Abstract
UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase is an α2β2γ2 hexamer that mediates the first step in the synthesis of the mannose 6-phosphate recognition marker on lysosomal acid hydrolases. Using a multifaceted approach, including analysis of acid hydrolase phosphorylation in mice and fibroblasts lacking the γ subunit along with kinetic studies of recombinant α2β2γ2 and α2β2 forms of the transferase, we have explored the function of the α/β and γ subunits. The findings demonstrate that the α/β subunits recognize the protein determinant of acid hydrolases in addition to mediating the catalytic function of the transferase. In mouse brain, the α/β subunits phosphorylate about one-third of the acid hydrolases at close to wild-type levels but require the γ subunit for optimal phosphorylation of the rest of the acid hydrolases. In addition to enhancing the activity of the α/β subunits toward a subset of the acid hydrolases, the γ subunit facilitates the addition of the second GlcNAc-P to high mannose oligosaccharides of these substrates. We postulate that the mannose 6-phosphate receptor homology domain of the γ subunit binds and presents the high mannose glycans of the acceptor to the α/β catalytic site in a favorable manner.
Keywords: Glycoproteins/Biosynthesis, Glycoproteins/Carbohydrates, Glycoproteins/Lysosomal, Phosphorylation/Enzymes, Protein/Post-translational Modification, Subcellular Organelles/Lysosomes
Introduction
In higher eukaryotes, newly synthesized acid hydrolases acquire mannose 6-phosphate (Man-6-P)3 residues on their N-linked glycans as they traverse the Golgi (1). These residues serve as high affinity ligands for binding to Man-6-P receptors in the trans-Golgi network. The hydrolase-receptor complexes are then packaged into transport carriers for delivery to endosomes and lysosomes. The Man-6-P recognition marker is synthesized in two steps. First, UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase (GlcNAc-1-phosphotransferase) binds to a conformation-dependent protein determinant on the acid hydrolase and transfers GlcNAc-1-P from UDP-GlcNAc to one or two of the mannose residues of the N-linked high mannose oligosaccharide. Second, N-acetylglucosamine 1-phosphodiester α-N-acetyglucosaminidase (“uncovering enzyme”) excises the N-acetylglucosamine to generate the Man-6-P monoester.
GlcNAc-1-phosphotransferase is a heterohexamer composed of three subunits (α2β2γ2) (2). The α and β subunits are encoded by a single gene GNPTAB, and the γ subunit is encoded by a separate gene GNPTG (3–5). Although it has been established that the α/β subunits contain the catalytic activity of the enzyme, the possible participation of these subunits in the recognition of the common protein determinant of the acid hydrolases has not been explored. Furthermore, the role(s) of the γ subunit is poorly understood. The initial insight into the function of the subunits came from studies of patients with the autosomal recessive lysosomal storage disorders termed mucolipidosis II (I-cell disease) and mucolipidosis III (pseudo-Hurler polydystrophy), the latter being the less severe of the two (6). These disorders arise from mutations in the genes encoding GlcNAc-1-phosphotransferase, resulting in impaired synthesis of the Man-6-P recognition marker and missorting of the acid hydrolases from the lysosomal pathway to the secretory pathway. Mucolipidosis II and IIIA are caused by mutations in the GNPTAB gene, whereas mucolipidosis IIIC, the mildest of the disorders, arises from mutations in the GNPTG gene (3, 4, 7, 8). Fibroblast extracts of patients with mutations in GNPTAB exhibit either no (ML II) or barely detectable (ML IIIA) GlcNAc- 1-phosphotransferase activity toward acid hydrolase substrates or the simple sugar α-methylmannoside, indicating impaired catalytic function. However, these assays did not address the issue of whether the α/β subunits have a role in the recognition of the protein determinant of the acid hydrolases. The GlcNAc-1-phosphotransferase activity in the fibroblasts of patients with GNPTG mutations (ML IIIC) had impaired activity toward two acid hydrolase substrates but normal activity toward α-methylmannoside (3, 9). These results indicated that the γ subunit has a role in facilitating the phosphorylation of protein substrates, but it is not required for the catalytic activity toward α-methylmannoside.
It has recently been reported that mice with disruption of the Gnptab gene exhibit many of the characteristic findings of patients with ML II, whereas mice with inactivation of the Gnptg gene have a much milder phenotype (10, 11). Biochemical analysis showed that loss of the Gnptab gene products totally abolished the formation of the Man-6-P recognition marker on the acid hydrolases, whereas loss of the γ subunit decreased phosphorylation of the hydrolases to a variable extent, but in no instance was phosphorylation completely lost (12). These findings confirm that the α/β subunits mediate the catalytic function of GlcNAc-1-phosphotransferase, whereas the γ subunit serves to enhance the phosphorylation of glycoprotein substrates.
Although these studies have provided important insight into the roles of the subunits of GlcNAc-1-phosphotransferase, several fundamental issues remain unresolved. First, do the α/β subunits have a role in the recognition of the protein determinant of acid hydrolase substrates in addition to mediating the catalytic function of the transferase? Second, how does the γ subunit enhance phosphorylation of acid hydrolases? Does it participate in the recognition of the protein determinant of the substrates? Does it bind the high mannose oligosaccharides of these substrates and present them in a favorable manner to the catalytic site on the α/β subunits? Does it do both? The γ subunit contains a mannose 6-phosphate receptor homology (MRH) domain that in other proteins has been shown to bind to high mannose oligosaccharides (13). In the case of the endoplasmic reticulum-localized glucosidase II, the MRH domain on the β subunit is required for the catalytic activity of the α subunit toward glycoprotein substrates (14, 15). The γ subunit of GlcNAc-1-phosphotransferase could potentially have a similar function.
In this study, we have taken a multifaceted approach to answer these questions. We have used mice lacking the γ subunit to determine the extent of phosphorylation of almost all the acid hydrolases relative to that observed in wild-type animals. This provides information about the variable requirement of the γ subunit for acid hydrolase phosphorylation. Fibroblasts from these mice were then used to examine in more detail the phosphorylation of the glycans of DNase I. These in vivo studies have been complemented by in vitro studies using recombinant α2β2γ2 and α2β2 forms of human GlcNAc-1- phosphotransferase. The findings demonstrate that the α/β subunits do recognize the protein determinant of the acid hydrolases in addition to mediating the catalytic function of the transferase. The γ subunit increases the activity of the α/β subunits toward protein acceptors to variable extents. In addition, it facilitates the addition of the second GlcNAc-P to high mannose oligosaccharides of protein substrates. A model that attempts to integrate these findings is proposed.
EXPERIMENTAL PROCEDURES
Materials
UDP-[6-3H]GlcNAc (20 Ci/mmol) was purchased from PerkinElmer Life Sciences. [β-32P]UDP-GlcNAc was prepared as described previously (2). [2-3H]Mannose was purchased from MP Biomedicals (Irvine, CA). Concanavalin A-Sepharose was obtained from GE Healthcare; protein A-agarose was from Repligen (Waltham, MA), and α-methylmannoside and QAE-Sephadex were from Sigma. Recombinant endoglycosidase H fused to maltose-binding protein was from New England Biolabs (Ipswich, MA). Calf intestinal phosphatase was from Fisher. Sulfo-N-hydroxysuccinimide acetate (SNA) was from Pierce. Soluble CI-MPR was purified from fetal calf serum and immobilized as reported previously (16). All other reagents were of the highest quality available and were purchased from Sigma or Fisher. Rabbit anti-bovine DNase I serum was prepared as described previously (17).
Protein/Oligosaccharide Acceptors
Cathepsin D, uteroferrin, ribonuclease B, and soybean agglutinin were purified as described previously (18). The Man9GlcNAc1 oligosaccharide was released from the soybean agglutinin with endoglycosidase H according to manufacturer's protocol. The released Man9GlcNAc1 was desalted on a Sephadex G-15 column, lyophilized, and resuspended in distilled water. The concentration of the oligosaccharide was determined by the phenolsulfuric acid assay for hexoses (19) using mannose as a standard.
The production of recombinant human NPC2 (20) and tripeptidyl peptidase 1 (21) has been described previously. The NPC2 was dephosphorylated using calf intestinal phosphatase according to the protocol of the manufacturer. Following an overnight incubation at room temperature, the sample was dialyzed against 50 mm Tris-HCl, pH 7.4, overnight at 4 °C.
