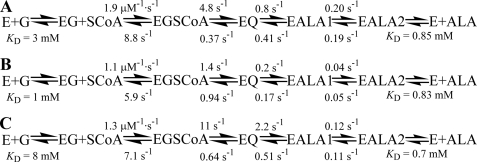Abstract
5-Aminolevulinate synthase (EC 2.3.1.37) (ALAS), a pyridoxal 5′-phosphate (PLP)-dependent enzyme, catalyzes the initial step of heme biosynthesis in animals, fungi, and some bacteria. Condensation of glycine and succinyl coenzyme A produces 5-aminolevulinate, coenzyme A, and carbon dioxide. X-ray crystal structures of Rhodobacter capsulatus ALAS reveal that a conserved active site serine moves to within hydrogen bonding distance of the phenolic oxygen of the PLP cofactor in the closed substrate-bound enzyme conformation and within 3–4 Å of the thioester sulfur atom of bound succinyl-CoA. To evaluate the role(s) of this residue in enzymatic activity, the equivalent serine in murine erythroid ALAS was substituted with alanine or threonine. Although both the KmSCoA and kcat values of the S254A variant increased, by 25- and 2-fold, respectively, the S254T substitution decreased kcat without altering KmSCoA. Furthermore, in relation to wild-type ALAS, the catalytic efficiency of S254A toward glycine improved ∼3-fold, whereas that of S254T diminished ∼3-fold. Circular dichroism spectroscopy revealed that removal of the side chain hydroxyl group in the S254A variant altered the microenvironment of the PLP cofactor and hindered succinyl-CoA binding. Transient kinetic analyses of the variant-catalyzed reactions and protein fluorescence quenching upon 5-aminolevulinate binding demonstrated that the protein conformational transition step associated with product release was predominantly affected. We propose the following: 1) Ser-254 is critical for formation of a competent catalytic complex by coupling succinyl-CoA binding to enzyme conformational equilibria, and 2) the role of the active site serine should be extended to the entire α-oxoamine synthase family of PLP-dependent enzymes.
Keywords: Enzymes, Enzymes/Heme, Enzymes/Pyridoxal Phosphate, Metabolism/Heme, Vitamins and Cofactors/Heme, Vitamins and Cofactors/Porphyrin, Vitamins and Cofactors/Pyridoxal Phosphate, Aminolevulinate Synthase
Introduction
5-Aminolevulinate synthase (EC 2.3.1.37; ALAS)2 is a homodimeric PLP-dependent enzyme that catalyzes the first and key regulatory step of the heme biosynthetic pathway in non-plant eukaryotes and the α-subclass of purple bacteria, involving the condensation of glycine and succinyl-CoA to produce CoA, carbon dioxide, and ALA (1). Animal genomes encode two highly conserved but differentially expressed ALAS genes, one of which is expressed in all tissues (ALAS1), and the other of which is erythroid-specific (ALAS2) (2). In humans, mutations in the ALAS2 gene can result in X-linked sideroblastic anemia (3), a hypochromic and microcytic anemia characterized by iron accumulation within erythroblast mitochondria (4). Approximately one-third of X-linked sideroblastic anemia patients are pyridoxine-responsive, and in these patients the ALAS mutations are commonly observed in the PLP-binding site (5, 6).
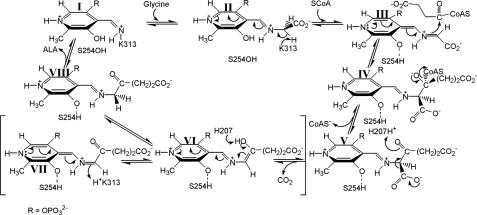
The ALAS chemical mechanism (Scheme 1) is complex and involves a high degree of stereoelectronic control, with individual steps, including the following: (i) binding of glycine; (ii) transaldimination with the active site lysine (Lys-313, murine ALAS2 numbering) to yield an external aldimine; (iii) abstraction of the pro-R proton of glycine; (iv) condensation with succinyl-CoA; (v) CoA release to generate an α-amino-β-ketoadipate intermediate; (vi) decarboxylation resulting in an enol-quinonoid rapid equilibrium; (vii) protonation of the enol to give an aldimine-bound molecule of ALA; and (viii) ultimately release of the product (ALA) (7). This mechanistic complexity is manifested structurally as an enzyme with an unusually high degree of sequence conservation, as exemplified by the observation that the catalytic cores of human ALAS2 and Rhodobacter capsulatus ALAS are 49% identical and 70% similar (8).
SCHEME 1.
PLP-dependent enzymes are classified according to structural and mechanistic similarities (9). ALAS is evolutionarily related to transaminases and is grouped within class II of the fold type I PLP-dependent enzyme superfamily, for which the prototypical enzyme is generally considered to be aspartate aminotransferase (10–12). ALAS is most closely related to the three other members of the α-oxoamine synthase subfamily, each of which also catalyzes a reaction involving a small amino acid, a CoA ester, and a 1,3-aminoketone (13, 14).
Aspartate aminotransferase exists in two predominant conformations, “open” and “closed” (10–12), and this property has been hypothesized to be a general feature of all PLP-dependent enzymes (15, 16). Catalysis occurs in the closed conformation, consistent with the induced fit hypothesis, where electrostatic and hydrophobic interactions between the substrates, cofactor, and amino acids comprising the active site provide the energetic impetus to stabilize a catalytically optimal conformation (17).
