected using ninhydrin staining, and radiolabeled phosphoamino acids were detected by autoradiography.
In Vitro Kinase Assay
GST-PDX1-WT fusion protein was expressed using BL21-RIPL cells (Stratagene, La Jolla CA) and purified using glutathione beads according to the manufacturer's instructions (Amersham Biosciences). Glutathione S-transferase-bound PDX1-WT was incubated with recombinant GSK3β (New England Biolabs) for 20 min at 30 °C in kinase assay buffer (200 μm ATP, 20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm dithiothreitol) using [γ-32P]ATP as described previously (30). Following SDS-PAGE, phosphoproteins were detected by autoradiography, followed by two-dimensional tryptic mapping as described above.
Radiolabel Pulse-Chase Studies
Min6 cells were washed with phosphate-buffered saline and incubated for 1 h in Met/Cys-free DMEM supplemented with 4% dialyzed serum. The medium was then replaced with Met/Cys-free DMEM containing 0.25 mCi of 35S-Met/35S-Cys/ml and 4% dialyzed serum. After a 4-h incubation, the samples were chased with complete nonradioactive DMEM and harvested every 6 h for 24 h. Cells were lysed and immunoprecipitated using anti-PDX1 protein A-Sepharose, subjected to SDS-PAGE, and detected by autoradiography.
Cycloheximide Chase
30 h after transfection, Min6 cells were cultured in DMEM containing 100 μg/ml cycloheximide for the indicated times. For low glucose, cells were starved in 4 mm glucose for 6 h and replaced with medium containing either low glucose (4 mm glucose) or high glucose (24 mm glucose) prior to addition of cycloheximide. Chase experiments were performed twice in triplicate.
Static Culture with Low Glucose
30 h after transfection, Min6 cells were starved in DMEM containing 4 mm glucose followed by either 4 or 24 mm glucose for 20 h in the presence or absence of GSK3β inhibitor. HA immunoprecipitates from transfected cells were resolved by SDS-PAGE and Western blotting for PDX1. Human and mouse islets were cultured in RPMI 1640 medium supplemented with 10% FBS and treated with GSK3β, or AKT inhibitor, or both for 20 h and processed for Western blotting as described below.
Glucose and Insulin Stimulation
Min6 cells were incubated with HEPES-balanced Krebs-Ringer buffer (KRBH), pH 7.4 (15 mm HEPES, 120 mm NaCl, 4.7 mm KCl, 1.2 mm MgCl2, 1.2 mm KH2PO4, 20 mm NaHCO3, 2 mm CaCl2), supplemented with 0.1% bovine serum albumin and no glucose for 30 min, and 4 mm glucose for a further 4 h, followed by a 20-min stimulation with either 24 mm glucose or 100 nm insulin in the presence or absence of AKT inhibitor. The cells were immediately harvested and subjected to SDS-PAGE and Western blot analysis.
Immunoprecipitation and Western Blotting
Mammalian expression vectors encoding HA-tagged fusion proteins were expressed in Min6, and the immunoprecipitation was performed in NEHN (0.5% Nonidet P-40, 0.5 mm EDTA, 40 mm HEPES, 150 mm NaCl with a complete complement of protease and phosphatase inhibitors) using HA beads or anti-PDX1 protein A-Sepharose. Following SDS-PAGE and transfer, immunoblots were probed as indicated. Equal loading was confirmed by probing for co-transfected internal controls as indicated. For immunoblots of islet or Min6 cells, the cells were lysed in 50 mm HEPES, 1% SDS, 6 m urea containing a mixture of protease and phosphatase inhibitors, normalized for total protein using a micro-BCA assay, and used for Western blotting analysis as indicated.
Statistics
Densitometry and quantification were performed using GelEval 1.22 (FrogDance, Dundee, Scotland, United Kingdom) or ImageQuant (Amersham Biosciences). Statistical analysis was performed using the Student's unpaired t test on data generated from at least three separate experiments. In all cases, p values less than or equal to 0.05 were considered significant. Analysis was performed using StatView 5.0 statistical software.
RESULTS
Glucose Modulates Phosphorylation of PDX1
Glucose-dependent phosphorylation of PDX1 has been suggested by multiple studies (summarized in Ref. 31), but in vivo mapping of a specific phosphorylation site(s) in PDX1 has not been reported. Using [32P]orthophosphate, we metabolically labeled rat islets and examined whether PDX1 phosphorylation status is altered under low (4 mm) and high (24 mm) glucose conditions. Immunoprecipitates of endogenous PDX1 from the radiolabeled islets were resolved by SDS-PAGE. The band containing PDX1 was excised from the gel and digested with trypsin, and the resulting peptides were subjected to two-dimensional separation using thin layer electrophoresis followed by thin layer chromatography. Autoradiographs from two-dimensional peptide mapping of PDX1 revealed a distinct pattern of phosphopeptide spots with two major tryptic phosphopeptides showing a high level of radiolabel incorporation (Fig. 1A, panels i and ii, termed spots A and B). Interestingly, a 45-min stimulation of islets with 24 mm glucose (Fig. 1A, panel ii) resulted in reduced incorporation of radiolabel in both spots A and B compared with results from islets incubated with 4 mm glucose (panel i). These data suggest that glucose either inhibits phosphorylation or promotes dephosphorylation of PDX1.
FIGURE 1.
