Abstract
FIP200 (FAK family-interacting protein of 200 kDa) is a conserved protein recently identified as a potential mammalian counterpart of yeast autophagy protein Atg17. However, it remains unknown whether mammalian FIP200 regulates autophagy in vivo. Here we show that neural-specific deletion of FIP200 resulted in cerebellar degeneration accompanied by progressive neuronal loss, spongiosis, and neurite degeneration in the cerebellum. Furthermore, deletion of FIP200 led to increased apoptosis in cerebellum as well as accumulation of ubiquitinated protein aggregates without any deficiency in proteasome catalytic functions. We also observed an increased p62/SQSTM1 accumulation in the cerebellum and reduced autophagosome formation as well as accumulation of damaged mitochondria in the mutant mice. Lastly, analysis of cerebellar neurons in vitro showed reduced JNK activation and increased susceptibility to serum deprivation-induced apoptosis in cerebellar neurons from the mutant mice. Taken together, these results provide strong genetic evidence for a role of FIP200 in the regulation of neuronal homeostasis through its function in autophagy in vivo.
Keywords: Ataxia, Autophagy, Mouse, Neurodegeneration, Ubiquitin, Autophagy, Cerebellar Ataxia, Conditional KO Mice, Neurodegeneration, Ubiquitinated Protein Aggregates
Introduction
The cerebellum provides central control of posture, balance, and coordination of movements (1–3). Damages to cerebellum caused by either injury or neurodegeneration are associated with various degrees of ataxia phenotype both in humans as well as mouse models. Previous studies have identified various gene mutations underlying the different neurodegenerative disorders and also revealed a potentially common cellular mechanism of these diseases, i.e. the accumulation of abnormal proteins aggregation. Protein quality control mechanisms, including ubiquitin-proteasome system and autophagy, have been shown to play crucial roles in the homeostasis of neurons and in neurodegeneration (4–8). Nevertheless, our understanding of the molecular and cellular mechanisms by which key signaling molecules and pathways regulate these cellular processes in neurodegeneration is still incomplete.
FIP200 (FAK family-interacting protein of 200 kDa)4 was initially identified as a protein that interacts with and inhibits the kinases Pyk2 and FAK (9). Subsequent studies showed that FIP200 also associates with other cellular proteins and regulates several signaling pathways (10). It was recently reported that RNA interference-mediated knockdown of FIP200 caused neurite atrophy and apoptosis in a neuroblastoma cell line Neuro-2a (11, 12), suggesting that FIP200 may play a role in homeostasis of neurons. Interestingly, recent studies identified FIP200 as a potential mammalian counterpart of yeast autophagy protein Atg17 (13, 14). FIP200 was shown to be a component of the ULK1-Atg13-FIP200 complex in fibroblasts (15–17) and to redistribute to autophagosomes upon induction of autophagy by starvation. Although the primary function of autophagy is the supply of amino acids as a response to starvation conditions in many organisms, a low level of basal (or constitutive) autophagy also plays an important role in maintaining cellular homeostasis, particularly in quiescent cells such as neurons. Neural-specific conditional KO of Atg5 or Atg7, both essential components of mammalian autophagy, led to abnormal accumulation of ubiquitinated protein aggregates, increased apoptosis, and neurodegeneration (18, 19). Despite these interesting observations, however, whether FIP200 plays a role in the regulation of basal autophagy and homeostasis of neurons in vivo remains totally unknown.
To explore a potential role of FIP200 in neurodegeneration in vivo, we generated and analyzed neural-specific conditional FIP200 KO mice using the Cre-loxP approach. Our results reveal a role of FIP200 in the regulation of neuronal homeostasis, and deletion of FIP200 led to cerebellar degeneration, which is caused by axon degeneration and loss of Purkinje cells as well as increased apoptosis of cerebellar granule cells.
EXPERIMENTAL PROCEDURES
Animals and Genotyping
FIP200f/f mice were described previously (20). nestin-Cre mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were housed and handled according to local, state, and federal regulations, and all experimental procedures were carried out according to the guidelines of Institutional Animal Care and Use Committee at University of Michigan. Mice genotyping for FIP200 and Cre alleles were performed by PCR analysis of tail DNA, essentially as described previously (20, 21).