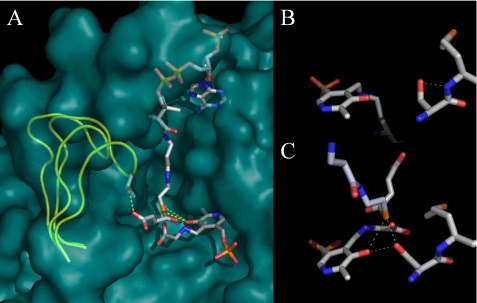
Prior to solution of an ALAS crystal structure, kinetic data led to the proposal that the ALAS catalytic cycle is dominated by a rate-limiting conformational change associated with release of ALA (7, 18–20). The crystal structures of R. capsulatus ALAS in holoenzymic and substrate-bound forms confirmed that the enzyme adopts open and closed conformations, respectively, and allowed discrimination of the specific regions and residues of the enzyme involved in protein dynamics (8). Although the structure in general collapses slightly around the bound substrates, a more conformationally mobile loop of amino acids located between two β-sheets at the subunit interface closes directly over the active site channel, which extends ∼20 Å down into the core of the enzyme to the PLP-binding site (Fig. 1) (8). A conserved threonine at the apex of the mobile loop forms a strong hydrogen bond (∼2.5 Å) with the carboxylate tail of succinyl-CoA in the substrate-bound structure and appears to simultaneously provide molecular recognition for succinyl-CoA while helping to lock this substrate into optimal stereoelectronic position for catalysis (21). Coincident with these changes, the side chain of Ser-189 (R. capsulatus numbering) migrates from noncovalently associating with the peptide macroskeleton to within hydrogen bonding distance of the PLP phenolic oxygen, as well as the sulfur atom of succinyl-CoA (8, 22, 23). These interactions suggest that this serine residue may be an important determinant in conformer equilibrium and catalysis by providing orientational binding energy between the enzyme, cofactor, and substrate, exclusively, in the closed Michaelis complex conformation. The conservation of this residue in ALAS and the other α-oxoamine synthases further suggests an important functionality that may be general to these enzymes (Fig. 2). Here, we present experiments aimed at probing the role of this serine in catalysis by murine ALAS2. We have generated and purified the positionally equivalent S254A and S254T variants and investigated the effects of these mutations on the kinetic and spectroscopic properties of the enzyme. The results support the postulate that this serine residue couples succinyl-CoA binding to enzyme conformational equilibria and catalysis.
FIGURE 1.
Structural models for murine erythroid ALAS based on the R. capsulatus crystal structures. A, Michaelis complex modeled by alignment of open holoenzyme and closed glycine- and succinyl-CoA-bound monomeric structures. Serine 254 is hidden by the succinyl-CoA ester in this view from the perspective of the adjacent subunit, which has been removed. The active site loop is shown in a yellow cartoon for the open and closed conformations, and all other structural features are for the closed conformation. B, serine 254 in the open conformation. C, serine 254 in the closed conformation with succinyl-CoA bound.
FIGURE 2.
Multiple sequence alignment of phylogenetically diverse members of the α-oxoamine synthase family in the region of murine ALAS2 serine 254. The amino acid sequences were retrieved from public data bases (NCBI) and aligned using ClustalW (42). The conserved serine residue is boxed and in boldface. The amino acid numbering (i.e. 254) refers to that of murine erythroid ALAS (mALAS2). Represented proteins are as follows: M. mus. AL2, Mus musculus erythroid ALAS (156255176); H. sap. AL2, Homo sapiens erythroid ALAS (28586); H. sap. AL1, H. sapiens housekeeping ALAS (40316939); S. cer. ALA, Saccharomyces cerevisiae ALAS (151942209); R. cap. ALA, R. capsulatus ALAS (974202); A. nig. AON, Aspergillus niger AONS (61696868); A. tha. AON, Arabidopsis thaliana AONS (42573269); M. mar. AON, Methanococcus maripaludis AONS (1599054); E. col. AON, E. coli AONS (85674759); H. sap. KBL, H. sapiens KBL (3342906); C. kor. KBL, Candidatus korarchaeum cryptofilum (17017433); E. col. KBL, E. coli KBL (169753078); H. sap. SPT, H. sapiens SPT (4758668); A. tha. SPT, A. thaliana SPT (17221603); S. cer. SPT, S. cerevisiae SPT (706828), E. col. SPT, E. coli SPT (170517920). AONS, 8-amino-7-oxononanoate synthase; SPT, serine palmitoyltransferase; KBL, 2-amino-3-oxobutyrate CoA ligase.
EXPERIMENTAL PROCEDURES
Reagents are listed under supplemental “Experimental Procedures.”
Mutagenesis
The pGF23 expression plasmid encoded the full-length sequence for the mature murine ALAS2 (24). Site-directed mutagenesis for the S254A and S254T mouse ALAS variants was performed on the whole plasmid pGF23 using a previously described method (25). Experimental details are described in the supplemental “Experimental Procedures.”
Protein Purification, SDS-PAGE, Protein Determination, and Steady-state Analysis
Recombinant murine ALAS2 and the S254 variants were purified from DH5α Escherichia coli bacterial cells containing the overexpressed protein as described previously (24). Purity was assessed by SDS-PAGE (26) and protein concentration determined by the bicinchoninic acid method using bovine serum albumin as the standard (27). All protein concentrations are reported on the basis of a subunit molecular mass of 56 kDa. Enzymatic activity was determined by a continuous spectrophotometric assay at 30 °C (see supplemental material) (28).
Structural Analyses
The Protein Data Base files 2BWN, 2BWO, and 2BWP, corresponding to the holoenzyme, succinyl-CoA-bound, and glycine-bound R. capsulatus ALAS crystal structures, were used as templates to model the PLP-binding core of the murine ALAS2 (8). Hydrogen bond determinations were accomplished using Deepview/Swiss-PdbViewer software (29, 30).
Spectroscopic Measurements
All spectroscopic measurements were performed with dialyzed enzyme in 20 mm HEPES, pH 7.5, with 10% (v/v) glycerol to remove free PLP. CD spectra were collected using an AVIV CD spectrometer calibrated for both wavelength maxima and signal intensity with an aqueous solution of D-10 camphorsulfonic acid (31). CD spectra recorded from 190 to 240 nm were analyzed by the ridge regression method using a modified version of the computer program CONTIN developed by Provencher and Glöckner (32). Protein concentrations were 10 and 100 μm for the near-UV-visible and far-UV CD spectra, respectively. The final concentration of succinyl-CoA was 100 μm, giving a 1:1 molar ratio of enzyme to ligand for collection of the latter spectra. At least three CD spectra were collected per experiment and averaged, using a 0.1-cm path length cuvette with a total volume of 300 μl. Blank CD spectra contained all components of the solution except enzyme. CD spectra containing the enzyme samples were collected immediately after adding the enzyme. The spectra of the samples containing enzyme were analyzed after subtracting the blank spectra. Co-factor fluorescence spectra were collected on a Shimadzu RF-5301 PC spectrofluorophotometer using protein concentrations of 2 μm. Spectra were measured at pH 7.5, 50 mm HEPES, and 20% (v/v) glycerol. Excitation was at 331 nm, and the excitation and emission slit widths were each set to 10 nm. Emission was measured over the wavelength range 350–600 nm. Buffer blanks were subtracted from the spectra.