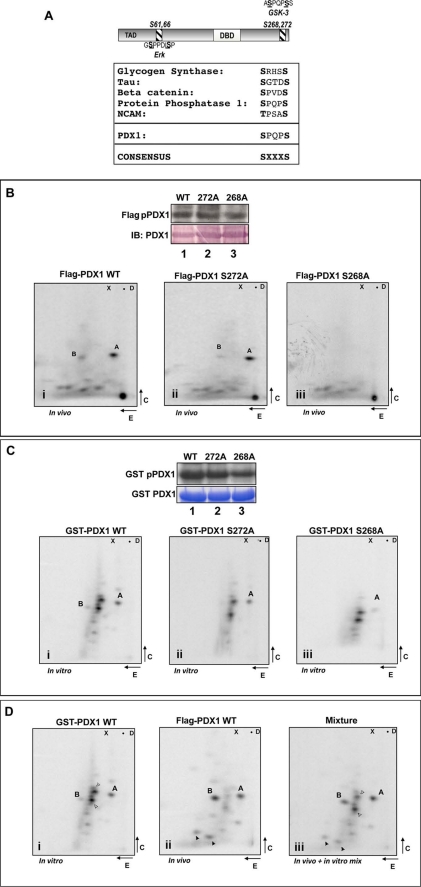
Mapping of glucose-regulated PDX1 phosphorylation sites using metabolically labeled rat islets and Min6 cells. A and B, PDX1 immunoprecipitates from [32P]orthophosphate-labeled rat islets (A) and Min6 cells (B) treated with 4 mm glucose (LG) or 24 mm glucose (HG) as indicated were resolved using SDS-PAGE. Phosphorylated PDX1 (pPDX) was identified by autoradiography, and tryptic peptide mapping was performed on the excised labeled bands. Two prominent phosphopeptides labeled as spots A and B showed a higher incorporation of phosphate in the presence of low glucose compared with high glucose (panels i and ii). C, phosphopeptide A and B localize to the C terminus of PDX1. FLAG immunoprecipitates from [32P]orthophosphate-labeled Min6 cells were transfected with FLAG-PDX1-WT (panel i) or FLAG-PDX1-C-term (amino acids 213–284) (panel ii). Phosphoamino acid analysis revealed spots A (panel iii) and B (panel iv) to be phosphoserines (pS), as inferred by the co-localization of counts/min from the phosphopeptide hydrolysates with unlabeled ninhydrin-stained phosphoamino acid standards (indicated by circles). Loading dye migration is represented by X (xylene cyanol) and D (dinitrophenyl-lysine); loading origin is marked by the closed circle; top edge of the map marks chromatography front.
Metabolic labeling of Min6 insulinoma cells revealed the same pattern of radiolabel incorporation into spots A and B as well as decreased incorporation of phosphate after incubation in high glucose (Fig. 1B, panels i and ii). To localize the phosphorylation event to a specific domain, we performed similar mapping experiments with transfected FLAG-tagged constructs encoding PDX1 or its subdomains in metabolically labeled cells as above. Full-length PDX1 and the PDX1 C terminus (amino acids 213–284) both showed similar patterns in their respective phosphopeptide maps, suggesting that the two prominent phosphorylation sites are located in the C terminus of the PDX1 protein (Fig. 1C, panels i and ii). The two phosphopeptides A and B were retrieved from the silica plates, acid-hydrolyzed, and resolved using two-dimensional electrophoresis as described previously (Fig. 1C, panels iii and iv). Spots A and B were both revealed to be phosphoserines, indicating that glucose regulates the phosphorylation status of two serine residues located in the C terminus of PDX1.
These findings contrast with previous reports suggesting that glucose induces phosphorylation of PDX1 (15, 16, 20). However, none of the studies have mapped the glucose-induced in vivo PDX1 phospho-acceptor sites, as phosphorylation occurs within the cell. ERK1 was found to phosphorylate PDX1 in vitro, at serines 61 and 66 (20). Using a similar in vitro phosphorylation approach, we also observed that ERK1 increased radiolabel incorporation into GST-PDX1 and that mutation of serines 61 and 66 into alanines significantly reduced incorporation (supplemental Fig. S1). Furthermore, two-dimensional peptide maps of ERK1-phosphorylated PDX1 revealed a distinct pattern of phosphopeptide spots that did not overlap with the spots observed after labeling of PDX1 in Min6 cells (supplemental Fig. S2). These data indicate that glucose-stimulated phosphorylation of the PDX1 C terminus is unlikely to be mediated by ERK1, and the role of ERK1 and -2 in modulating PDX1 phosphorylation in vivo merits further investigation.
PDX1 Phosphorylation Maps to a C-terminal GSK3 Consensus Sequence
A closer scrutiny of the potential phosphoserines in the C terminus of PDX1 revealed a consensus site for GSK3 (glycogen synthase kinase-3) at positions 268 and 272. This site is evolutionarily conserved in all mammalian homologs of PDX1 and is highly homologous to phosphorylation sites in other known GSK3 substrates (Fig. 2A). FLAG-tagged WT and mutant PDX1 constructs were transfected into Min6 cells, the cells metabolically labeled, and PDX1 immunoprecipitates separated by SDS-PAGE. PDX1 bands were ascertained by autoradiography, and equal loading of protein was confirmed by Western blotting using color detection (Fig. 2B, lanes 1–3). Finally, the PDX1 bands were excised and processed as described. Mutation of serine 272 resulted in the loss of spot B; however, mutation of serine 268 resulted in loss of both spots A and B (Fig. 2B, panels i–iii). Based on the predicted trypsin cleavage sites, serines 268 and 272 are contained within the same peptide. Our data are therefore consistent with spot B representing a dually phosphorylated peptide as judged by increased acidity and migration toward the cathode at pH 8.9. Spot A likely represents phosphorylation at a single site. Interestingly, mutation of serine 268 was sufficient to abolish the phosphorylation of both sites suggesting a connection between phosphorylation events at each site.