Bovine pancreatic DNase I, obtained from Sigma, was further purified and enriched for species with high mannose glycans as follows. The material was dissolved in 10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm MnCl2, 1 mm CaCl2 and applied to a column of concanavalin A-Sepharose. The column was washed with 30 ml of the same buffer and eluted with 30 ml of the buffer containing 100 mm α-methylmannoside. The fractions containing the DNase I in the α-methylmannoside eluate were pooled and dialyzed against 50 mm Tris-HCl, pH 7.4, overnight at 4 °C.
Preparation of Deglycosylated Cathepsin D
Cathepsin D (0.5 mg) was incubated with 5000 units of endoglycosidase H in G5 reaction buffer (50 mm sodium citrate, pH 5.5) in a final volume of 70 μl for 1 h at 37 °C. The reaction mixture was loaded on a 1-ml QAE-Sephadex column equilibrated in 2 mm Tris base. The column was washed three times with 1 ml of 2 mm Tris base, and the deglycosylated cathepsin D was eluted five times with 4 ml of 10 mm NaCl in 2 mm Tris base. Each fraction was concentrated using Microcon Centriplus concentrator (Millipore, Billerica, MA), and aliquots were analyzed by 12% SDS-PAGE. The fractions with deglycosylated cathepsin D were pooled and dialyzed against 50 mm Tris-HCl, pH 7.4, at 4 °C overnight. The pooled fractions were free of endoglycosidase H.
Acetylation of Protein Acceptors
All acceptor glycoproteins were dialyzed against phosphate-buffered saline at 4 °C overnight prior to acetylation. SNA was added at a 75-fold molar excess (calculated on basis of lysine content) followed by incubation at 4 °C for 90 min. The reaction mixture was dialyzed against 50 mm Tris-HCl, pH 7.4, at 4 °C overnight to remove unreacted SNA.
Recombinant α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase
Recombinant soluble forms of human α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase were expressed in stably transfected CHO-K1 cells and purified as described previously (22). The molecular masses for α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase are 330 and 270 kDa, respectively, because both enzymes are missing the transmembrane segments.
GlcNAc-1-phosphotransferase Assay
The reaction mixture contained various concentrations of the substrates and either 1.2 μg of the α2β2γ2 or 0.97 μg of the α2β2 GlcNAc-1-phosphotransferase in 50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 10 mm MnCl2, 75 μm UDP-[3H]GlcNAc (1 μCi), 2 mg/ml bovine serum albumin in a final volume of 50 μl. Incubations were for 1 h at 37 °C. For glycoprotein substrates, the reactions were terminated by addition of 300 μl of 1.5% phosphotungstic acid, 0.75 n HCl, and 100 μl of 1 mg/ml bovine serum albumin. Samples were vortexed and centrifuged at 14,000 rpm for 10 min at 4 °C. The pellets were washed three times with 1 ml of the acid mixture and resuspended in 1 ml of 50 mm Tris-HCl, pH 11. The incorporated [3H]GlcNAc-P was determined following addition of 10 ml of EcoLite scintillation fluid (MP Biomedicals Inc., Irvine, CA). For α-methylmannoside and Man9GlcNAc1 substrates, reactions were terminated by addition of 1 ml of 5 mm EDTA, pH 8.0. The sample was applied to a 1-ml column of QAE-Sephadex equilibrated with 2 mm Tris base. The column was washed with 1 ml of 2 mm Tris base five times, and the phosphorylated products were eluted with 1 ml of 30 mm NaCl in 2 mm Tris base five times. The radioactivity in all the fractions was determined by scintillation counting. In the experiments examining the transfer of 1 or 2 mol of GlcNAc-1 to the Man9GlcNAc1 acceptor, the column was eluted with 30 mm NaCl in 2 mm Tris base followed by 70 mm NaCl in Tris base (to elute molecules with two GlcNAc-P moieties).
Mice
Mice with a mutation in the Gnptab gene (GenBankTM accession number AK173132) were obtained from the OMNIBANK gene trap library as described previously (10). Mice with a mutation in the Gnptg gene (GenBankTM accession number AK078230) were obtained with a directed gene deletion approach that deleted exons 4–11 of the gene (12). The complete characterization of the mice is presented elsewhere (11). All experiments and procedures involving the mice were conducted in compliance with approved Institutional Animal Care and Use Committee guidelines. Skin fibroblasts were prepared from wild-type and mutant mice and maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 μg/ml penicillin, and 100 units/ml streptomycin.
MPR Affinity Purification and Analysis
Brains from four mice of a given genotype were combined to yield 1.3 g of total tissue wet weight. Each of the three genotypes were processed in parallel essentially as described previously (23). Tissue was diluted into 9 volumes (w/v) of homogenization buffer (20 mm sodium phosphate, pH 6.8, 150 mm NaCl, 2.5 mm EDTA, 5 mm β-glycerophosphate, 1 μg/ml leupeptin, 1 μg/ml pepstatin A) containing 1 mm Pefabloc and 1% Triton X-100 and disrupted using a Brinkmann Polytron. The homogenate was centrifuged at 25,000 × g at 4 °C for 1 h, and the supernatant was saved. The pellet was re-homogenized in 4.5 volumes of the same buffer, and centrifugation was repeated. Supernatants were combined and loaded onto a 2-ml bed volume column of immobilized CI-MPR (2.3 mg/ml). The column was sequentially washed with 5 column volumes of homogenization buffer containing 1% Triton X-100, 5 column volumes of homogenization buffer without Triton X-100, and 5 column volumes of homogenization buffer containing 10 mm glucose 6-phosphate. Man-6-P-containing glycoproteins were then specifically eluted with 5 column volumes of homogenization buffer containing 10 mm Man-6-P. Total protein in the Man-6-P eluates were 70.1, 13.7, and 33.2 μg for the wild-type, α/β-, and γ-knock-out samples, respectively.
Lysosomal enzyme activity assays were conducted to determine the fraction of material specifically eluted in a Man-6-P-dependent manner (Man-6-P eluate divided by the sum of the flow-through, washes, glucose 6-phosphate, and Man-6-P eluates). The reaction conditions for β-galactosidase, β-glucuronidase, β-hexosaminidase, β-mannosidase, α-fucosidase, and α-mannosidase were as described previously (16). Tripeptidyl peptidase 1 was assayed with 0.25 mm Ala-Ala-Phe 7-amido-4-methylcoumarin in 100 mm acetate buffer, 150 mm NaCl, 0.1% Triton X-100, pH 4.5, using the previously described end point assay (25). α-Glucosidase was assayed with 1 mm 4-methylumbelliferyl-α-d-glucoside in 100 mm acetate buffer, 150 mm NaCl, 0.1% Triton X-100, pH 4.0. β-Glucosidase was assayed with 5 mm 4-methylumbelliferyl-β-d-glucoside in 200 mm acetate buffer, 150 mm NaCl, 0.1% Triton X-100, pH 5.0, containing 0.25% sodium taurocholate. Recovery (sum of all column fractions divided by load) was essentially ∼100% for all activities except for tripeptidyl peptidase 1. The recovery of the latter enzyme was ∼50% and can be attributed to the processed form of tripeptidyl peptidase 1 tightly adhering to the column and requiring glycine for complete elution (23).
Column fractions eluted with Man-6-P were concentrated and buffer exchanged to 100 mm ammonium bicarbonate using ultrafiltration (Ultra-4, Millipore). Protein concentration was determined using the Bradford method with bovine serum albumin as a standard (26). Aliquots were stored at −80 °C until use. Equivalent amounts of soluble extracts (40 μg) or Man-6-P-eluted column fractions (200 ng) for each genotype were fractionated by SDS-PAGE and transferred to nitrocellulose. Man-6-P-containing glycoproteins were detected using 125 I-labeled CI-MPR as described previously (27).