Stopped-flow Spectroscopy
All of the experiments were carried out at 30 °C in 100 mm HEPES, pH 7.5, and 10% (v/v) glycerol. Because of the difference in Km for succinyl-CoA between the two variant enzymes, two different succinyl-CoA concentrations were used to ensure the identification of a single enzyme-catalyzed event. For the S254A-catalyzed reaction, the final concentrations were as follows: 120 μm S254A, 130 mm glycine, and 30 μm succinyl-CoA. The final concentrations of the reactants for S254T were as follows: 50 μm S254T, 130 mm glycine, and 10 μm succinyl-CoA. Rapid scanning stopped-flow kinetic measurements were conducted using an OLIS model RSM-1000 stopped-flow spectrophotometer. The dead time of this instrument is ∼2 ms, and the observation chamber optical path length is 4.0 mm. Scans covering the wavelength region 270–550 nm were acquired at a rate of either 62 or 31 scans/s to condense the resulting data files to a tractable size for data fitting analysis. An external water bath was utilized to maintain constant temperature of the syringes and observation chamber. Specifically, spectral data covering the 270–550 nm range were analyzed by global fitting to a triple exponential using the SIFIT program supplied with the stopped-flow instrument (OLIS, Inc.) (33). The quality of fits was judged by visual analysis of the calculated residuals in conjunction with the Durbin-Watson statistic (34). Single turnover data were interpreted using a three kinetic step mechanism as described by Reaction 1.
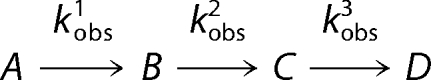 |
The observed rate constants were determined from at least three replicate experiments, and the reported values represent the average and standard error of measurement for each experimental condition. The forward and reverse rate constants depicted in the kinetic mechanism (Fig. 7) were obtained by modeling single wavelength kinetic traces at 510 nm with KinTekSim (Austin, TX) kinetic simulation software (35). The eight interior rate constants were allowed to float, althoughthe KD values were held constant as determined separately.
FIGURE 7.
Kinetic mechanisms of the Ser-254 variant enzymes. The single turnover quinonoid formation and decay reaction kinetics of the variant enzymes and wild-type ALAS and the reactions of the enzymes with glycine and ALA were used to model their kinetic mechanisms. The fits for S254A (A), S254T (B), and wild-type ALAS (C) (7) were indistinguishable from those completed based on the global fit of spectral data. E, ALAS; G, glycine; EG, ALAS-glycine complex; SCoA, succinyl-CoA; EGSCoA, ALAS-glycine-succinyl-CoA complex; EQ, observable quinonoid intermediate; EALA1, ALAS-ALA internal aldimine with active site loop closed; and EALA2, ALAS-ALA internal aldimine with active site loop open.
Transient Kinetics of the Reaction of Glycine with the Variant Enzymes
The reactions of the murine ALAS2 variants with glycine were performed using the same instrument as was described for the single turnover reactions with the enzyme-glycine complex and succinyl-CoA. The final enzyme concentration was 60 μm. The glycine concentration was always at least 10-fold greater than the enzyme concentration to ensure that pseudo-first order kinetics were observed. The treatment of the data was performed using the fitting software supplied with the instrument. The time courses at 420 nm were fitted to Equation 1,
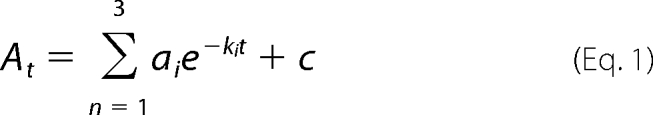 |
where At is the absorbance at time t; a is the amplitude of each phase; k is the observed rate constant for each phase, and c is the final absorbance. The quality associated with the fit was determined by the calculated residuals and from the Durbin-Watson ratio (34). The observed rate constants were plotted against increasing concentrations of glycine, and the resulting data were fitted to a two-step reaction process represented by Reaction 2. Data fitting to Equation 2 used the nonlinear regression analysis program SigmaPlot10 (Systat, San Jose, CA),
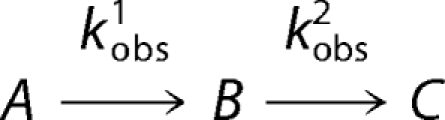 |
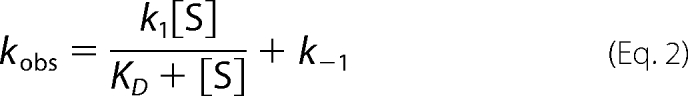 |
where S is the concentration of substrate; k1 and k−1 are the forward and reverse rate constants; KD is the dissociation constant, and kobs, the observed rate constant.
Intrinsic Protein Fluorescence Quenching
The presteady-state kinetics of the product binding reaction of ALAS and the two serine variants were examined by measuring changes in the intrinsic protein fluorescence intensity. An OLIS RSM-1000F rapid mixing spectrofluorimeter, equipped with a high intensity xenon arc lamp, was used to monitor the reaction. The enzyme and ligand in 20 mm HEPES, pH 7.5, and 10% glycerol were maintained at 30 °C in separate syringes prior to their mixing in the reaction chamber. The intrinsic protein fluorescence, as measured with 5 μm enzyme, was evaluated in the presence of increasing concentrations of the product ALA. The excitation wavelength and the slit width were 280 and 5 mm, respectively. The emitted light was filtered using a cutoff filter (WG 320; 80% transmittance at 320 nm (Edmund Optics, Barrington, NJ)). Typically, 500 time points were collected for varying lengths of time, and three or more experiments were averaged. Each averaged data set was then fitted to Equation 3, using the Global fitting software provided with the instrument.
where Fobs(t) is the observed fluorescence change (in arbitrary units) at time t; kobs is the observed first order rate constant; A1 is the pre-exponential factor, and A0 is the offset. The observed rate constants were then plotted against ligand concentration, and the data were fitted to Equation 4 by nonlinear regression. The rates of dissociation (koff) and association (kon) as well as the ligand binding constants (KD) were calculated from the asymptotic maximal observed rate, the ordinate intercept, and the ligand concentration (x) in Equation 4.