FIGURE 2.
PDX1 phosphorylation maps to a C-terminal GSK3 consensus site. A, schematic of PDX1 protein highlighting the ERK (Ser-61 and -66) and newly identified GSK3 (Ser-268 and -272) site (top panel). Also shown is a comparison of the PDX1 phosphorylation site with known GSK3 sites (bottom panel). B, PDX1 one-dimensional autoradiography (FLAG-pPDX) and Western immunoblotting (IB; alkaline phosphatase-based detection (top panel)). Lower panels show PDX1 tryptic peptides derived from FLAG-PDX-WT (lane 1 and panel i), FLAG-PDX-S272A (lane 2 and panel ii), and FLAG-PDX-S268A (lane 3 and panel iii). C, one-dimensional autoradiography (GST pPDX1), Coomassie staining (GST PDX1), and phosphopeptide maps of bacterially expressed GST-PDX1 WT (lane 1 and panel i), GST-PDX1-S272A (lane 2 and panel ii), and GST-PDX1-S268A (lane 3 and panel iii) phosphorylated in vitro using purified GSK3β and [γ-32P]ATP. Note loss of spot B in PDX-S272A mutant (panel ii in B and C) and loss of both spots A and B in PDX-S268A mutant (panel iii in B and C). D, tryptic maps of PDX1 phosphopeptides from in vitro GSK3-labeled GST-PDX1 (panel i), in vivo labeled FLAG-PDX1-WT (panel ii), or a mixture of equal counts from both (panel iii). Note the co-migration of spot A and B from the two reactions in panel iii. Open arrowheads indicate points of reference from in vitro maps and closed arrowheads from in vivo maps.
In vitro, at high concentrations, although inefficient, GSK3 is capable of phosphorylating both target sites (32). Therefore, we examined whether bacterially expressed GST-WT PDX1, or S268A, or S272A-PDX1 mutants can be directly phosphorylated in vitro using purified GSK3 and [γ-32P]ATP. Coomassie staining of total bacterial protein used in each reaction and a one-dimensional autoradiograph of the kinase reaction is shown (Fig. 2C, lanes 1–3). As seen in Fig. 2C (panels i–iii), phosphopeptide mapping revealed GSK3-induced phosphate incorporation into two tryptic peptides that resembled the migration of spots A and B. To ascertain that tryptic peptides generated from direct phosphorylation of PDX1 by GSK3 in vitro were identical to those generated from Min6 cells, we resolved a mix of equal counts of in vitro and in vivo labeled PDX1 peptides by two-dimensional mapping. As shown in Fig. 2D, spots from the in vitro reaction co-migrated with spots A and B from the in vivo reaction, indicating that PDX1 may indeed be a substrate for GSK3 in Min6 cells.
AKT Modulates PDX1 Phosphorylation in Min6 Cells
GSK3 is activated during fasting and inhibited upon refeeding by a direct AKT-mediated phosphorylation of GSK3. Therefore, we examined whether PDX1 phosphorylation was affected by modulating AKT activity. Min6 cells were transfected with FLAG-PDX1 along with kinase-dead (KD) or constitutively active myristoylated (Myr) AKT, followed by two-dimensional mapping of FLAG immunoprecipitates from metabolically labeled Min6 cells. One-dimensional autoradiograph and Western blot of the equal PDX1 immunoprecipitated from each treatment are shown (Fig. 3A, lanes 1–4). The two-dimensional maps reveal that in the presence of AKT2-KD incorporation of radiolabel was higher in both spots (Fig. 3A, compare 1st and 2nd panels). Importantly, treatment with a cell-permeable inhibitor of GSK3 that specifically competes for the ATP-binding site in GSK3 suppressed phosphate incorporation compared with AKT2-KD alone (Fig. 3A, compare 1st and 2nd panels). Furthermore, constitutively active Myr-AKT attenuated incorporation into spot B relative to both vector control and AKT2-KD (Fig. 3A, compare 1st, 3rd, and 4th panels). These data support a model in which AKT modulates the activity of GSK3, which is then reflected in phosphorylation of PDX1.
FIGURE 3.
AKT and GSK3β kinases regulate the phosphorylation and total protein levels of PDX1. A, one-dimensional autoradiography (FLAG pPDX), Western immunoblotting (IB), and phosphopeptide maps of FLAG-PDX1 immunoprecipitates from Min6 cells overexpressing AKT-KD in the absence (lane 1 and panel i) or presence of GSK3β inhibitor (lane 2 and panel ii) or overexpressing the vector control (lane 3 and panel iii) or Myr-AKT (lane 4 and panel iv). B–E, Western blots and densitometry of PDX1 protein levels from extracts of mouse (B and C) and human islets (D and E) in the presence of vehicle (NT, lane 1), AKT inhibitor (AKT-I, lane 2), GSK3β inhibitor (GSK-I, lane 3), or both (AKT-I+ GSK-I, lane 4). Histone H3 was used as a loading control. Densitometric data from three independent experiments were normalized to vehicle-treated controls (NT) and presented as the mean ± S.D. fold difference (*, p < 0.05 compared with untreated islets).
GSK3 and AKT Inversely Regulate Total PDX1 Protein Levels in Mouse and Human Islets
Although its functional role has not been clarified, the C terminus of PDX1 has been linked with familial diabetes (33, 34). To determine the functional impact of PDX1 phosphorylation at serine residues 268 and 272 in the C terminus, we first examined whether phosphomutants impacted the subcellular localization or the transactivation potential of PDX1 in transfected Min6 cells. Under both low and high glucose conditions, PDX1 WT and phosphomutants showed nuclear localization. Similarly, when assessed in Gal4 transactivation assays, the transactivation potential of the phosphomutants was not different from that of PDX1 WT (data not shown).