Mass spectrometry and spectral count analysis were as described previously (23) with minor modifications. Tryptic digests of reduced, carboxyamidomethylated proteins eluted from the affinity columns with Man-6-P were analyzed by nanospray LC-MS/MS using an LTQ linear ion trap mass spectrometer (Thermo Electron). LC-MS/MS runs (0.5 μg of sample/LC run) were conducted for each sample type using a gradient of 2–45% acetonitrile in 0.1% formic acid in 135 min. Three replicate runs for each sample were used for subsequent data base searching. Spectra were assigned to proteins in the ENSEMBL NCBIm37.55 protein data base and a list of contaminants commonly associated with proteomics experiments using the X!Tandem search engine (28) (local implementation of gpm-xe-tornado-20090501-win64).
Retroviral Vector
To prepare the bovine DNase I expression retroviral vector, two different BstXI restriction sites were introduced into bovine DNase I cDNA in pSVK3 expression vector by PCR (29). The sequence of N-terminal BstXI was CCAGTGTGCTGG and that of C-terminal BstXI was CCATCACACTGG. ΔU3 nlsLacZ was kindly provided by D. Ory (see Ref. 30). The nlsLac region of ΔU3 nlsLacZ construct was replaced with BstXI fragment containing bovine DNase I cDNA.
Virus Production
To produce the virus expressing bovine DNase I, transient transfections were accomplished with 293GPG cells in 100-mm plates. The day before the transfection, 1.3 × 107 293GPG cells were plated. Eleven μg of DNA were mixed with 42.7 μl of PLUS reagent (Invitrogen) and diluted with DMEM to give the final volume of 400 μl. After 15 min of incubation at room temperature, 32 μl of Lipofectamine diluted into 368 μl of DMEM was mixed with the PLUS/DNA mixture and incubated another 15 min at room temperature. The PLUS-DNA-Lipofectamine complex was layered on the top of the 293GPG cells that had been washed with DMEM and placed with 3.2 ml of DMEM. Seven hours post-transfection, 4 ml of DMEM containing 20% fetal calf serum was added. The next day, the media were replaced with 8 ml of DMEM containing 10% fetal calf serum, and the viral supernatant was harvested at 96 h.
Assay for Virus Activity
To determine viral activity, cells were stained for β-galactosidase activity (Mirus Bio Corp., Madison, WI) and Western blot analysis was accomplished. To see the transduction efficiency of β-galactosidase, 2 × 106 cells were placed in 6-well plates the day before infection. The cells were treated with 2 ml of viral supernatant for 7–8 h and replaced with fresh DMEM containing 10% fetal calf serum. The next day, cells were harvested, resuspended with 0.1 m Tris buffer containing 150 mm NaCl, 2 mm EDTA, and 1% Triton X-100, sonicated, and spun at 16,000 rpm for 10 min. Sixty μg of the supernatant was loaded on 13% SDS-polyacrylamide gel and probed with polyclonal anti-bovine DNase I antibody in nonreducing condition.
Labeling of Mouse Fibroblasts
Mouse fibroblasts infected with wild-type or mutated bovine DNase I viral supernatant in 60-mm plates were labeled 4 h with 1 ml of DMEM containing 5 mm glucose, 10% dialyzed fetal calf serum, 10 mm NH4Cl, and 200 μCi [2-3H]mannose and chased 4 h by addition of 300 μl of chase medium containing 10 mm glucose and 10 mm mannose to stop mannose uptake (31).
Immunoprecipitation and Oligosaccharide Analysis
Bovine DNase I in the media was immunoprecipitated with a polyclonal anti-bovine DNase I antibody. The immunoprecipitates eluted from protein A-Sepharose beads were treated with endoglycosidase H, and oligosaccharide analysis was carried out according to the previously described Experimental Procedures in Ref. 32.
RESULTS
Phosphorylation of Brain Acid Hydrolases in the Absence of the γ Subunit
In our previous study we passed brain extracts from wild-type and the α/β and γ gene knock-out mice over a CI-MPR affinity column and followed the binding and Man-6-P specific elution of six acid hydrolases as a measure of their degree of phosphorylation (12). Brains were utilized in these experiments because brain cells dephosphorylate the Man-6-P-containing oligosaccharides of the acid hydrolases much slower than occurs in other tissues (16). We found that 41–82% of each acid hydrolase of wild-type brain bound specifically to the affinity column (the exception was acid β-glucosidase, which is known to lack Man-6-P residues), whereas 6–42% of the same acid hydrolases of the γ gene knock-out samples bound and less than 2% of these enzymes from the α/β gene knock-out brain bound. These results illustrate the usefulness of this approach for giving an overview of acid hydrolase phosphorylation in the mutant mice. However, the limitation of the procedure is the need to have accurate activity assays for the acid hydrolases that total over 50. Therefore, we turned to a technique that uses LC-MS/MS to simultaneously identify and quantitate the amount of all the acid hydrolases in the various fractions of the affinity columns.
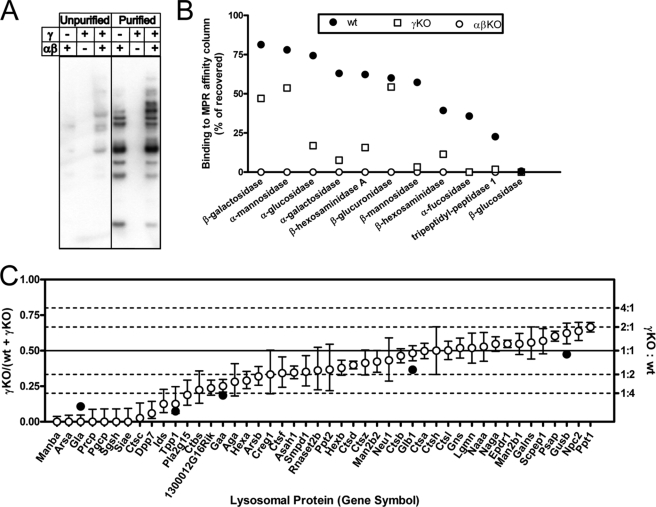
Equal aliquots of wild-type and γ gene knock-out brain homogenates were applied to a column containing immobilized soluble CI-MPR. After extensive washing, the column was mock-eluted with 10 mm glucose 6-phosphate and then specifically eluted with 10 mm Man-6-P. For a preliminary analysis, aliquots of the starting material and the Man-6-P eluates were analyzed by Western blotting using 125I-labeled soluble CI-MPR as the probe. Fig. 1A shows that the soluble CI-MPR affinity column was enriched for the Man-6-P-containing glycoproteins in both the wild-type and γ-KO extracts. Most of the bands detected in wild-type brain were also present in the γ-KO brains at comparable intensities, indicating substantial phosphorylation of the acid hydrolases in these mice. The brain of mice lacking the α/β subunits of GlcNAc-1-phosphotransferase served as a control for nonspecific binding and elution with Man-6-P. This sample had no detectable signal in the Western blot. The fractions were next assayed for a number of acid hydrolase activities to determine the percent of each enzyme that bound to the column and was specifically eluted with Man-6-P (Fig. 1B). With the wild-type extract, 20–80% of the various hydrolases bound specifically to the affinity column with the exception of acid β-glucosidase. Likewise, other than acid β-glucosidase, all the acid hydrolases in the γ gene knock-out sample bound specifically to the affinity column, although the extent of the binding was lower (2–55%). As found in the Western blot assay, none of the acid hydrolases of the α/β gene knock-out sample bound detectably to the column. These results are mostly in line with our previous analysis involving assays of only five acid hydrolases plus acid β-glucosidase (12).
FIGURE 1.
Analysis of acid hydrolase phosphorylation in wild-type and phosphotransferase-deficient mouse brain. A, Man-6-P-containing glycoproteins in unpurified brain homogenates or in affinity column Man-6-P eluates. B, analysis of binding to CI-MPR column using enzyme activity assays. C, comparison of relative levels of Man-6-P-containing lysosomal proteins in wild-type and γ-knock out samples. Open circles and error bars represent the point estimate and 95% confidence intervals for the proportion of each indicated gene product determined by LC-MS/MS analysis and spectral counting of the affinity-purified samples. Only proteins of interest with ≥30 spectral counts that have been classified as lysosomal Man-6-P-containing glycoproteins or likely candidates are shown (35). Data for all proteins identified and statistical analyses are presented in supplemental Table 1. Filled circles represent the relative proportion (% bound for γ-KO/(% bound for wild type + % bound for γ-KO)) for select enzymes in B that can be attributed to a given gene product. Note that equivalent amounts of affinity-purified samples were used for spectral count analysis, yet the yield of protein eluted from the column was ∼2-fold greater for the wild-type compared with the γ-knock-out sample. Thus, the spectral count analysis overestimates (or represents an upper limit for) the relative amount of a given protein in the γ-knock-out sample.