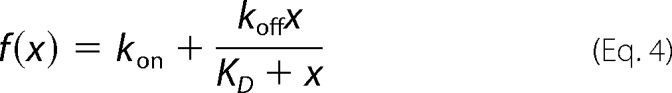 |
RESULTS
Kinetic Characterization of the Ser-254 Variants
The steady-state kinetic parameters of the Ser-254 variants were determined, and the results are summarized in Table 1. The mutation of serine 254 to alanine resulted in a kcat ∼2-fold higher than that of the wild-type ALAS value. The Km value for succinyl-CoA was increased ∼25-fold relative to ALAS, although the Km value for glycine was not significantly affected. The overall catalytic efficiency for succinyl-CoA decreased 13-fold, although the value for glycine remained virtually unchanged as compared with ALAS values. The replacement of serine 254 with threonine caused a 2.8-fold decrease in the wild-type kcat value. The Michaelis constants for both substrates were indistinguishable from those of the wild-type enzyme.
TABLE 1.
Summary of steady-state kinetic parameters
| Enzyme | KmGly | KmSCoA | kcat | kcat/KmGly | kcat/KmSCoA |
|---|---|---|---|---|---|
| mm | μm | s−1 | mm−1·s−1 | μm−1 ·s−1 | |
| Wild type | 25 ± 4 | 1.3 ± 0.9 | 0.14 ± 0.02 | 5.6 × 10−3 | 1.1 × 10−1 |
| S254A | 18 ± 2 | 32 ± 7 | 0.27 ± 0.01 | 1.5 × 10−2 | 8.4 × 10−3 |
| S254T | 27 ± 3 | 1.2 ± 0.3 | 0.050 ± 0.004 | 1.9 × 10−3 | 4.2 × 10−2 |
Spectroscopy
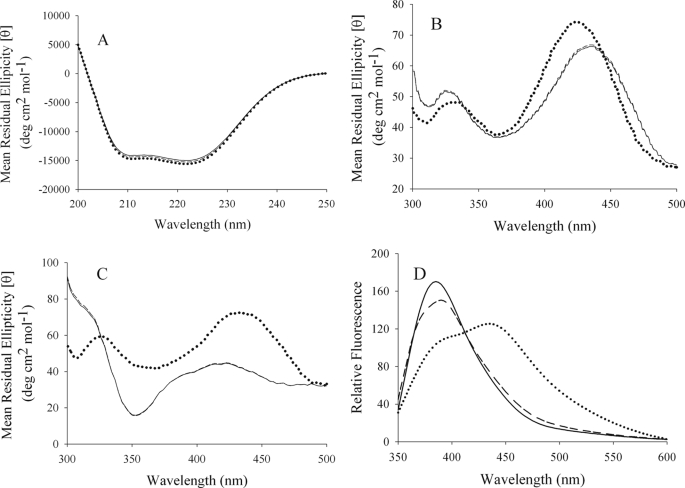
The orientation and average positioning of the PLP cofactor relative to the conserved serine and the active site were perturbed by the replacement of the serine with either an alanine or a threonine. Gross structural changes evidenced by changes in the α-helix and β-sheet content of proteins can be identified from CD spectroscopic changes in the far-UV (36). The analysis of the UV CD spectra (Fig. 3A) by CONTIN-CD indicates that any changes in the secondary structure between wild-type ALAS and the two ALAS variants are negligible. Locally chiral substructures that include the PLP microenvironment modulate the visible CD characteristics of the chromophore. Spectra for the wild-type and variant enzymes, as holo- and succinyl-CoA-bound enzymes, were collected (Fig. 3). The spectra for wild-type ALAS and S254T holoenzymes displayed positive dichroic bands at 330 and 420 nm. However, in relation to the wild-type ALAS, the S254A variant showed a decrease in amplitude of the 330-nm band, whereas the amplitude in the 420-nm region increased (Fig. 3B). Wild-type ALAS and variant enzymes responded differently to the presence of 100 μm ligand (Fig. 3C). Comparison of the CD spectra for the holoenzymes with succinyl-CoA showed that spectra of wild-type ALAS and S254T changed in the presence of substrate. In contrast, the spectrum of succinyl-CoA-bound S254A exhibited only slightly broader bands and a small decrease in the ratio of the dichroic intensities of the two bands when compared with those observed under ligand-free conditions (i.e. (I426 nm/I330 nm)ScoA-bound = 1.24 versus (I426 nm/I330 nm) = 1.54).
FIGURE 3.
Circular dichroism and fluorescence emission spectra of ALAS and the Ser-254 variants. Spectra of wild-type ALAS (–), S254A (···) and S254T (- - -). A and B, ligand-free holoenzymes; C, in the presence of 100 μm succinyl-CoA; D, upon excitation of the cofactor at 331 nm.
Specifically, for wild-type ALAS and S254T, the amplitude of both dichroic bands decreased and the 330-nm peak red-shifted to ∼350 nm. The cofactor fluorescence emission spectra obtained at pH 7.5 for the S254T variant enzyme with excitation at 331 nm is similar to that of the wild-type enzyme with a well formed emission maximum at ∼385 nm (Fig. 3D). A notable deviation in emission maximum is observed for the S254A variant. Specifically, excitation of S254A at 331 nm results in a broader fluorescence emission band centered around 450 nm. The changes in the fluorescence emission profile for the alanine variant suggest that the tautomeric equilibrium between at least two forms of the PLP cofactor aldimine linkage with the catalytic lysine is perturbed.