Histology and Immunohistochemistry
Mice were euthanized using CO2, and a complete tissue set was harvested during necropsy. Fixation was carried out for 16 h at 4 °C using freshly made, pre-chilled (4 °C) PBS-buffered formalin. The brain tissues were all sagittal sectioned and then embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin for routine histological examination or left unstained for immunohistochemistry. Hematoxylin- and eosin-stained sections were examined under a BX41 light microscope (Olympus America, Inc., Center Valley, PA), and images were captured with an Olympus digital camera (model DP70) using DP Controller software (Version 1.2.1.10 8). For immunohistochemistry, unstained tissue sections were first deparaffinized in 3 washes of xylene (3 min each) and then were rehydrated in graded ethanol solutions (100, 95, 70, 50, and 30%). After heat-activated antigen retrieval (Retriever 2000, PickCell Laboratories B.V., Amsterdam, Holland) according to the manufacturer's specifications, sections were treated with blocking solutions; first with Avidin-Biotin Block (Dako Corp., Carpinteria, CA) then with Protein Block Serum Free (Dako Corp.). Sections were then incubated with the primary antibodies at 37 °C for 1 h in a humid chamber, washed in PBS 3 times (5 min each), then incubated with the biotinylated secondary antibodies (1:200 dilution, Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in a humidified chamber for 1 h at 37 °C, and washed in PBS similarly as before. Finally, sections were incubated with horseradish peroxidase-streptavidin (ABC Elite kit, Vector Laboratories) for 15 min at room temperature in a humid chamber and washed with PBS similarly as before. As the last staining step, 3,3′-diaminobenzidine (SIGMA FAST® DAB with Metal Enhancer, Sigma-Aldrich) was added to the sections, and the mixture was incubated at room temperature until a macroscopically appreciable light brown color developed in the sections (generally 30 s to 5 min). After incubation with 3,3′-diaminobenzidine, sections were lightly counterstained will Gill's hematoxylin. Histological examination and digital photography were carried out as described previously (20, 21).
Transmission Electron Microscopy
Samples were fixed in 2.5% glutaraldehyde in 0.1 m Sorensen's buffer, pH 7.4, overnight at 4 °C. After several rinses with buffer, they were postfixed in 1% osmium tetroxide in the same buffer. They were then rinsed in double distilled water to remove phosphate salt and then stained with aqueous 3% uranyl acetate for 1 h. The samples were dehydrated in ascending concentrations of ethanol, rinsed two times in propylene oxide, and embedded in epoxy resin. They were Ultrathin-sectioned at 70 nm in thickness and stained with uranyl acetate and lead citrate. The sections were examined using a Philips CM100 electron microscope at 60 kV. Images were recorded digitally using a Hamamatsu ORCA-HR digital camera system operated using AMT software (Advanced Microscopy Techniques Corp., Danvers, MA).
In Situ Detection of Apoptosis
Cerebellum sections from various time points (6 μm) were deparaffinized, incubated in methanol containing 0.3% H2O2 for 30 min, washed, and incubated with proteinase K (20 μg/ml) in PBS for 15 min at room temperature. Apoptotic cells were detected as described in the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore, Billerica, MA). Sections were counterstained with methyl green.
Protein Extraction, SDS-PAGE, and Western Blotting
The mouse brain tissues were collected from control or FIP200f/f;nestin-Cre mice at postnatal day 0 (P0). The protein lysates were prepared by homogenization in modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, 0.2% SDS, 1 mm sodium EDTA) supplemented with protease inhibitors (5 μg/ml leupeptin, 5 μg/ml aprotinin, and 1 mm phenylmethylsulfonyl fluoride). Tissue and cell debris were removed by centrifugation, and cleared supernatants were further transferred into a clean microcentrifuge tube. Protein concentration was determined using Bio-Rad protein assay reagent. In other experiments, lysates were prepared from cerebellar neuron cultures in vitro using similar methods. The lysates were boiled for 5 min in 1 × SDS sample buffer (50 mm Tris-HCl, pH 6.8, 12.5% glycerol, 1% SDS, 0.01% bromphenol blue) containing 5% β-mercaptoethanol and were then resolved with 6% polyacrylamide gel electrophoresis. After the SDS-PAGE were transferred onto a nitrocellulose membrane, membranes were incubated with antibody against FIP200 (1:1000) for 1 h at room temperature, washed using TBST, and incubated with the horseradish peroxidase anti-rabbit secondary antibodies (1:5000, Jackson ImmunoResearch Laboratories, Inc.). An Enhanced Chemiluminescent (ECL) kit (Pierce) was used to detect the immunoreactivity signal.