GSK3-mediated, phosphorylation-dependent proteasomal degradation is a prevalent mechanism in mammalian cells (35–38). Therefore, we next examined whether PDX1 protein levels are affected by pharmacologic inhibition of GSK3 and its upstream regulator kinase AKT. We found that PDX1 protein levels were markedly reduced in mouse (Fig. 3, B and C) as well as in human islets (Fig. 3, D and E) upon treatment with an allosteric inhibitor of AKT kinase. By contrast, pharmacologic inhibition of GSK3 increased PDX1 levels under basal conditions and also rescued the loss of PDX1 protein observed upon pharmacologic inhibition of AKT. These findings in both mouse and human primary islets provide strong support for a model in which AKT regulates PDX1 protein levels through its inhibitory effect on GSK3.
AKT Stabilizes and GSK3 Induces Degradation of PDX1 Protein
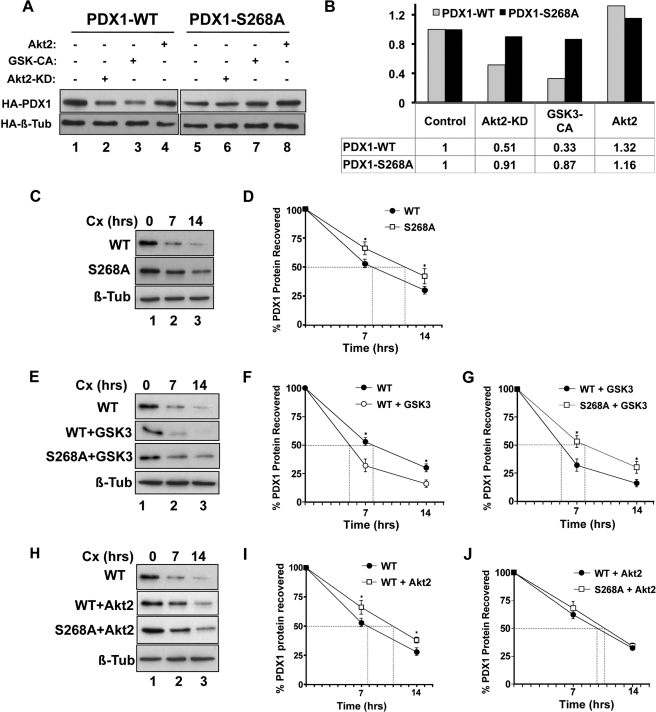
AKT has been shown to augment Pdx1 transcription by relieving the repressive effects of FOXO1 (39). To demonstrate that AKT also directly impacts PDX1 protein levels, Min6 cells were transfected with cytomegalovirus promoter-driven WT and S268A PDX1 constructs along with AKT and GSK3 kinases (Fig. 4A). Because mutation of serine 268 resulted in the loss of radiolabel incorporation into both spots A and B, the S268A PDX1 mutant was used for these assays. Consistent with a model of reciprocal AKT-GSK3 action, constitutively active GSK3 resulted in a nearly 70% decrease in PDX1 protein levels, whereas overexpression of AKT2 resulted in a 30% increase compared with PDX1 levels in the presence of control vector (Fig. 4, A and B). Accordingly, AKT2-KD led to a 50% decline in PDX1 protein levels. Notably, PDX1-S268A was resistant to the modulation of kinase activity, suggesting that the AKT-GSK3 axis regulates PDX1 protein levels via Ser-268 phosphorylation.
FIGURE 4.
AKT and GSK3β regulate PDX1 stability via the newly identified C-terminal phosphorylation site. A, Western blotting of anti-HA immunoprecipitates from Min6 cells expressing HA-PDX1-WT (lanes 1–4) or HA-PDX1-S268A (lanes 5–8) in the presence of control vector (lanes 1 and 5) or kinase-dead AKT2 (AKT2-KD, lanes 2 and 6), constitutively active GSK3β (GSK-CA, lanes 3 and 7), or AKT2 (lanes 4 and 8). Densitometric quantification shown as fold difference in PDX1 protein levels normalized to HA-PDX1-WT (gray bars) or HA-PDX1-S268A (black bars) in the presence of control vector. C–J, HA immunoprecipitates from Min6 cells expressing HA-PDX1-WT or HA-PDX1-S268A alone (C and D), or in the presence of GSK3β (E–G) or AKT2 (H–J) following a time-dependent cycloheximide (Cx) chase were probed using anti-PDX1 antibodies. Tubulin (Tub) was used as a control. Densitometric quantification (D, F, and G, and I and J) demonstrates the mean ± S.D. (n = 3) of protein loss at each time point (*, p ≤ 0.05 between treatments at the indicated time point).
We next assessed the impact of phosphorylation on PDX1 half-life. We first compared the half-life of PDX1 in Min6 cells using cycloheximide to inhibit translation (Fig. 4, C and D) or by pulse-chase labeling with [35S]methionine (supplemental Fig. S3). The calculated half-life by both methods was comparable and assessed to be ∼8 h. Hence, all subsequent assessments of PDX1 half-life were made using cycloheximide chase experiments. S268A was assessed to be more stable than PDX1-WT with a half-life of ∼11.5 h (Fig. 4, C and D). GSK3 overexpression reduced PDX1 half-life to 5 h (Fig. 4, E and F), whereas the PDX1-S268A mutant again was more stable, with an average half-life of 8 h even with overexpression of GSK3 (Fig. 4, E and G). By contrast, overexpression of AKT2 increased the half-life of PDX1 to ∼11 h (Fig. 4, H and I), which was comparable with the previously observed half-life S268A mutant. AKT2 overexpression had no further stabilizing effect on PDX1-S268A (Fig. 4, H and J).