Reaction of Glycine with the Ser-254 Variants
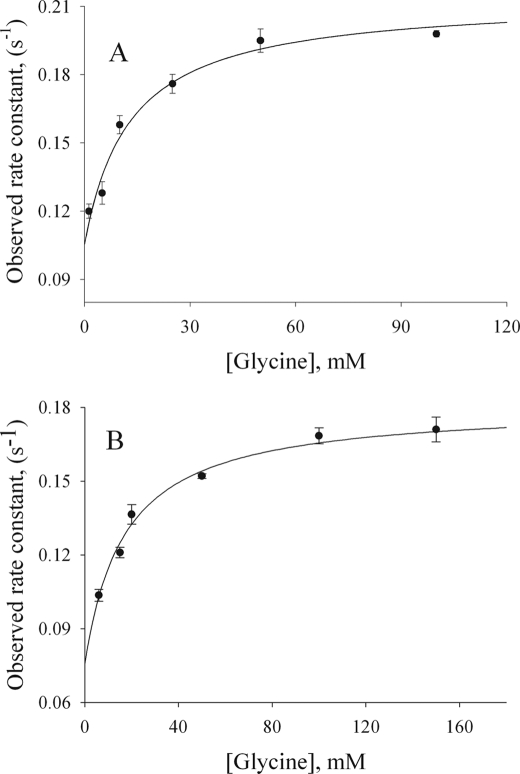
The reaction of 60 μm S254 variants with increasing amounts of glycine resulted in an increased absorbance at 420 nm (Fig. 4). A global fit of the data for the reaction of S254A with glycine yielded values for the reaction rates (k1 and k−1) with k1 = 0.159 ± 0.04 s−1 and k−1 = 0.072 ± 0.001 s−1 (Fig. 4B) and for the dissociation constant (KD) with KD = 6.6 ± 0.57 mm. The fitting of the data corresponding to the reaction of the S254T variant with glycine provided values of 0.11 ± 0.01 s−1 and of 0.070 ± 0.004 for k1 and k−1, respectively, and a KD of 1.5 ± 0.39 mm (Fig. 4A).
FIGURE 4.
Reaction of the Ser-254 variants (60 μm) with increasing concentrations of glycine. Data were fitted to Reaction 2 for a two-exponential process, yielding equilibrium and rate constants for S254T (KD = 1.5 ± 0.4 mm, k1 = 0.11 ± 0.01 s−1, and k−1 = 0.070 ± 0.004 s−1) (A) and S254A (KD = 6.6 ± 0.6 mm, k1 = 0.159 ± 0.04 s−1, and k−1 = 0.072 ± 0.001 s−1) (B). The fitted rate constants for wild-type ALAS yielded an overall binding KD of 8.0 ± 0.1 mm (18).
ALA Binding Kinetics Monitored by Intrinsic Protein Fluorescence
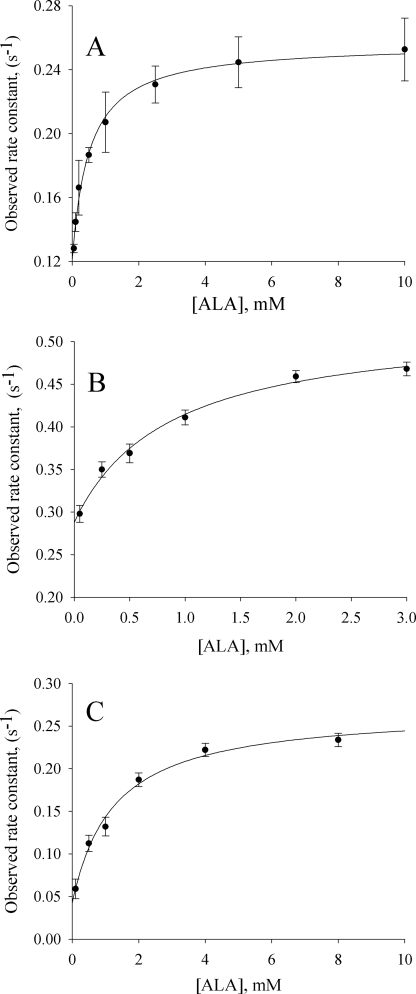
The observed rates of intrinsic protein fluorescence quenching upon ALA binding were determined as a function of ALA concentration. The results are presented in Fig. 5. The hyperbolic nature of the binding data indicate a two-step kinetic process and may be ascribed to formation of a collision complex followed by the conformational change associated with ALA binding (7). Significantly, for each of the three enzymes, the resolved rate constants for the off-rate conformational change (k−1) coincide with the kcat values determined through steady-state kinetics. The dissociation constants of the variants for ALA are increased by ∼60% over that of the wild-type enzyme.
FIGURE 5.
Reaction of wild-type ALAS and the Ser-254 variants (5 μm) with ALA. The observed rate constants were calculated by fitting the decrease in intrinsic protein fluorescence over time to Equation 2 for a single exponential process. A, resolved equilibrium and rate constants for wild-type ALAS are as follows: KD = 500 ± 16 μm, k1 = 0.120 ± 0.015 s−1, and k−1 = 0.140 ± 0.05 s−1. B, for the S254A variant, the constants are as follows: KD = 855 ± 66 μm, k1 = 0.235 ± 0.006 s−1, and k−1 = 0.29 ± 0.02 s−1. C, for the S254T variant, the constants are as follows: KD = 832 ± 49 μm, k1 = 0.19 ± 0.09 s−1, and k−1 = 0.057 ± 0.007 s−1.