Proteasomal Catalytic Activity Assay
Proteasomal catalytic activity was analyzed for the lysates using the synthetic peptide substrates linked to the fluorometric receptor and aminomethylcoumarin (AMC, Proteasome Substrate Pack from Biomol, Plymouth Meeting, PA). Mice cerebella were dissected, placed on ice, and homogenized in proteolysis activity buffer (0.5 mm dithiothreitol, 5 mm ATP, 5 mm MgCl2). The 250-μl aliquots containing equal amounts of protein were incubated for 30 min at 37 °C with 2.5 μl of Ac-Gly-Pro-Leu-Asp-AMC (5 mm), Z-Leu-Leu-Glu-AMC (5 mm), Suc-Leu-Leu-Val-Tyr-AMC (5 mm), Ac-Arg-Leu-Arg-AMC (5 mm), or Boc-Leu-Arg-Arg-AMC (5 mm) for caspase-like, chymotrypsin-like, or trypsin-like activity, respectively. The reaction was stopped by adding 252.5 μl of ice-cold ethanol (96%). The proteasomal catalytic activity was determined by measuring the increase of fluorescence from AMC hydrolysis (380 nm excitation and 460 nm emission; Craiu et al. (54)).
Preparation and Analysis of Cerebellar Neuron Culture in Vitro
Cerebellar neuronal cell cultures were prepared from cerebellum of P7 mice. Isolated cerebella were removed of meninges and cut into 0.5-mm3 pieces. Tissue was dissociated for 15 min at 37 °C in 0.02% trypsin. Trypsinization was stopped by the addition of an equal volume of Dulbecco's modified Eagle's medium containing 0.0016% DNase and 10% horse serum (Invitrogen). Cells were further dissociated by trituration. After passing through a 70-μm filter to remove debris, the cell suspension was centrifuged at ∼200 × g for 5 min at 4 °C. The resulting pellet was then re-suspended in Eagle's basel medium (Invitrogen, Carlsbad, CA) supplemented with 1% Pen/Strep (Invitrogen), 0.5 mm glutamine, 25 mm KCl, 10% horse serum. Cells were plated in poly-l-lysine-coated plates at a density of 1000 cells/mm2. To induce neuronal cell death, cultures on in vitro day 5 were treated by replacing growth media with BME media without horse serum for 6 h. They were then stained with propidium iodide to detect cell death in the culture. In some experiments, 20 μm caspase inhibitor Z-VAD-FMK (Santa Cruz Biotechnology) or 10 μm necroptosis inhibitor necrostatin-1 (Alexis Biochemicals) were included in the incubation with media without serum.