Inhibition of Phosphorylation at Serine 268 Renders PDX1 Refractory to the Destabilizing Effects of Low Glucose
To determine whether the ability of glucose to modulate phosphorylation of PDX1 at serine 268 is mediated by the reciprocal actions of AKT and GSK3, we first examined whether glucose stimulation leads to phosphorylation of AKT and its downstream substrate GSK3. Min6 cells were preincubated in low glucose to reduce basal insulin secretion and AKT activation. As shown in Fig. 5A, a 20-min stimulus with high glucose (24 mm) increased phosphorylation of both AKT and GSK3 (lanes 1 and 2). Both phosphorylation events were inhibited by pharmacologic inhibition of AKT kinase activity (Fig. 5A, lane 3). The effects of glucose stimulation on AKT and GSK3 phosphorylation were comparable with the effect of direct insulin stimulation (Fig. 5A, lane 4). These data suggest that glucose stimulation may activate the AKT kinase pathway through the autocrine effect of insulin on the beta cell.
FIGURE 5.
Glucose treatment modulates activity of AKT and GSK kinases to regulate half-life of PDX1 protein. A, Western blotting of protein extracts from Min6 cells incubated in 4 mm glucose (L, lane 1), 24 mm glucose (H, lanes 2 and 3), or 4 mm glucose + 100 nm insulin (lanes 4 and 5) in the absence (lanes 2 and 4) or presence of AKT inhibitor (lanes 3 and 5). Cellular extracts were probed as indicated. B, Min6 cells transfected with HA-PDX1-WT (lanes 1–4), along with control vector (lanes 1–3) or AKT2 (lane 3), or with HA-PDX1-S268A (lanes 5 and 6) were incubated in 4 mm glucose (L, lanes 1, 3, 4, and 5), in the presence or absence of GSK3β inhibitor (GSK-I) or 24 mm glucose (H, lanes 2 and 6), followed by immunoprecipitation using anti-HA antibodies and Western blotting for PDX1 protein. Tubulin was used as a control. C, densitometric quantitation of bands (n = 3) was normalized to PDX1-WT in 4 mm glucose (LG) and presented as the mean ± S.D. fold difference (*, p < 0.05 compared with PDX1-WT in 4 mm glucose; ns, not significant). D–F, time-dependent cycloheximide chase of HA-immunoprecipitates from Min6 cells transfected with either HA-PDX1-WT or HA-PDX1-S268A and incubated in 4 mm glucose (LG) or 24 mm glucose (HG) as indicated. Tubulin (Tub) was used as a control. t½ was determined from densitometric quantification (E and F) of time-dependent protein loss presented as the mean ± S.D. (n = 3). Closed and open circles, PDX1-WT in HG and LG, respectively; closed squares, PDX1-S268A in LG; *, p < 0.05 between treatments at the indicated time point.
Next we examined whether the glucose-stimulated changes in AKT and GSK3 also impacted total PDX1 protein levels. To isolate the effect of glucose on PDX1 protein stability from its reported effect on Pdx1 gene transcription (40), we transfected early passage Min6 cells with cytomegalovirus promoter-driven expression vectors encoding HA-tagged WT and S268A PDX1. HA immunoprecipitation followed by Western blotting showed that compared with incubation in high glucose, a 20-h static incubation with low glucose resulted in a dramatic decrease in PDX1 protein level (Fig. 5B, lanes 1 and 2). These effects were reversed by preincubation with the GSK3 kinase inhibitor (Fig. 5B, lane 4), thus linking the loss of PDX1 protein under low glucose conditions to the activation of GSK3. Furthermore, co-transfection of AKT2 under low glucose conditions prevented the decrease in PDX1 protein and even boosted the levels beyond those observed with high glucose (Fig. 5B, lane 3). Unlike PDX1-WT, protein levels of the S268A PDX1 mutant are maintained even in low glucose conditions and are not significantly different under high glucose conditions (Fig. 5B, lanes 5 and 6). These data, presented in a graphic form in Fig. 5C, underscore the important role of Ser-268 phosphorylation in regulating PDX1 stability in a glucose-dependent manner.
Finally, we examined whether low glucose culture conditions that result in decreased AKT activation and increased GSK3 activity also impact the half-life of PDX1 protein (Fig. 5D). Low glucose culture conditions resulted in the rapid loss of PDX1 protein and a calculated half-life of ∼4.8 h, nearly 40% lower than the observed PDX1 half-life under high glucose conditions (Fig. 5, D and E). Remarkably, the half-life of the PDX1-S268A mutant remained unchanged (Fig. 5, D and F). Altering the glucose level in the culture medium produces changes that are likely to be complex and to extend beyond the involvement of AKT and GSK3 alone. However, the important role of serine 268 in modulating the PDX1 half-life with GSK3 overexpression or under low glucose conditions suggests that GSK3 regulates at least a subset of the effects produced by low glucose conditions. Our results clearly demonstrate that phosphorylation of PDX1 on the C-terminal site mapped here has a pivotal role in regulating PDX1 stability under biologically relevant conditions.
DISCUSSION
In this study, we demonstrate that within the beta cell, glucose regulates stability of the PDX1 protein via novel phosphorylation sites. Using an unbiased approach employing [32P]orthophosphate labeling of cells, we found that glucose regulates phosphorylation of serines 268 and 272 in the C terminus of the PDX1 protein. Furthermore, PDX1 phosphorylation mapped to a consensus site for GSK3, and both GSK3 activity and PDX1 phosphorylation were reduced upon glucose stimulation. Consistent with the ability of AKT kinase to inhibit GSK3, conditions that increased AKT activity also stabilized the PDX1 protein. The overall conclusion that we draw from this study is that glucose stimulation of beta cells is required to maintain steady-state levels of PDX1 within a narrow range, and this regulation of PDX1 is coordinated by the GSK3/AKT axis (see Fig. 6 schematic).