The Man-6-P eluates from the wild-type- and γ-deficient brain samples were then digested with trypsin and subjected to LC-MS/MS. These data were analyzed using the spectral count method (33). The open circles in Fig. 1C show the proportion of spectral counts of γ-KO/wild type + γ-KO with upper and lower 95% confidence intervals for 46 identified acid hydrolases. The ratio of spectral contents of the two samples is indicated in the right axis of Fig. 1C. This agrees well with the trend found in measuring the relative amount of a given lysosomal protein retained on the CI-MPR column as determined by enzyme activity assays (Fig. 1C, filled circles). It is apparent that about one-third of the acid hydrolases of the γ-deficient brain were phosphorylated at levels equivalent to that in wild-type brain, although about 25% were very poorly phosphorylated compared with their wild-type counterparts. The rest of the acid hydrolases of the γ-deficient sample were phosphorylated at an intermediate level. These data clearly show that some acid hydrolases are highly dependent on the presence of the γ subunit to acquire the Man-6-P, although others are well phosphorylated by the α/β subunits alone.
Activity of α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase toward Substrates in Vitro
To complement the analysis of acid hydrolase phosphorylation in brain tissue, we determined the ability of recombinant human α2β2γ2 and α2β2 GlcNAc-1- phosphotransferase to phosphorylate a variety of high mannose-containing glycoproteins, including acid hydrolases, a Man9GlcNAc1 oligosaccharide, and the simple sugar α-methylmannoside. The recombinant α2β2γ2 GlcNAc-1-phosphotransferase was prepared as described previously (22), whereas the α2β2 species was prepared in a similar manner except that the CHO-K1 cells were transfected with the α/β cDNA alone. Analysis of the purified α2β2 product by SDS-PAGE established that it lacked the γ subunit (data not shown).
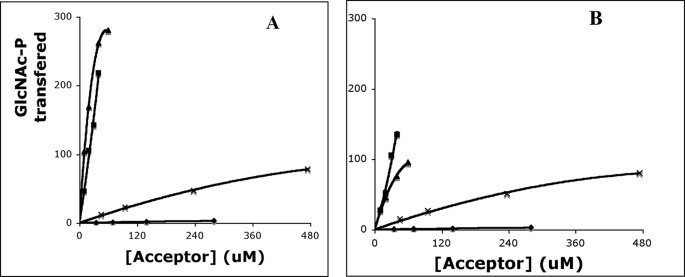
In these experiments, equivalent amounts of recombinant α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase were incubated with 75 μm UDP-[3H]GlcNAc and various concentrations of the substrates for 1 h, and the transfer of [3H]GlcNAc-P to the acceptors was measured as described under “Experimental Procedures.” Representative examples of these data are shown in Fig. 2, and the apparent Km and kcat values derived from the data are summarized in Table 1, along with the relative catalytic efficiency (kcat divided by apparent Km) with the values normalized to α-methylmannoside.
FIGURE 2.
GlcNAc-1-phosphotransferase activity toward various acceptors. Cathepsin D (▴), NPC2 (■), RNase B (×), and soybean agglutinin (♦) were compared as acceptors for α2β2γ2 (A) and α2β2 forms (B) of GlcNAc-1-phosphotransferase. Various concentrations of the substrates and either 1.2 μg of the α2β2γ2 or 0.97 μg of the α2β2 GlcNAc-1-phosphotransferase were incubated in 50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 10 mm MnCl2, 75 μm UDP-[3H]GlcNAc (1 μCi), 2 mg/ml bovine serum albumin in a final volume of 50 μl for 1 h at 37 °C. The incorporated [3H]GlcNAc-P was determined by scintillation counting. The activity of the GlcNAc-1-phosphotransferase was expressed as moles of [3H]GlcNAc-P transferred per h/mol of the GlcNAc-1-phosphotransferase.
TABLE 1.
Kinetic parameters of α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase toward various acceptors
The acceptors were incubated with the two forms of the GlcNAc-1-phosphotransferase under identical conditions, and the products were isolated as described under “Experimental Procedures.” The Man9GlcNAc1 was derived from soybean agglutinin by endoglycosidase H treatment. Apparent Km and kcat values were generated from double-reciprocal plots using a least square approximation for the best fit line. The values are the average of two to five separate determinations.
| Acceptor |
Km,app |
kcat |
kcat/Km,app |
Relative catalytic efficiency |
||||
|---|---|---|---|---|---|---|---|---|
| αβγ | αβ | αβγ | αβ | αβγ | αβ | αβγ | αβ | |
| μm | min−1 | μm−1min−1 | ||||||
| α-Methylmannoside | 48,000 | 33,000 | 45.1 | 30.2 | 0.001 | 0.001 | 1 | 1 |
| Man9GlcNAc1 | 5700 | 3300 | 93.5 | 45 | 0.016 | 0.014 | 16 | 14 |
| Soybean agglutinin | 1000 | 1000 | 0.30 | 0.25 | 0.0003 | 0.0003 | 0.3 | 0.3 |
| RNase B | 556 | 500 | 3.08 | 2.52 | 0.0055 | 0.005 | 6 | 5 |
| DNase I | 30 | 60 | 0.27 | 0.30 | 0.009 | 0.005 | 9 | 5 |
| Pro-tripeptidyl peptidase | 110 | 110 | 2.32 | 0.53 | 0.022 | 0.0045 | 22 | 5 |
| Cathepsin D | 25 | 25 | 6.6 | 1.8 | 0.267 | 0.072 | 267 | 72 |
| Uteroferrin | 33 | 25 | 1.98 | 0.68 | 0.06 | 0.027 | 60 | 27 |
| NPC2 | 367 | 352 | 37.4 | 26.55 | 0.102 | 0.075 | 102 | 75 |
Both forms of the GlcNAc-1-phosphotransferase exhibited similar catalytic efficiencies toward α-methylmannoside, the Man9GlcNAc1 oligosaccharide, and the two nonlysosomal glycoproteins, soybean agglutinin and RNase B. Man9GlcNAc1 and RNase B are better substrates than α-methylmannoside because each contains a Manα1,2 Man sequence in its high mannose unit that is preferred by the transferase (22). Interestingly, the kcat value for the Man9GlcNAc1 oligosaccharide is much greater than that obtained with soybean agglutinin from which it was derived. This may indicate that the high mannose glycan is less accessible on the intact protein. However, all these substrates had a relatively poor Km,app compared with the values obtained with the acid hydrolase substrates (with the exception of NPC2), reflecting the lack of the conformation-dependent protein recognition site. These data show that the γ subunit does not enhance the ability of the α/β subunits to phosphorylate high mannose oligosaccharides that are either in solution or covalently bound to nonlysosomal glycoproteins.
In contrast, substantial differences were observed in the ability of the two forms of GlcNAc-1-phosphotransferase to phosphorylate acid hydrolase substrates. Although both enzymes had similar apparent Km values toward pro-tripeptidyl peptidase 1, cathepsin D, uteroferrin, and NPC2, the catalytic efficiencies of the α2β2 transferase were decreased by 78, 73, 55, and 25%, respectively, compared with the values obtained with the α2β2γ2 enzyme. Thus, as found in the analysis of acid hydrolase phosphorylation in the brain of γ gene knock-out mice, the requirement for the γ subunit to achieve efficient phosphorylation differs among the various acid hydrolases. Furthermore, the relative efficiency of phosphorylation of tripeptidyl peptidase 1, cathepsin D, and NPC2 in the brain and in the in vitro assays was similar, indicating that the in vitro studies are a good reflection of what is occurring in vivo.