RESULTS
Ablation of FIP200 in the Neuronal Precursors Leads to Severe Neurological Defects in Mice
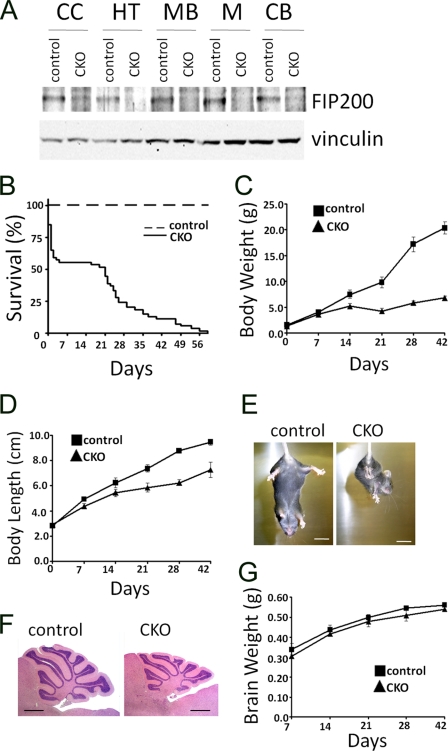
To study the potential role of FIP200 in the central nervous system, the floxed FIP200 (FIP200f/f) mice, in which exons 4 and 5 of FIP200 gene are flanked by two loxP sequences (20), were crossed with the nestin-Cre transgenic mice, which express Cre recombinase in neural precursors from embryonic day 10.5 (E10.5) (22–24). Cre-mediated deletion of exons 4 and 5 leads to a frameshift mutation because of direct splicing from exon 3 (containing ATG codon) to exon 6, producing a small truncated and nonfunctional peptide. In contrast to embryonic lethality of total KO of FIP200 (20), FIP200f/f;nestin-Cre mice were born at the expected Mendelian ratio. To evaluate deletion of FIP200 in the mutant mice, lysates were prepared from various regions of the brain of the mutant and control mice and subjected to Western blotting analysis. Fig. 1A shows that FIP200 was efficiently deleted in several regions of the mutant brain, including cerebral cortex, hypothalamus, midbrain, medulla, and cerebellum.
FIGURE 1.
Conditional ablation of FIP200 by nestin-Cre causes early death, growth retardation, and cerebellar ataxia. A, lysates were prepared from cerebral cortex (CC), hypothalamus (HT), midbrain (MB), medulla (M), and cerebellum (CB) of control or FIP200f/f;nestin-Cre (CKO) mice at P0 and then analyzed by Western blotting using anti-FIP200 (upper) or anti-vinculin (lower) antibodies. B, Kaplan-Meier survival curve of control (n = 30) and FIP200f/f;nestin-CRE mice (n = 53). CKO versus control: p < 0.01 by the log-rank test. C and D, body weight (C) and length (D) of control and FIP200f/f;nestin-CRE mice at various days after birth. E, control mice (left) extend their hind limbs and bodies, whereas FIP200f/f;nestin-CRE mice (right) show abnormal limb-clasping reflexes by crossing their limbs, when suspended by tail. Bar = 1 cm. F, histological analysis of sagittal sections of cerebellum from control and FIP200f/f;nestin-CRE mice at P28. Note the slightly smaller cerebellum and thinner granule cell layer in CKO mice. Bar = 1 mm. G, brain weight of control and FIP200f/f;nestin-CRE mice at various days after birth.
Although neural-specific deletion of FIP200 did not lead to embryonic lethality, ∼45% of the mutant mice died shortly after birth within the first few days. The remaining mice also showed significantly reduced survival rate after P14, and all mutant mice died by 60 days (Fig. 1B). The mutant mice showed growth retardation as compared with the control littermates starting from P7. By P21, the average weight and length of the mutant mice reached only ∼50 and 75%, respectively, of the control (i.e. FIP200f/f) mice (Fig. 1, C and D). Moreover, the mutant mice exhibited deficits in motor coordination starting around P14, which progressed to a more severe extent with tremors and stiff movement. When they are suspended by their tails, the mutant mice showed the hind-limb crosses phenotype, whereas the control mice had the normal extended limbs (Fig. 1E). Consistent with the ataxia phenotype, histological analysis of the cerebella showed a smaller size for the mutant mice compared with the controls (Fig. 1F). The reduced cerebellum size is not a consequence of the smaller body size, because the total size of the brain of the mutant mice was comparable to the controls (Fig. 1G). The foliation and fissuration appeared to be slightly less developed, and the granular cell layer was thinner in the mutant cerebellum compared with the controls. Together, these results suggest that deletion of FIP200 by nestin-Cre led to the development of cerebellar ataxia in mice.