Presteady-state Reaction of the Variant Enzyme-Glycine Complexes with Succinyl-CoA
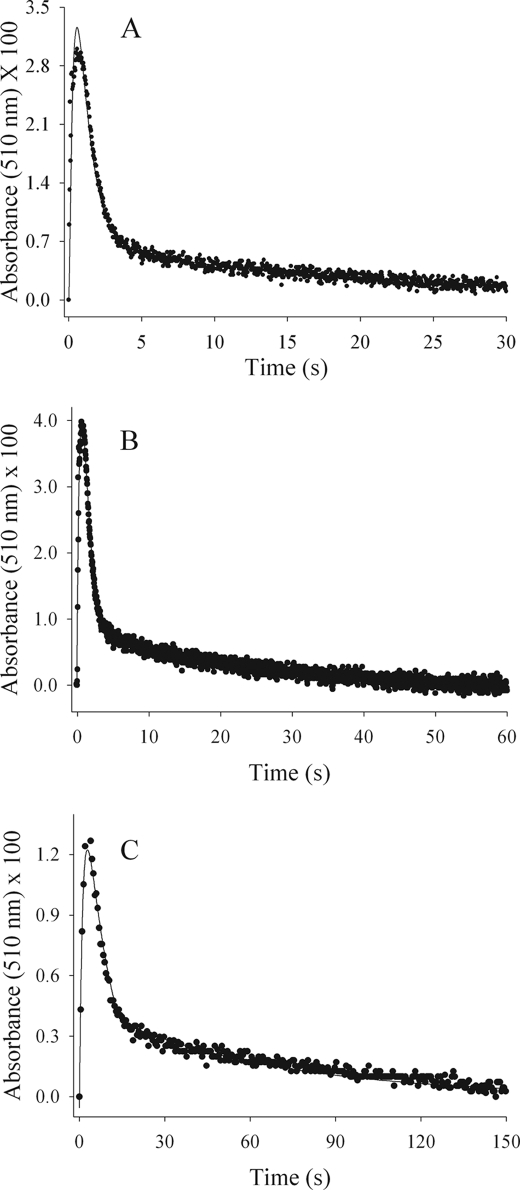
ALAS catalysis involves the sequential binding of glycine first (I and II), then succinyl-CoA (III and IV), leading to an enol-quinonoid equilibrium (VI–VII) after the liberation of CO2 (Scheme 1) (7). To determine the microscopic rates associated with the lifetime of the quinonoid intermediate, we monitored the time course of the ALAS-catalyzed reaction under single turnover conditions. The time courses of the absorbance change were best fit to a sequential, three-step kinetic mechanism outlined by Reaction 1. Among all the enzymes tested, a single step assigned to formation of a quinonoid intermediate followed by a biphasic step of its decay were observed (Fig. 6). For each enzyme, the global fit of the spectral data collected at 510 nm, which has been assigned to the wavelength of the quinonoid intermediate absorbance maximum (18, 20), is shown in Fig. 6 as a solid line overlaid with the time course data at 510 nm (dots). The rate constants associated with quinonoid intermediate formation (kQf) differ between the two variants. For S254A, the value is 4.8 ± 0.2 s−1, a rate similar to that of the wild-type enzyme (7). However, the rate value for the S254T variant was decreased, showing an ∼4-fold reduction with a rate of 1.4 ± 0.3 s−1. These data suggest that the loss of the hydroxyl group at position 254 has only a modest effect on quinonoid intermediate formation, and therefore it does not appear to play any obvious role in active site chemistry during catalysis. The rates assigned to the two-step quinonoid intermediate decay (kQd1 and kQd2) also differ among the variants. Both variants have initial quinonoid intermediate decay rates that are at least 2-fold lower than that of the wild-type enzyme (i.e. 0.8 and 0.25 s−1 versus 2.0 s−1). However, the second step of quinonoid intermediate decay, which most closely approximates kcat for the overall reaction and was previously assigned to a protein conformation change associated with product release (18, 20), is 40% faster for the S254A variant, in comparison with that of the wild-type enzyme. This increased value agrees with the greater kcat that was determined from the steady-state kinetic analysis (Fig. 6 versus Table 1). As such, the rates for S254A observed during the lifetime of the quinonoid intermediate (kQf and kQd1) indicate fast transformation of the enzyme-substrate complex to the enzyme-product complex, followed by a comparatively faster rate of product dissociation and the return of the conformational equilibrium to the catalytically favored open conformation (kQd2).
FIGURE 6.
Reaction of wild-type ALAS- and Ser-254 variant-glycine complexes with succinyl-CoA under single turnover conditions. The data for single turnover quinonoid intermediate formation and decay reaction kinetics were fitted with the SIFIT program (OLIS, Inc.). The data (●) are overlaid with the line representing the fitted data. A, rate constants for the three-step sequence in the wild-type enzyme are as follows: 6.0, 2.0, and 0.075 s−1. B, rate constants for the three steps in S254A-catalyzed reaction are as follows: 4.8, 0.8, and 0.19 s−1. C, rate constants for the three steps in S254T-catalyzed reaction are as follows: 1.4, 0.2, and 0.037 s−1.
DISCUSSION
The spectroscopic and kinetic properties of the S254A and S254T murine ALAS variants were examined and compared with those of the wild-type enzyme in an effort to better understand the role of Ser-254 in the catalytic process. Mutation of serine to alanine removes the side chain oxygen atom and eliminates the possibility of hydrogen bond formation with the PLP cofactor and succinyl-CoA. The conservative S254T mutation adds a methyl group and is predicted to allow for hydrogen bonding, although some steric constraints may be introduced due to the tight packing in this region of the active site. Comparison of the steady-state kinetic parameters of the variants with those of the wild-type enzyme determined under similar conditions hints at the unusual kinetic significance of Ser-254 (Table 1). Point mutations typically result in increased Michaelis constants and decreased turnover numbers, but strikingly, in the S254A variant both of these parameters were increased. The Km value for succinyl-CoA was increased 25-fold over that of the wild-type enzyme, whereas the kcat value was increased 2-fold. Although the physiological significance remains to be established, it is worth noting that mutation of this evolutionarily conserved residue in such a metabolically important enzyme led to enhanced values for both kcat and catalytic efficiency toward glycine (kcat/KmGly).
The kcat value for murine ALAS2 has been proposed to be defined by a conformational change of the enzyme accompanying ALA release (7). This is further supported here by the studies involving quenching of intrinsic protein fluorescence upon product binding with both wild-type ALAS and the Ser-254 variants (Fig. 5). Indeed, the protein fluorescence-associated ALA off-rates (k−1) were indistinguishable from the steady-state kcat values (Table 1). These data also indicated that the rate-limiting step of the reaction cycle was unaltered in the mutated enzymes. The increase in kcat value observed for the S254A variant may be attributable to diminished stability of the closed conformation, leading to faster reversion to the open conformation and product release. In this interpretation, the large concomitant increase in the Km value for succinyl-CoA in the S254A variant would result not only from the direct effect of loss of hydrogen bonding to the substrate but also from the less direct effect of impaired enzyme conformational change as triggered by the substrate, which is believed to be required for optimal binding (7, 18). The 3-fold decrease in the kcat value without any effect on the Km value for succinyl-CoA resulting from the more conservative S254T mutation would then arise primarily from enhanced stability of the closed conformation. Both mutations substantially diminished the catalytic efficiency with succinyl-CoA (kcat/KmSCoA) but in different ways, and although the effects of these very conservative mutations on the steady-state kinetic parameters may appear relatively modest, they are presumably sufficient to have metabolically harmful effects in vivo, given the strong evolutionary selection for serine at this position.