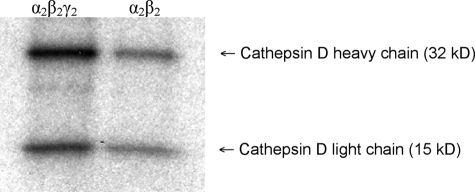
One potential explanation for the decreased phosphorylation of the acid hydrolases by the α2β2 enzyme is that this form of the transferase is unable to phosphorylate specific glycans that are acted upon by the α2β2γ2 enzyme. To explore this possibility, the two forms of the transferase were incubated with cathepsin D in the presence of [β-32P]UDP-GlcNAc. Cathepsin D contains two high mannose glycans, one on the heavy chain and the other on the light chain. Following the incubation, the labeled cathepsin D was subjected to SDS-PAGE and autoradiography. Fig. 3 shows that both forms of the transferase acted upon each glycan of cathepsin D, with the α2β2 transferase exhibiting decreased activity toward both glycans compared with that of the α2β2γ2 enzyme. Thus, at least in the case of cathepsin D, the decreased rate of transfer of GlcNAc-P to its glycans by the α2β2 enzyme cannot be accounted for by an inability to phosphorylate glycans located at certain positions on the protein backbone.
FIGURE 3.
Phosphorylation of individual glycosylation sites of cathepsin D. α2β2γ2 (left) and α2β2 forms (right) of GlcNAc-1-phosphotransferase (2.5 μg of each) were incubated with 5 μm cathepsin D in the presence of 75 μm [β-32P]UDP-GlcNAc (10 μCi) in a volume of 50 μl. After 30 min, the reaction mixtures were diluted to 500 μl of Tris-buffered saline, 1% Triton X-100 followed by immunoprecipitation with anti-cathepsin D antibody, SDS-PAGE, and autoradiography. The upper band represents the cathepsin D heavy chain, and the lower band represents the light chain.
We next asked whether the two forms of GlcNAc-1-phosphotransferase differ in their ability to add a second GlcNAc-P to the high mannose glycans present on the acid hydrolases. For this experiment, NPC2 that had been treated with alkaline phosphatase to remove any pre-existing Man-6-P residues was utilized as the acceptor. Following incubation with UDP-[3H]GlcNAc and the two transferases, the high mannose N-linked glycans were released from the NPC2 with endoglycosidase H and fractionated on QAE-Sepharose to separate the oligosaccharides with one or two [3H]GlcNAc-P residues. Table 2 shows the results of two separate experiments. As in the kinetic experiment, the α2β2γ2 GlcNAc-1-phosphotransferase transferred more [3H]GlcNAc-P to the acceptor than did the enzyme lacking the γ subunit. More strikingly, the α2β2γ2 transferase synthesized substantially more of the diphosphorylated oligosaccharide (average 2Pi/1Pi ratio of 1.1 in α2β2γ2 transferase product versus 0.61 in the α2β2 transferase product). This indicates that the γ subunit facilitates the addition of the second GlcNAc-P to the oligosaccharides of acceptor proteins.
TABLE 2.
Effect of the γ subunit on addition of second GlcNAc-P to NPC2 glycans
The assays contained either 1.2 μg of α2β2γ2 or 0.97 μg of α2β2 GlcNAc-1- phosphotransferase and 20 μm NPC2 that had been treated with intestinal alkaline phosphatase to remove pre-existing Man-6-P residues. After a 1-h incubation, the NPC2 was precipitated and treated with endoglycosidase H to release its high mannose glycans that were fractionated on QAE-Sephadex into oligosaccharides with one or two GlcNAc-P-man residues as described under “Experimental Procedures.” The ratio of these two species was calculated. The data are from two independent experiments.
| Exp. | α2β2γ2 |
α2β2 |
||
|---|---|---|---|---|
| GlcNAc-P transferred | Ratio 2Pi/1Pi | GlcNAc-P transferred | Ratio 2Pi/1Pi | |
| pmol | pmol | |||
| 1 | 179 | 0.91 | 110 | 0.63 |
| 2 | 146 | 1.25 | 83 | 0.59 |
In addition to the known acid hydrolases, bovine pancreatic DNase I was tested as a substrate because it has been shown to be phosphorylated by GlcNAc-1-phosphotransferase in vivo (34). It interacted with both forms of the transferase with good affinity (Km,app of 30 μm for α2β2γ2 and 60 μm for α2β2) and similar kcat values, although these values were considerably lower than those obtained with the acid hydrolases (Table 1).
Taken together, these data provide strong evidence that the α/β subunits have the ability to recognize the common protein determinant expressed on acid hydrolases, although the γ subunit enhances the rate of GlcNAc-P transfer to the high mannose glycans of a subset of these acceptors.
Deglycosylated Cathepsin D Inhibits the Activity of Both Forms of GlcNAc-1-phosphotransferase toward Cathepsin D
Previously, we reported that deglycosylated acid hydrolases are specific inhibitors of the phosphorylation of intact acid hydrolases in in vitro assays, as would be expected if the protein portion of the hydrolases contains a recognition determinant that mediates high affinity binding to GlcNAc-1-phosphotransferase (18). Table 3 shows that deglycosylated cathepsin D inhibits the phosphorylation of intact cathepsin D by both α2β2γ2 and α2β2 GlcNAc-1-phosphotransferase to the same extent, providing additional support for the conclusion that the α2β2 species binds the protein recognition domain of the acid hydrolases. The deglycosylated cathepsin D was a very poor acceptor indicating that the removal of its N-linked oligosaccharides by endoglycosidase H had been complete (data not shown).
TABLE 3.
Effect of deglycosylated cathepsin D on GlcNAc-1-phosphotransferase
The assays contained either 1.2 μg of α2β2γ2 or 0.97 μg of α2β2 GlcNAc-1- phosphotransferase and the indicated amounts of cathepsin D, deglycosylated cathepsin D, or α-methylmannoside under standard assay conditions. The cathepsin D was deglycosylated by endoglycosidase H treatment as described under “Experimental Procedures.”
| Enzyme source | Acceptor |
GlcNAc-P transferred |
|||
|---|---|---|---|---|---|
| Cathepsin D (10 μm) | α-Methylmannoside (20 mm) | Deglycosylated cathepsin D (56 μm) | pmol | +dG CD/−dG CDa | |
| α2β2γ2 | + | − | − | 180 | 0.40 |
| + | − | + | 72 | ||
| − | + | − | 276 | 2.22 | |
| − | + | + | 612 | ||
| α2β2 | + | − | − | 116 | 0.41 |
| + | − | + | 48 | ||
| − | + | − | 97 | 3.20 | |
| − | + | + | 310 | ||
a dG means deglycosylated cathepsin D.
In contrast to its effect on the phosphorylation of intact cathepsin D, the deglycosylated cathepsin D stimulated the phosphorylation of α-methylmannoside by both forms of the transferase (Table 3). A similar effect was obtained with DNase I. Because DNase I interacts with α2β2γ2 and α2β2 GlcNAc-1- phosphotransferase with high affinity (Km,app values of 30 and 60 μm, respectively) but is poorly phosphorylated, we tested the intact molecule in the assay. The workup of the α-methylmannoside-P-GlcNAc product removes any phosphorylated DNase I that may be produced. Table 4 demonstrates that 16 μm DNase I strongly stimulated the phosphorylation of α-methylmannoside by both of the transferases. The DNase I also stimulated the phosphorylation of the Man9GlcNAc1 oligosaccharide by 6-fold (data not shown). Thus, binding of the protein determinant of acid hydrolases to the α2β2 subunits (or the α2β2γ2 subunits) of GlcNAc-1-phosphotransferase appears to induce a conformational change that enhances the catalytic activity toward the sugar acceptor.
TABLE 4.