Deletion of FIP200 Results in Progressive Neuronal Loss, Spongiosis, and Neurite Degeneration in the Cerebellum
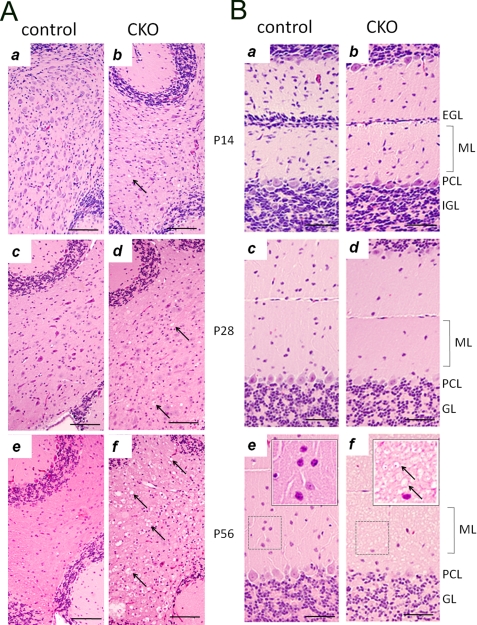
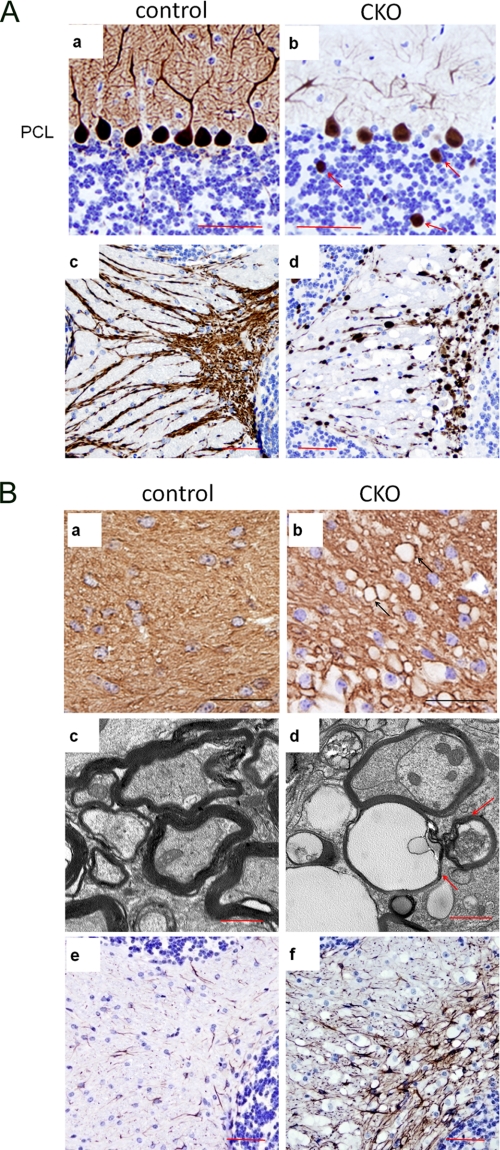
To study the cerebellum defects in FIP200f/f;nestin-Cre mice, histological analyses were performed on cerebella of control and mutant mice at various stages of postnatal development. At P7, the cellular organization of the cerebella were comparable between FIP200f/f;nestin-Cre and control mice, although the external granular layer appeared slightly thinner in the mutant mice (data not shown). At P14 and thereafter, FIP200f/f;nestin-Cre mice showed a progressive spongiosis in the cerebellar white matter (Fig. 2A, arrows in right panels) as well as a decreased thickness of the molecular layer (Fig. 2B). We also observed a decrease in the number of Purkinje cells, neurons in the molecular layer and granular cells in the mutant mice. At P56, severe spongiosis was also found in the molecular layer of the mutant cerebellum (Fig. 2B, panel f). We next examined cerebellar sections by immunohistochemistry using antibody against calbindin, a marker for Purkinje cells. These analyses revealed loss of Purkinje cells and extensive degeneration of their dendrites in FIP200f/f;nestin-Cre mice compared with the control mice (Fig. 3A, top panels). Furthermore, calbindin-positive cellular elements (red arrows, Fig. 3A, panel b) were also found in the granular layer of the mutant cerebellum, which could be segments of swollen (degenerating and/or abnormal) Purkinje cell axons traversing the granular layer. Examination of the cerebellar white matter showed extensive axonal degeneration associated with swelling of labeled axons and vacuoles typical of spongiosis in the mutant mice (Fig. 3A, bottom panels). Cerebellar sections from FIP200f/f;nestin-Cre and control mice were also analyzed by immunohistochemistry using antibody against myelin basic protein (anti-MBP). Anti-MBP staining appeared equally robust in the white matter of FIP200f/f;nestin-Cre and control mice, suggesting that myelination was not significantly affected by FIP200 deletion (Fig. 3B, panels a and b). Furthermore, similar staining was found for anti-olig2 (marker for oligodendrocytes) in FIP200f/f;nestin-Cre and control mice (data not shown). These data suggest that axonal degeneration of Purkinje cells is unlikely a secondary consequence caused by deficiency of myelination in the mutant mice.