Significant effects on the cofactor microenvironment of ALAS were induced with the S254A mutation, as determined using fluorescence and CD spectroscopies. The differences in the fluorescence spectra reflected alterations in cofactor tautomeric equilibria, which must have occurred upon loss of hydrogen bonding interactions between the phenolic oxygen of PLP and the side chain of Ser-254 (Fig. 3D). We previously assigned the 330- and 410-nm absorption species to a substituted aldamine and a ketoenamine, respectively (37). The assignment of the 330-nm absorption species to a substituted aldamine, and not the enolimine form of the Schiff base, was due to the intense emission fluorescence of this species at 385 nm and not at the characteristic 510-nm emission wavelength of an enolimine (37, 38). The diminished substituted aldamine tautomer emission at 386 nm for the S254A variant is consistent with loss of hydrogen bonding at the cofactor phenolic oxygen. An increased proportion of the cofactor is probably in the ketoenamine tautomer, which we previously ascribed to the active form of the coenzyme (37), as the 410-nm absorption species appears to be more prominent in the S254A variant than in the wild-type ALAS and S254T variant (data not shown). CD spectroscopic examination of the S254A and S254T variants in the far-UV region confirmed that the mutations did not significantly alter the overall secondary structure of the enzymes (Fig. 3A). Any alterations in the conformational equilibria of the active site loop, which adopts an extended conformation and is thus not CD active, were not apparent in these spectra, but the visible CD spectra of the S254A variant do diverge substantially from those of the wild-type and the S254T variant. The S254A holoenzyme maximum at 435 nm was blue-shifted toward ∼420 nm, and the ratio of the mean residual ellipticity at this wavelength to the one at 330 nm was increased (Fig. 3B). These ellipticities arise from Cotton effects associated with the ketoenamine (435 nm) and substituted aldamine (330 nm) tautomers of the PLP Schiff base and are indicative of the microenvironment surrounding this linkage between the cofactor and the active site lysine (39–41). Succinyl-CoA binding to the wild-type and S254T variants induced decreases in asymmetry of the cofactor, whereas the S254A variant was relatively unchanged under similar conditions (Fig. 3C). A logical interpretation is that the decrease in asymmetry observed for the wild-type and S254T variants arose from partial conversion of the internal aldimine to free PLP aldehyde bound at the active site, as was observed in three of four R. capsulatus crystal structure active sites upon succinyl-CoA binding (8). In the crystal structures these events were accompanied by transition to a closed conformation, from which it might be concluded that the S254A variant retained the internal aldimine in the presence of succinyl-CoA, and it may not have been induced to adopt a closed conformation upon binding of this substrate.
Monitoring of the formation and decay of the quinonoid reaction intermediate under single turnover conditions indicated that the two Ser-254 variants follow a kinetic mechanism similar to that of wild-type ALAS (Fig. 6, A and B). A rapid step of ALA-bound quinonoid intermediate formation upon decarboxylation is followed by two successively slower decay steps, associated with protonation of the ALA-quinonoid intermediate and ALA release, respectively (7). The quinonoid intermediate formation rate decreased 4-fold for the S254T variant-catalyzed reaction, which could be explained by a change in the flow of electrons from the site of bond scission throughout the cofactor, precipitated by a shift in the conformational equilibrium toward the closed state. In the ALAS crystal structure, PLP is noted to change position by 15° when substrate is bound (8). Changes to the stereoelectronic relationship between the cofactor and the α-carbon bonds of the external aldimine can influence the chemical mechanism dramatically (7). Therefore, it is possible that the hydrogen bond between serine 254 and the phenolic oxygen of PLP may be an influential part of maintaining the angle of PLP during not only formation and decay of the quinonoid intermediate but also during the complete reaction cycle. The increase observed in the second step of quinonoid intermediate decay in the S254A variant is also consistent with the increases in kcat and the ALA off-rate determined from enzyme fluorescence quenching.
By utilizing the microscopic parameters obtained from the transient kinetics for the reactions catalyzed by the variant enzymes under single turnover conditions, coupled with the rate constants and dissociation constants determined for the reactions between the product or substrate with the enzymes, we were able to model their kinetic mechanisms as shown in Fig. 7. The simulations of the kinetic pathways revealed that the S254T mutation significantly retards the kinetic mechanism in relation to that of the wild-type ALAS. Conversely, the modeled pathway for S254A highlights the increases observed in the kinetics associated with the variant. Overall, the mechanistic data for both variants support the hypothesis that the interaction between Ser-254 and the O3′ of PLP is a limiting factor in enforcing an induced fit mechanism by coupling substrate recognition to conformational equilibria. However, how the structural differences between the variant enzymes with ligand bound accomplish the conformational transition awaits three-dimensional structural and protein dynamics information.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK63191 (to G. C. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and an additional reference.
- ALAS
- 5-aminolevulinate synthase
- ALA
- 5-aminolevulinate
- PLP
- pyridoxal 5′-phosphate.