FIGURE 6.
Schematic representation of AKT and GSK3β in glucose-regulated stability of PDX1 protein.
The identification of GSK3 as the C-terminal PDX1 kinase satisfies multiple criteria that are significant for a protein to be a substrate of a kinase, including the following: in vitro phosphorylation of Ser-268–272 by GSK3; co-migration of phosphopeptides in two-dimensional maps from in vitro and in vivo reactions; inhibition of in vivo phosphorylation by pharmacologic inhibition of GSK3; and the failure of GSK3 to phosphorylate GSK3 consensus site mutants of PDX1 both in vivo and in vitro (Fig. 2B). Most importantly, phosphorylation-dependent degradation of PDX1 by GSK3 is consistent with the biologic role of both proteins in the beta cell. Two-dimensional tryptic phosphopeptide mapping was the preferred method because it is both sensitive and reproducible. Also, as shown in this study, in the absence of a phospho-specific antibody, this method affords the flexibility of examining phosphorylation under varying biologic conditions. This method is also more successful at identifying sites that are phosphorylated at sub-stoichiometric levels, which can be a limitation with mass spectrometry (41).
Our results of PDX1 phosphorylation and phosphopeptide mapping differ from published studies that link glucose-stimulated increase in PDX1 transcriptional activity with in vitro phosphorylation of PDX1 (20). Unlike other studies (14, 15, 20), we found that phosphorylation of PDX1 was reduced in response to glucose stimulation. These comparisons are somewhat limited because glucose-induced PDX1 phosphoacceptor sites were not mapped in the above-mentioned studies. Serines 268 and 272 together constitute a consensus site for GSK3. In this regard, a previous report also identified GSK3 as a PDX1 N-terminal kinase (Ser-61 and -66) under conditions of oxidative stress (42). Under the conditions used in our study, the two-dimensional phosphopeptide maps do not indicate the presence of an additional GSK3 phosphorylation site in PDX1. Compared with a canonical GSK3 consensus (see Fig. 2B), the presence of an extra amino acid between Ser-61 and -66 may suggest an opportunistic GSK3-dependent phosphorylation of PDX1 that is specifically associated with severe oxidative stress.
Our studies show that PDX1 phosphorylation at Ser 268–272 hastens its degradation. Phosphorylation events expand the repertoire of structural changes, which in turn serve as conformational switches for regulated recruitment of proteins. Thus, the possibility exists for as yet unidentified protein(s) to cooperate with Ser-268–272 in regulating PDX1 protein stability. A shift in PDX1 half-life from 7 to 8 h in high glucose conditions to 4–5 h under low glucose conditions, or an increase in half-life to nearly 11 h upon overexpression of AKT, would suggest that PDX1 levels may be profoundly impacted under pathophysiologic states that compromise beta cell glucose uptake. PDX1 function may also be affected by shifts in glucose levels under physiologic conditions such as overnight fasting.
In the beta cell, effects of glucose and insulin show extensive overlap (43). Such an overlap can be explained by the fact that glucose activates the insulin receptor-IRS1/2 pathway by the autocrine effect of glucose-stimulated insulin secretion (24, 44). Glucose stimulation has been shown to promote phosphorylation of AKT (24, 45) and, consequently, AKT-dependent inhibition of its downstream target GSK3 (46). Similar to PDX1, MafA, and MCL-1, protein stability is also regulated by glucose-modulated GSK3 activity (47–49), suggesting that PDX1 protein turnover is regulated by a prevalent cellular mechanism.
The reciprocal relationship between AKT and GSK3 highlights the biologic significance of PDX1 phosphorylation by GSK3. Indeed, PDX1 levels are reduced in mice with beta cell-specific overexpression of GSK3 (46) or severe loss of AKT (50). Similarly, PDX1 levels are augmented with loss of GSK3 (51) and AKT overexpression as shown here. These stabilizing effects of AKT on PDX1 would be in addition to relief of forkhead transcription factor FOXO1-mediated Pdx1 gene repression. Functionally, insulin-IRS-AKT signaling is required for a normal beta cell glucose response and is also critical for beta cell survival and growth (50, 52–56). Because PDX1 action is central to expression of genes involved in GSIS, and is required for beta cell survival and growth, it is reasonable to suggest that AKT mediates some of its effects on beta cell via PDX1. The interdependence between PDX1 and insulin-AKT action in the beta cell is supported by two studies, showing that beta cell failure observed in Irs2−/− mice can be rescued either by PDX1 overexpression (57) or by haploinsufficiency of FoxO1 (39).
PDX1 is abundantly expressed in the beta cell, yet even small changes in PDX1 protein levels impact its function. Brissova et al. (58) elegantly demonstrate that islets from Pdx1+/− mice retain 68% expression of PDX1 and yet show a clear and significant compromise in glucose metabolism and calcium mobilization. Taken together, our results suggest the presence of a self-sustaining loop between glucose stimulation and genes required for glucose sensing via insulin signaling and PDX1.