Effect of DNase I on α2β2γ2 and α2β2 forms of GlcNAc-1-phosphotransferase toward α-methylmannoside
The assays contained either 1.2 μg of α2β2γ2 or 0.97 μg of α2β2 GlcNAc-1- phosphotransferase and the indicated amounts of α-methylmannoside and DNase I under standard assay conditions.
| Enzyme source | Acceptor |
GlcNAc-P transferred |
||
|---|---|---|---|---|
| α-Methylmannoside | DNase I (16 μm) | pmol | + DNase I/− DNase I | |
| α2β2γ2 | + (4 mm) | − | 722 | 2.34 |
| + (4 mm) | + | 1688 | ||
| + (8 mm) | − | 1449 | 3.23 | |
| + (8 mm) | + | 4678 | ||
| α2β2 | + (4 mm) | − | 220 | 3.14 |
| + (4 mm) | + | 690 | ||
| + (8 mm) | − | 417 | 4.59 | |
| + (8 mm) | + | 1916 | ||
Effect of Lysine Modification on GlcNAc-1-phosphotransferase Activity
A number of reports have established that specific lysine residues on acid hydrolases facilitate the phosphorylation of these proteins (17, 31, 36–41). Although most of these studies have involved transfecting acid hydrolase cDNAs with lysine modifications into tissue culture cells, two studies showed that acetylation of lysines on cathepsin L and cathepsin D with SNA caused a drastic loss of acceptor activity in an in vitro assay utilizing partially purified rat liver GlcNAc-1- phosphotransferase (37, 38).
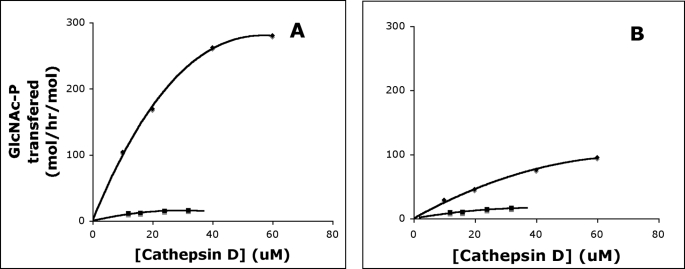
Because the α/β subunits of GlcNAc-1-phosphotransferase recognize the conformation-dependent protein determinant on the acid hydrolase substrates, we asked whether these subunits interact with the critical lysines or, alternatively, whether this is a function of the γ subunit. Fig. 4 shows that the activity of both α2β2γ2 and α2β2 GlcNAc-1-phosphotransferases toward cathepsin D is greatly decreased following acetylation of the lysines on this substrate. Double-reciprocal plots of these data reveal that the lysine modification has little or no effect on the apparent Km values for the acid hydrolase, while causing a major decrease in the kcat values (Table 5). These results clearly show that the α/β subunits respond to the lysine modification in the absence of the γ subunit. Similar results were obtained following acetylation of NPC2 (Table 5).
FIGURE 4.
Effect of lysine modification on cathepsin D phosphorylation. α2β2γ2 (A) and α2β2 forms (B) of GlcNAc-1-phosphotransferase were incubated with increasing concentrations of cathepsin D (♦) and SNA-treated cathepsin D (■) in 50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 10 mm MnCl2, 75 μm UDP-[3H]GlcNAc (1 μCi), 2 mg/ml bovine serum albumin in a final volume of 50 μl for 1 h at 37 °C. The incorporated [3H]GlcNAc-P was determined by scintillation counting. The activity of the GlcNAc-1-phosphotransferase was expressed as moles of [3H]GlcNAc-P transferred per h/mol of the GlcNAc-1-phosphotransferase.
TABLE 5.
Effect of substrate acetylation on GlcNAc-1 phosphotransferase activity
Aliquots of the various acceptors were treated with a 75-fold excess of SNA (based on lysine content) as described under “Experimental Procedures.” Control and SNA-treated acceptors were incubated with the two forms of GlcNAc-1-phosphotransferase under standard assay conditions. The apparent Km and kcat values were generated from double-reciprocal plots using a least square approximation for the best fit line.
| Acceptor | α2β2γ2 |
α2β2 |
||||||
|---|---|---|---|---|---|---|---|---|
|
Km,app |
kcat |
Km,app |
kcat |
|||||
| Control | Acetylated | Control | Acetylated | Control | Acetylated | Control | Acetylated | |
| μm | min−1 | μm | min−1 | |||||
| Cathepsin D | 25 | 20 | 6.6 | 0.42 | 25 | 26 | 1.8 | 0.38 |
| NPC2 | 367 | 367 | 37.4 | 10.45 | 352 | 333 | 26.6 | 6.3 |
| Soybean agglutinin | 1000 | 1000 | 0.3 | 0.3 | 1000 | 1000 | 0.25 | 0.25 |
| DNase I | 30 | 30 | 0.27 | 0.23 | 60 | 60 | 0.30 | 0.25 |
In contrast to these findings with acid hydrolase substrates, acetylation of the nonlysosomal glycoproteins soybean agglutinin and DNase I resulted in either no effect (soybean agglutinin) or a small inhibition (DNase I).
Function of γ Subunit in Fibroblasts
Another means of accessing the ability of the α/β and γ subunits of GlcNAc-1- phosphotransferase to respond to critical lysines on protein substrates is via transfection of cells with cDNAs encoding wild-type and mutant forms of bovine DNase I. In particular, we have reported that mutation of Asn-74 to a lysine enhances the extent of phosphorylation of the N-linked glycan at position 108 and greatly stimulates the addition of a second Man-6-P residue at that site in COS cells (17). The lysine effect was specific for this glycan as it had no effect on the phosphorylation of the N-linked glycan at Asn-18. To test these constructs in wild-type and γ-knock-out fibroblasts, we utilized a retroviral transfection system as it gave better levels of transfection compared with other reagents. The transfected fibroblasts were incubated with [2-3H]mannose to label the N-linked glycans followed by immunoprecipitation of the secreted DNase I. The N-linked glycans of the various forms of DNase I were then analyzed for the extent of phosphorylation and the ratio of one Man-6-P- versus two Man-6-P-containing high mannose oligosaccharides.
The total phosphorylation of wild-type DNase I by the control and γ-knock-out fibroblasts was similar at about 40% (Table 6). However, the percent of the total phosphorylated high mannose oligosaccharides that contained two Man-6-P residues was significantly reduced in the γ-deficient fibroblasts (18 ± 4% in the wild-type versus 11 ± 6% in the γ-knock-out cells, p < 0.05). Mutation of Asn-74 to lysine stimulated the formation of high mannose oligosaccharides with two Man-6-P residues in both cell lines, but the wild-type cells synthesized twice as much of this species compared with γ-deficient cells (38 ± 9 versus 20 ± 11% respectively, p < 0.01). The difference in the ability of the two cell types to form high mannose oligosaccharides with two Man-6-P residues was even more apparent when phosphorylation at the Asn-106 site was examined using the N18Q construct that eliminates glycosylation at this site. As with wild-type DNase I, the total phosphorylation at Asn-106 was similar in both types of fibroblasts (34% wild-type, 38% γ-knock-out), whereas the percent of phosphorylated glycans with two Man-6-P residues was significantly greater in the wild-type fibroblasts (24 ± 5 versus 14 ± 6% for the knock-out cells, p < 0.05). The DNase I with the double mutation (N18Q/N74K) was phosphorylated to a greater extent by both cell types, but the wild-type fibroblasts exhibited a much larger increase in the synthesis of glycans with two Man-6-P residues (from 24 ± 5 to 83 ± 13%) than occurred in the γ-deficient cells (14 ± 6 to 40 ± 19%). These results demonstrate that the γ subunit is not essential for the addition of the first Man-6-P residue to the N-linked glycans of DNase I, but it serves to facilitate the addition of the second Man-6-P residue, particularly at position 106.
TABLE 6.
The function of the γ subunit in mouse fibroblasts
| DNase I construct | Wild type phosphorylation | γ-KO phosphorylation | WT (2Pi/1Pi + 2Pi) | γ-KO (2Pi/1Pi + 2Pi) | p value (2Pi/1Pi + 2Pi) |
|---|---|---|---|---|---|
| % | % | ||||
| Wild type | 38 ± 21 (n = 5) | 43 ± 20 (n = 5) | 18 ± 4 | 11 ± 6 | <0.05 |
| N74K | 54 ± 10 (n = 5) | 46 ± 19 (n = 5) | 38 ± 9 | 20 ± 11 | <0.01 |
| N18Q | 34 ± 15 (n = 3) | 38 ± 19 (n = 3) | 24 ± 5 | 14 ± 6 | <0.05 |
| N18Q/N74K | 65 ± 16 (n = 4) | 49 ± 9 (n = 4) | 83 ± 13 | 40 ± 19 | <0.01 |
DISCUSSION
The data presented in this study provide a number of new insights into the role of the α/β and γ subunits of GlcNAc-1- phosphotransferase. It has previously been established both by in vitro studies and analysis of mutations in patients with ML II and ML III that the α/β subunits contain the catalytic function of the transferase. However, it was unknown whether these subunits also have the ability to recognize the conformation-dependent protein determinant expressed by the acid hydrolase substrates. Our findings provide strong evidence that the α/β subunits do indeed recognize the protein determinant of the acid hydrolases. Studies of the extent of mannose phosphorylation of the acid hydrolases present in the brain of mice with a disruption of the gene encoding the γ subunit showed that all hydrolases had detectable phosphorylation, and most importantly, about one-third of them were phosphorylated to about the same extent as occurred in wild-type mice. This phosphorylation was specific for the acid hydrolases, as acid β-glucosidase, a glycoprotein known not to be a substrate for GlcNAc-1-phosphotransferase, had no detectable phosphorylation.