REFERENCES
- 1.Akhtar M., Abboud M. M., Barnard G., Jordan P., Zaman Z. (1976) Philos. Trans. R. Soc. Lond. B Biol. Sci. 273, 117–136 [DOI] [PubMed] [Google Scholar]
- 2.May B. K., Dogra S. C., Sadlon T. J., Bhasker C. R., Cox T. C., Bottomley S. S. (1995) Prog. Nucleic Acid Res. Mol. Biol. 51, 1–51 [DOI] [PubMed] [Google Scholar]
- 3.May A., Bishop D. F. (1998) Haematologica 83, 56–70 [PubMed] [Google Scholar]
- 4.Bottomley S. S. (2006) Curr. Hematol. Rep. 5, 41–49 [PubMed] [Google Scholar]
- 5.Shoolingin-Jordan P. M., Al-Daihan S., Alexeev D., Baxter R. L., Bottomley S. S., Kahari I. D., Roy I., Sarwar M., Sawyer L., Wang S. F. (2003) Biochim. Biophys. Acta 1647, 361–366 [DOI] [PubMed] [Google Scholar]
- 6.Bottomley S. S. (2004) in Wintrobe's Clinical Hematology ( Greer J., Foerrster J., Lukens J. N., Rodgers G. M., Paraskevas R., Glader B. eds) pp. 1012–1033, Lippincott, Williams & Wilkins, Philadelphia [Google Scholar]
- 7.Hunter G. A., Zhang J., Ferreira G. C. (2007) J. Biol. Chem. 282, 23025–23035 [DOI] [PubMed] [Google Scholar]
- 8.Astner I., Schulze J. O., van den Heuvel J., Jahn D., Schubert W. D., Heinz D. W. (2005) EMBO J. 24, 3166–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliot A. C., Kirsch J. F. (2004) Annu. Rev. Biochem. 73, 383–415 [DOI] [PubMed] [Google Scholar]
- 10.Jäger J., Moser M., Sauder U., Jansonius J. N. (1994) J. Mol. Biol. 239, 285–305 [DOI] [PubMed] [Google Scholar]
- 11.Picot D., Sandmeier E., Thaller C., Vincent M. G., Christen P., Jansonius J. N. (1991) Eur. J. Biochem. 196, 329–341 [DOI] [PubMed] [Google Scholar]
- 12.McPhalen C. A., Vincent M. G., Picot D., Jansonius J. N., Lesk A. M., Chothia C. (1992) J. Mol. Biol. 227, 197–213 [DOI] [PubMed] [Google Scholar]
- 13.Christen P., Mehta P. K. (2001) Chem. Rec. 1, 436–447 [DOI] [PubMed] [Google Scholar]
- 14.Alexander F. W., Sandmeier E., Mehta P. K., Christen P. (1994) Eur. J. Biochem. 219, 953–960 [DOI] [PubMed] [Google Scholar]
- 15.Jansonius J. N., Eichele G., Ford G. C., Kirsch J. F., Picot D., Thaller C., Vincent M. G., Gehring H., Christen P. (1984) Biochem. Soc. Trans. 12, 424–427 [DOI] [PubMed] [Google Scholar]
- 16.Jansonius J. N., Eichele G., Ford G. C., Kirsch J. F., Picot D., Thaller C., Vincent M. G., Gehring H., Christen P. (1984) Prog. Clin. Biol. Res. 144, 195–203 [PubMed] [Google Scholar]
- 17.Koshland D. E., Jr., Neet K. E. (1968) Annu. Rev. Biochem. 37, 359–410 [DOI] [PubMed] [Google Scholar]
- 18.Hunter G. A., Ferreira G. C. (1999) J. Biol. Chem. 274, 12222–12228 [DOI] [PubMed] [Google Scholar]
- 19.Ferreira G. C., Zhang J. S. (2002) Cell. Mol. Biol. 48, 827–833 [PubMed] [Google Scholar]
- 20.Zhang J., Ferreira G. C. (2002) J. Biol. Chem. 277, 44660–44669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lendrihas T., Zhang J., Hunter G. A., Ferreira G. C. (2009) Protein Sci. 18, 1847–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gregoret L. M., Rader S. D., Fletterick R. J., Cohen F. E. (1991) Proteins 9, 99–107 [DOI] [PubMed] [Google Scholar]
- 23.Rajagopal S., Vishveshwara S. (2005) FEBS J. 272, 1819–1832 [DOI] [PubMed] [Google Scholar]
- 24.Ferreira G. C., Dailey H. A. (1993) J. Biol. Chem. 268, 584–590 [PubMed] [Google Scholar]
- 25.Miyazaki K., Takenouchi M. (2002) BioTechniques 33, 1033–1034, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 27.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 28.Hunter G. A., Ferreira G. C. (1995) Anal. Biochem. 226, 221–224 [DOI] [PubMed] [Google Scholar]
- 29.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 31.Chen G. C., Yang J. T. (1977) Anal. Lett. 10, 1195–1207 [Google Scholar]
- 32.Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 33.Tsai M. D., Weintraub H. J., Byrn S. R., Chang C., Floss H. G. (1978) Biochemistry 17, 3183–3188 [DOI] [PubMed] [Google Scholar]
- 34.Durbin J., Watson G. S. (1950) Biometrika 37, 409–428 [PubMed] [Google Scholar]
- 35.Barshop B. A., Wrenn R. F., Frieden C. (1983) Anal. Biochem. 130, 134–145 [DOI] [PubMed] [Google Scholar]
- 36.Kelly S. M., Price N. C. (2000) Curr. Protein Pept. Sci. 1, 349–384 [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Cheltsov A. V., Ferreira G. C. (2005) Protein Sci. 14, 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikushiro H., Hayashi H., Kagamiyama H. (2004) Biochemistry 43, 1082–1092 [DOI] [PubMed] [Google Scholar]
- 39.Futaki S., Ueno H., Martinez del Pozo A., Pospischil M. A., Manning J. M., Ringe D., Stoddard B., Tanizawa K., Yoshimura T., Soda K. (1990) J. Biol. Chem. 265, 22306–22312 [PubMed] [Google Scholar]
- 40.Kallen R. G., Korpela T., Martell A. E., Matsushima Y., Metzler C. M., Metzler D. E., Morozov Y. V., Ralston I. M., Savin F. A., Torchinsky Y. M., Ueno H. (1985) in Transaminases ( Christen P., Metzler D. E. eds) pp. 37–108, John Wiley & Sons, Inc., New York [Google Scholar]
- 41.Cellini B., Bertoldi M., Montioli R., Borri Voltattorni C. (2005) Biochemistry 44, 13970–13980 [DOI] [PubMed] [Google Scholar]
- 42.Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.