Supplementary Material
Acknowledgments
We are deeply grateful to Dr. Marc Montminy (The Salk Institute for Biological Studies, La Jolla, CA) for providing guidance and resources during the PDX1 phosphopeptide mapping studies. We also thank Dr. Greg Korbutt (Alberta Diabetes Institute, Canada) for help with islet isolation, Jill Meisenhelder and the laboratory of Dr. Tony Hunter (The Salk Institute, La Jolla CA) for use of phosphopeptide mapping facilities, and Dr. Doris Stoffers for a critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK080147 (to U. S. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ERK
- extracellular signal-regulated kinase
- HA
- hemagglutinin
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- WT
- wild type.
REFERENCES
- 1.Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 2.Stoffers D. A., Zinkin N. T., Stanojevic V., Clarke W. L., Habener J. F. (1997) Nat. Genet. 15, 106–110 [DOI] [PubMed] [Google Scholar]
- 3.Stoffers D. A., Ferrer J., Clarke W. L., Habener J. F. (1997) Nat. Genet. 17, 138–139 [DOI] [PubMed] [Google Scholar]
- 4.Dutta S., Bonner-Weir S., Montminy M., Wright C. (1998) Nature 392, 560. [DOI] [PubMed] [Google Scholar]
- 5.Ahlgren U., Jonsson J., Jonsson L., Simu K., Edlund H. (1998) Genes Dev. 12, 1763–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J. D., Bernal-Mizrachi E., Alejandro E. U., Han Z., Kalynyak T. B., Li H., Beith J. L., Gross J., Warnock G. L., Townsend R. R., Permutt M. A., Polonsky K. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19575–19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulkarni R. N., Jhala U. S., Winnay J. N., Krajewski S., Montminy M., Kahn C. R. (2004) J. Clin. Invest. 114, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peers B., Leonard J., Sharma S., Teitelman G., Montminy M. R. (1994) Mol. Endocrinol. 8, 1798–1806 [DOI] [PubMed] [Google Scholar]
- 9.Ohlsson H., Karlsson K., Edlund T. (1993) EMBO J. 12, 4251–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watada H., Kajimoto Y., Umayahara Y., Matsuoka T., Kaneto H., Fujitani Y., Kamada T., Kawamori R., Yamasaki Y. (1996) Diabetes 45, 1478–1488 [DOI] [PubMed] [Google Scholar]
- 11.Waeber G., Thompson N., Nicod P., Bonny C. (1996) Mol. Endocrinol. 10, 1327–1334 [DOI] [PubMed] [Google Scholar]
- 12.Brissova M., Shiota M., Nicholson W. E., Gannon M., Knobel S. M., Piston D. W., Wright C. V., Powers A. C. (2002) J. Biol. Chem. 277, 11225–11232 [DOI] [PubMed] [Google Scholar]
- 13.Gauthier B. R., Brun T., Sarret E. J., Ishihara H., Schaad O., Descombes P., Wollheim C. B. (2004) J. Biol. Chem. 279, 31121–31130 [DOI] [PubMed] [Google Scholar]
- 14.Wu H., MacFarlane W. M., Tadayyon M., Arch J. R., James R. F., Docherty K. (1999) Biochem. J. 344, 813–818 [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen H. V., Peshavaria M., Pedersen A. A., Philippe J., Stein R., Madsen O. D., Serup P. (1998) FEBS Lett. 431, 362–366 [DOI] [PubMed] [Google Scholar]
- 16.Macfarlane W. M., McKinnon C. M., Felton-Edkins Z. A., Cragg H., James R. F., Docherty K. (1999) J. Biol. Chem. 274, 1011–1016 [DOI] [PubMed] [Google Scholar]
- 17.Mosley A. L., Corbett J. A., Ozcan S. (2004) Mol. Endocrinol. 18, 2279–2290 [DOI] [PubMed] [Google Scholar]
- 18.Stanojevic V., Habener J. F., Thomas M. K. (2004) Endocrinology 145, 2918–2928 [DOI] [PubMed] [Google Scholar]
- 19.Mosley A. L., Ozcan S. (2004) J. Biol. Chem. 279, 54241–54247 [DOI] [PubMed] [Google Scholar]
- 20.Khoo S., Griffen S. C., Xia Y., Baer R. J., German M. S., Cobb M. H. (2003) J. Biol. Chem. 278, 32969–32977 [DOI] [PubMed] [Google Scholar]
- 21.Lu M., Miller C., Habener J. F. (1996) Endocrinology 137, 2959–2967 [DOI] [PubMed] [Google Scholar]
- 22.Peshavaria M., Cissell M. A., Henderson E., Petersen H. V., Stein R. (2000) Mol. Endocrinol. 14, 1907–1917 [DOI] [PubMed] [Google Scholar]
- 23.Liu A., Desai B. M., Stoffers D. A. (2004) Mol. Cell. Biol. 24, 4372–4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assmann A., Ueki K., Winnay J. N., Kulkarni R. N. (2009) Mol. Biol. Cell 29, 3219–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma S., Jhala U. S., Johnson T., Ferreri K., Leonard J., Montminy M. (1997) Mol. Cell. Biol. 17, 2598–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow E., Lane D. (2006) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27.Shapiro A. M., Hao E., Rajotte R. V., Kneteman N. M. (1996) Cell Transplant. 5, 631–638 [DOI] [PubMed] [Google Scholar]
- 28.Meisenhelder J., Hunter T., van der Geer P. (2001) Current Protocols Protein Science, ( Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. eds) Vol. 2, pp. 13.9.1–13.9.27, John Wiley & Sons, New York: [DOI] [PubMed] [Google Scholar]
- 29.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 30.Parker D., Jhala U. S., Radhakrishnan I., Yaffe M. B., Reyes C., Shulman A. I., Cantley L. C., Wright P. E., Montminy M. (1998) Mol. Cell 2, 353–359 [DOI] [PubMed] [Google Scholar]