Further support for this conclusion comes from the in vitro assays comparing the activity of recombinant α2β2γ2 and α2β2 forms of GlcNAc-1-phosphotransferase toward a panel of lysosomal and nonlysosomal protein acceptors. In all instances, the apparent Km values were equivalent for both forms of the transferase, indicating that the γ subunit does not substantially enhance the binding of the α/β subunits to the protein acceptors. The difference between the two forms of the transferase was in the kcat values toward a subset of the acid hydrolase acceptors, with the α2β2 enzyme having significantly lower activity toward pro-tripeptidyl peptidase, cathepsin D, and uteroferrin. The kcat value of the α2β2 enzyme toward NPC2 was only decreased by 25% relative to the wild-type enzyme. These differences in kcat values are in line with the differences in the extent of phosphorylation of the same hydrolases observed in the brains of wild-type and γ-deficient mice. In contrast to these findings, a previous analysis of GlcNAc-1-phosphotransferase activity in fibroblast extracts from three patients classified as having ML IIIC indicated that the mutant enzyme had a reduced affinity for cathepsin D and uteroferrin as well as a decreased kcat value (42). The reason for this apparent discrepancy is not clear.
The finding that deglycosylated cathepsin D is a specific inhibitor of the phosphorylation of intact cathepsin D by both the α2β2γ2 and α2β2 forms of the transferase provides additional evidence that the α/β subunits interact with the protein determinant of cathepsin D. Interestingly, the deglycosylated cathepsin D as well as DNase I stimulated the phosphorylation of α-methylmannoside by both forms of the transferase. The simplest explanation is that binding of the α/β subunits to the protein domain of these hydrolases induces a conformational change that increases the catalytic activity toward the mannose acceptor.
A number of studies have implicated lysine residues as key determinants of the phosphorylation of acid hydrolases (17, 31, 36–41). Cell transfection studies have shown that mutation of specific lysines on acid hydrolases decreases the extent of phosphorylation, although insertion of lysines at specific positions on the nonlysosomal glycoprotein pepsinogen increases Man-6-P formation. In several instances, it was demonstrated that the critical lysine enhanced phosphorylation at a single N- linked glycosylated site. Furthermore, the Sahagian laboratory showed that biochemical modification of lysine residues of several acid hydrolases with sulfo-N-hydroxysuccinimide greatly decreased phosphorylation in in vitro assays (37, 38).
This study extends these previous studies by showing that the α/β subunits are sensitive to modification of the lysines on acceptor hydrolases. Both α2β2γ2 and α2β2 forms of GlcNAc-1- phosphotransferase exhibited decreased activity toward cathepsin D and NPC2 following modification of their lysine residues by SNA. Interestingly, kinetic analysis showed that the lysine modification did not alter the apparent Km values but greatly decreased the kcat values. In the case of bovine pancreatic DNase I (which lacks lysines 27 and 74), the treatment with SNA had only a slight inhibitory effect on the kcat of the reaction and no effect on the apparent Km. Together, these data are consistent with a model whereby the α/β subunits bind to a basal protein recognition domain followed by the interaction with critical lysines that direct the catalytic site to specific N-linked high mannose glycans.
The fibroblast transfection experiments using a variety of bovine DNase I constructs have provided additional insight into the function of the γ subunit. First, the overall phosphorylation of the DNase I in wild-type and γ-deficient fibroblasts is similar. However, when the synthesis of oligosaccharides with one versus two Man-6-P residues was determined, it was found that the formation of oligosaccharides with two Man-6-P residues was significantly impaired in the fibroblasts lacking the γ subunit. These results show that the γ subunit enhances the transfer of the second GlcNAc-P to some N-linked high mannose oligosaccharides, but it is not absolutely required for this reaction to occur. Similar results were obtained in the in vitro assays using NPC2 as acceptor (Table 2).
In summary, we present the following evidence. 1) The α/β subunits of GlcNAc-1-phosphotransferase recognize and bind the conformation-dependent protein determinant of acid hydrolases as well as critical lysines. 2) The subunits then mediate the catalytic function of the transferase. 3) Interaction of the α/β subunits with the protein determinant of acid hydrolases stimulates the catalytic function of the transferase. The γ subunit enhances the rate of GlcNAc-P transfer to the oligosaccharides of acid hydrolase acceptors. 4) The requirement for the γ subunit to achieve optimal levels of phosphorylation varies greatly among the different acid hydrolases, and in some instances the γ subunit is dispensable. Although the α/β subunits can transfer one or two GlcNAc-P residues to the high mannose oligosaccharide units of acceptor hydrolases, the γ subunit enhances the addition of the second GlcNAc-P.
These data lead us to propose the following model for how GlcNAc-1-phosphotransferase recognizes and phosphorylates acid hydrolases. The initial event is the binding of the α/β subunits to the protein determinant on the acid hydrolase substrate. This binding induces a conformational change in the α/β subunits that facilitates the catalytic activity. The α/β subunits also engage critical lysines that serve to direct the catalytic site toward the high mannose oligosaccharides of the hydrolase substrate, a step that is followed by transfer of GlcNAc-P from UDP-GlcNAc to specific mannose residues. The γ subunit serves at least two roles. The first is to interact with the high mannose oligosaccharide of the acceptor hydrolase in a manner that facilitates the addition of the second GlcNAc-P to the molecule. This GlcNAc-P is transferred to a specific mannose residue on the 3′ arm of the high mannose unit, whereas the first GlcNAc-P almost always is added to a mannose on the 6′ arm of the glycan (43). We postulate that the MRH domain of the γ subunit binds the high mannose oligosaccharide in such a way that the transfer of the second GlcNAc-P is enhanced, similar to the role of the MRH domain of the β subunit of glucosidase II in facilitating the ability of the catalytic site on the α subunit to cleave glucose residues from Glc2Man9GlcNAc2 units on glycoprotein substrates (14, 15). The second role of the γ subunit is to enhance the overall phosphorylation of a subset of the acid hydrolases. This could occur in several ways. Previous studies have reported that acid hydrolases with multiple N-linked glycans exhibit considerable variability in the extent of phosphorylation at the individual sites (44–48). It has also been found that the introduction of new glycosylation sites on several acid hydrolases results in variable degrees of phosphorylation (24, 49). These findings can be explained if the location of the N-linked glycans on the surface of the hydrolases varies relative to the position of the protein recognition domain. Therefore, one mechanism whereby the γ subunit could enhance phosphorylation would be to bind high mannose glycans with unfavorable positioning relative to the catalytic site on the α/β subunits and bring the glycans into a more favorable location. Alternatively, or in addition, the γ subunit could interact with the α/β subunits to induce a conformational change that facilitates the interaction of the α/β subunits with N-linked glycans that have suboptimal locations relative to the protein binding domain.
We are currently preparing α2β2γ2 forms of GlcNAc-1- phosphotransferase with mutations in the MRH domain of the γ subunit that should prevent binding to high mannose glycans. These constructs should help in testing the model.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA08759 (to S. K.) and DK054317 (to P. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- Man-6-P
- mannose 6-phosphate
- CI-MPR
- cation-independent mannose 6-phosphate receptor
- LC-MS/MS
- liquid chromatography-mass spectrometry/mass spectrometry
- DMEM
- Dulbecco's modified Eagle's medium
- SNA
- sulfo-N-hydroxysuccinimide acetate
- KO
- knock out
- MRH
- mannose 6-phosphate receptor homology
- ML
- mucolipidosis.
REFERENCES
- 1.Braulke T., Bonifacino J. S. (2009) Biochim. Biophys. Acta 1793, 605–614 [DOI] [PubMed] [Google Scholar]
- 2.Bao M., Booth J. L., Elmendorf B. J., Canfield W. M. (1996) J. Biol. Chem. 271, 31437–31445 [DOI] [PubMed] [Google Scholar]
- 3.Raas-Rothschild A., Cormier-Daire V., Bao M., Genin E., Salomon R., Brewer K., Zeigler M., Mandel H., Toth S., Roe B., Munnich A., Canfield W. M. (2000) J. Clin. Invest. 105, 673–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiede S., Storch S., Lübke T., Henrissat B., Bargal R., Raas-Rothschild A., Braulke T. (2005) Nat. Med. 11, 1109–1112 [DOI] [PubMed] [Google Scholar]
- 5.Kudo M., Bao M., D'Souza A., Ying F., Pan H., Roe B. A., Canfield W. M. (2005) J. Biol. Chem. 280, 36141–36149 [DOI] [PubMed] [Google Scholar]
- 6.Kornfeld S., Sly W. S. (2000) in The Metabolic and Molecular Bases of Inherited Disease ( Scriver C. R., Beaudet A. L., Sly W. S., Valle D. eds) pp. 3469–3483, McGraw-Hill, New York [Google Scholar]
- 7.Bargal R., Zeigler M., Abu-Libdeh B., Zuri V., Mandel H., Ben Neriah Z., Stewart F., Elcioglu N., Hindi T., Le Merrer M., Bach G., Raas-Rothschild A. (2006) Mol. Genet. Metab. 88, 359–363; Correction (2007) Mol. Genet. Metab.91, 299 [DOI] [PubMed] [Google Scholar]
- 8.Cathey S. S., Leroy J. G., Wood T., Eaves K., Simensen R. J., Kudo M., Stevenson R. E., Friez M. J. (2009) J. Med. Genet., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varki A. P., Reitman M. L., Kornfeld S. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7773–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelfman C. M., Vogel P., Issa T. M., Turner C. A., Lee W. S., Kornfeld S., Rice D. S. (2007) Invest. Ophthalmol. Vis. Sci. 48, 5221–5228 [DOI] [PubMed] [Google Scholar]
- 11.Vogel P., Payne B. J., Read R., Lee W. S., Gelfman C. M., Kornfeld S. (2009) Vet. Pathol. 46, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee W. S., Payne B. J., Gelfman C. M., Vogel P., Kornfeld S. (2007) J. Biol. Chem. 282, 27198–27203 [DOI] [PubMed] [Google Scholar]
- 13.Munro S. (2001) Curr. Biol. 11, R499–R501 [DOI] [PubMed] [Google Scholar]
- 14.Stigliano I. D., Caramelo J. J., Labriola C. A., Parodi A. J., D'Alessio C. (2009) Mol. Biol. Cell. 20, 3974–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe T., Totani K., Matsuo I., Maruyama J., Kitamoto K., Ito Y. (2009) Glycobiology 19, 834–840 [DOI] [PubMed] [Google Scholar]
- 16.Sleat D. E., Sohar I., Lackland H., Majercak J., Lobel P. (1996) J. Biol. Chem. 271, 19191–19198 [DOI] [PubMed] [Google Scholar]
- 17.Nishikawa A., Gregory W., Frenz J., Cacia J., Kornfeld S. (1997) J. Biol. Chem. 272, 19408–19412 [DOI] [PubMed] [Google Scholar]
- 18.Lang L., Reitman M., Tang J., Roberts R. M., Kornfeld S. (1984) J. Biol. Chem. 259, 14663–14671 [PubMed] [Google Scholar]
- 19.Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. (1956) Anal. Chem. 28, 350–356 [Google Scholar]
- 20.Liou H. L., Dixit S. S., Xu S., Tint G. S., Stock A. M., Lobel P. (2006) J. Biol. Chem. 281, 36710–36723 [DOI] [PubMed] [Google Scholar]
- 21.Lin L., Lobel P. (2001) Biochem. J. 357, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudo M., Canfield W. M. (2006) J. Biol. Chem. 281, 11761–11768 [DOI] [PubMed] [Google Scholar]
- 23.Qian M., Sleat D. E., Zheng H., Moore D., Lobel P. (2008) Mol. Cell. Proteomics 7, 58–70 [DOI] [PubMed] [Google Scholar]
- 24.Warner J. B., Thalhauser C., Tao K., Sahagian G. G. (2002) J. Biol. Chem. 277, 41897–41905 [DOI] [PubMed] [Google Scholar]
- 25.Sohar I., Lin L., Lobel P. (2000) Clin. Chem. 46, 1005–1008 [PubMed] [Google Scholar]
- 26.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27.Valenzano K. J., Kallay L. M., Lobel P. (1993) Anal. Biochem. 209, 156–162 [DOI] [PubMed] [Google Scholar]
- 28.Craig R., Beavis R. C. (2004) Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa A., Nanda A., Gregory W., Frenz J., Kornfeld S. (1999) J. Biol. Chem. 274, 19309–19315 [DOI] [PubMed] [Google Scholar]
- 30.Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steet R., Lee W. S., Kornfeld S. (2005) J. Biol. Chem. 280, 33318–33323 [DOI] [PubMed] [Google Scholar]
- 32.Dustin M. L., Baranski T. J., Sampath D., Kornfeld S. (1995) J. Biol. Chem. 270, 170–179 [DOI] [PubMed] [Google Scholar]
- 33.Liu H., Sadygov R. G., Yates J. R., 3rd (2004) Anal. Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 34.Cacia J., Quan C. P., Pai R., Frenz J. (1998) Biochemistry 37, 15154–15161 [DOI] [PubMed] [Google Scholar]
- 35.Lübke T., Lobel P., Sleat D. E. (2009) Biochim. Biophys. Acta 1793, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baranski T. J., Faust P. L., Kornfeld S. (1990) Cell 63, 281–291 [DOI] [PubMed] [Google Scholar]
- 37.Cuozzo J. W., Sahagian G. G. (1994) J. Biol. Chem. 269, 14490–14496 [PubMed] [Google Scholar]
- 38.Cuozzo J. W., Tao K., Wu Q. L., Young W., Sahagian G. G. (1995) J. Biol. Chem. 270, 15611–15619 [DOI] [PubMed] [Google Scholar]
- 39.Tikkanen R., Peltola M., Oinonen C., Rouvinen J., Peltonen L. (1997) EMBO J. 16, 6684–6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cuozzo J. W., Tao K., Cygler M., Mort J. S., Sahagian G. G. (1998) J. Biol. Chem. 273, 21067–21076 [DOI] [PubMed] [Google Scholar]
- 41.Yaghootfam A., Schestag F., Dierks T., Gieselmann V. (2003) J. Biol. Chem. 278, 32653–32661 [DOI] [PubMed] [Google Scholar]
- 42.Lang L., Takahashi T., Tang J., Kornfeld S. (1985) J. Clin. Invest. 76, 2191–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazzarino D. A., Gabel C. A. (1988) J. Biol. Chem. 263, 10118–10126 [PubMed] [Google Scholar]
- 44.Goldberg D. E., Kornfeld S. (1981) J. Biol. Chem. 256, 13060–13067 [PubMed] [Google Scholar]
- 45.Sonderfeld-Fresko S., Proia R. L. (1989) J. Biol. Chem. 264, 7692–7697 [PubMed] [Google Scholar]
- 46.Weitz G., Proia R. L. (1992) J. Biol. Chem. 267, 10039–10044 [PubMed] [Google Scholar]
- 47.Shipley J. M., Grubb J. H., Sly W. S. (1993) J. Biol. Chem. 268, 12193–12198 [PubMed] [Google Scholar]
- 48.Dittmer F., Pohlmann R., von Figura K. (1997) J. Biol. Chem. 272, 852–858 [DOI] [PubMed] [Google Scholar]
- 49.Cantor A. B., Kornfeld S. (1992) J. Biol. Chem. 267, 23357–23363 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.