- 31.Andrali S. S., Sampley M. L., Vanderford N. L., Ozcan S. (2008) Biochem. J. 415, 1–10 [DOI] [PubMed] [Google Scholar]
- 32.Thomas G. M., Frame S., Goedert M., Nathke I., Polakis P., Cohen P. (1999) FEBS Lett. 458, 247–251 [DOI] [PubMed] [Google Scholar]
- 33.Hani E. H., Stoffers D. A., Chèvre J. C., Durand E., Stanojevic V., Dina C., Habener J. F., Froguel P. (1999) J. Clin. Invest. 104, R41–R48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng J., Macfarlane W. M., Lehto M., Gu H. F., Shepherd L. M., Ivarsson S. A., Wibell L., Smith T., Groop L. C. (2001) Diabetologia 44, 249–258 [DOI] [PubMed] [Google Scholar]
- 35.Guo X., Ramirez A., Waddell D. S., Li Z., Liu X., Wang X. F. (2008) Genes Dev. 22, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang J. (2006) Cell Cycle 5, 2457–2463 [DOI] [PubMed] [Google Scholar]
- 37.Wu R. C., Feng Q., Lonard D. M., O'Malley B. W. (2007) Cell 129, 1125–1140 [DOI] [PubMed] [Google Scholar]
- 38.Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., Hung M. C. (2004) Nat. Cell Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 39.Kitamura T., Nakae J., Kitamura Y., Kido Y., Biggs W. H., 3rd, Wright C. V., White M. F., Arden K. C., Accili D. (2002) J. Clin. Invest. 110, 1839–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eto K., Kaur V., Thomas M. K. (2006) Endocrinology 147, 2923–2935 [DOI] [PubMed] [Google Scholar]
- 41.Arnott D., Gawinowicz M. A., Grant R. A., Neubert T. A., Packman L. C., Speicher K. D., Stone K., Turck C. W. (2003) J. Biomol. Tech. 14, 205–215 [PMC free article] [PubMed] [Google Scholar]
- 42.Boucher M. J., Selander L., Carlsson L., Edlund H. (2006) J. Biol. Chem. 281, 6395–6403 [DOI] [PubMed] [Google Scholar]
- 43.Ohsugi M., Cras-Méneur C., Zhou Y., Warren W., Bernal-Mizrachi E., Permutt M. A. (2004) Diabetes 53, 1496–1508 [DOI] [PubMed] [Google Scholar]
- 44.Da Silva Xavier G., Qian Q., Cullen P. J., Rutter G. A. (2004) Biochem. J. 377, 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivasan S., Bernal-Mizrachi E., Ohsugi M., Permutt M. A. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E784–E793 [DOI] [PubMed] [Google Scholar]
- 46.Liu Z., Tanabe K., Bernal-Mizrachi E., Permutt M. A. (2008) Diabetologia 51, 623–631 [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y., Altman B. J., Coloff J. L., Herman C. E., Jacobs S. R., Wieman H. L., Wofford J. A., Dimascio L. N., Ilkayeva O., Kelekar A., Reya T., Rathmell J. C. (2007) Mol. Cell. Biol. 27, 4328–4339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo S., Burnette R., Zhao L., Vanderford N. L., Poitout V., Hagman D. K., Henderson E., Ozcan S., Wadzinski B. E., Stein R. (2009) J. Biol. Chem. 284, 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han S. I., Aramata S., Yasuda K., Kataoka K. (2007) Mol. Cell. Biol. 27, 6593–6605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ueki K., Okada T., Hu J., Liew C. W., Assmann A., Dahlgren G. M., Peters J. L., Shackman J. G., Zhang M., Artner I., Satin L. S., Stein R., Holzenberger M., Kennedy R. T., Kahn C. R., Kulkarni R. N. (2006) Nat. Genet. 38, 583–588 [DOI] [PubMed] [Google Scholar]
- 51.Tanabe K., Liu Z., Patel S., Doble B. W., Li L., Cras-Méneur C., Martinez S. C., Welling C. M., White M. F., Bernal-Mizrachi E., Woodgett J. R., Permutt M. A. (2008) PLoS Biol. 6, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jhala U. S., Canettieri G., Screaton R. A., Kulkarni R. N., Krajewski S., Reed J., Walker J., Lin X., White M., Montminy M. (2003) Genes Dev. 17, 1575–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garofalo R. S., Orena S. J., Rafidi K., Torchia A. J., Stock J. L., Hildebrandt A. L., Coskran T., Black S. C., Brees D. J., Wicks J. R., McNeish J. D., Coleman K. G. (2003) J. Clin. Invest. 112, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernal-Mizrachi E., Fatrai S., Johnson J. D., Ohsugi M., Otani K., Han Z., Polonsky K. S., Permutt M. A. (2004) J. Clin. Invest. 114, 928–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernal-Mizrachi E., Wen W., Stahlhut S., Welling C. M., Permutt M. A. (2001) J. Clin. Invest. 108, 1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tuttle R. L., Gill N. S., Pugh W., Lee J. P., Koeberlein B., Furth E. E., Polonsky K. S., Naji A., Birnbaum M. J. (2001) Nat. Med 7, 1133–1137 [DOI] [PubMed] [Google Scholar]
- 57.Kushner J. A., Ye J., Schubert M., Burks D. J., Dow M. A., Flint C. L., Dutta S., Wright C. V., Montminy M. R., White M. F. (2002) J. Clin. Invest. 109, 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brissova M., Blaha M., Spear C., Nicholson W., Radhika A., Shiota M., Charron M. J., Wright C. V., Powers A. C. (2005) Am. J. Physiol. Endocrinol. Metab. 288, E707–E714 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.