FIGURE 2.
Progressive spongiosis and neuronal loss in cerebellum of FIP200f/f;nestin-Cre mice. A and B, cerebellar sections from control (a, c, and e) and FIP200f/f;nestin-Cre (CKO) (b, d, panel f) mice at different ages were stained by hematoxylin and eosin. External granular layer (EGL), molecular layer (ML), Purkinje cell layer (PCL), and internal granular layer (GL) are marked on the right. Insets show high magnification view of bracketed areas in e and f. Arrows mark spongiosis in the white matter (A) and molecular layer (B). Bars = 200 μm.
FIGURE 3.
Purkinje cell degeneration and reactive gliosis in cerebellum of FIP200f/f;nestin-Cre mice. A, cerebellum sections from control (a and c) and FIP200f/f;nestin-Cre (CKO) (b and d) mice at 6 weeks of age were analyzed by immunohistochemistry with anti-calbindin. Note the loss of Purkinje cells in the Purkinje cell layer (b) and degeneration of their axons (d) and dendrites (b) in cerebella from FIP200f/f;nestin-CRE mice compared with control mice. Arrows in panel b mark abnormal staining with anti-calbindin in the granular layer of the mutant mice. Scale bars = 100 μm. B, cerebellum sections from control (a, c, and e) and FIP200f/f;nestin-CRE (b, d, and f) mice at 4 weeks of age were analyzed by immunohistochemistry with anti-MBP (a and b), anti-GFAP (e and f), or by transmission electron microscopy (c and d). Note that the vacuoles are embedded in myelin-sheath (arrows in b and d) of the mutant mice. Scale bars = 100 μm (a, b, e, and f) and 1 μm (c and d), respectively.
Similar to hematoxylin and eosin and calbindin staining of the white matter (see Figs. 2A and 3A), significant spongiosis was also evident by anti-MBP staining. Interestingly, the vacuoles appear to be embedded in the circle of MBP staining (Fig. 3B, arrows in panel b), suggesting these vacuoles may be generated by degeneration of Purkinje cell axons, which occupied the now empty space encircled by myelination. The vacuoles are negative by stainings with Oil red O for lipids or periodic acid-Schiff for polysaccharides (data not shown). Further examination of the cerebellar white matter sections by transmission electron microscopy also showed presence of apparently normal myelination in the mutant mice (Fig. 3B, panels c and d). However, the axon bundles in the mutant mice were more rounded compared with the irregular shape of that in the control mice. Furthermore, some of the axon ensheathments are empty (red arrow in panel d) in the mutant mice. These results provide further evidence of extensive swelling and degeneration of Purkinje cell axons, which may be responsible for the spongiosis observed in the cerebellar white matter of FIP200f/f;nestin-Cre mice. Consistent with the extensive neural degeneration, immunostaining with antibody against GFAP (glial fibrillary acidic protein) showed significant reactive gliosis in the mutant cerebellum (Fig. 3B, panels e and f). Together, these studies suggested that degeneration of Purkinje cell axons and dendrites as well as reduced Purkinje cells and other cerebellar neurons are likely to be responsible for the loss of normal cerebellum functions in the motor coordination of FIP200f/f;nestin-Cre mice.
Increased Apoptosis and Accumulation of Ubiquitinated Protein Aggregates upon FIP200 Deletion
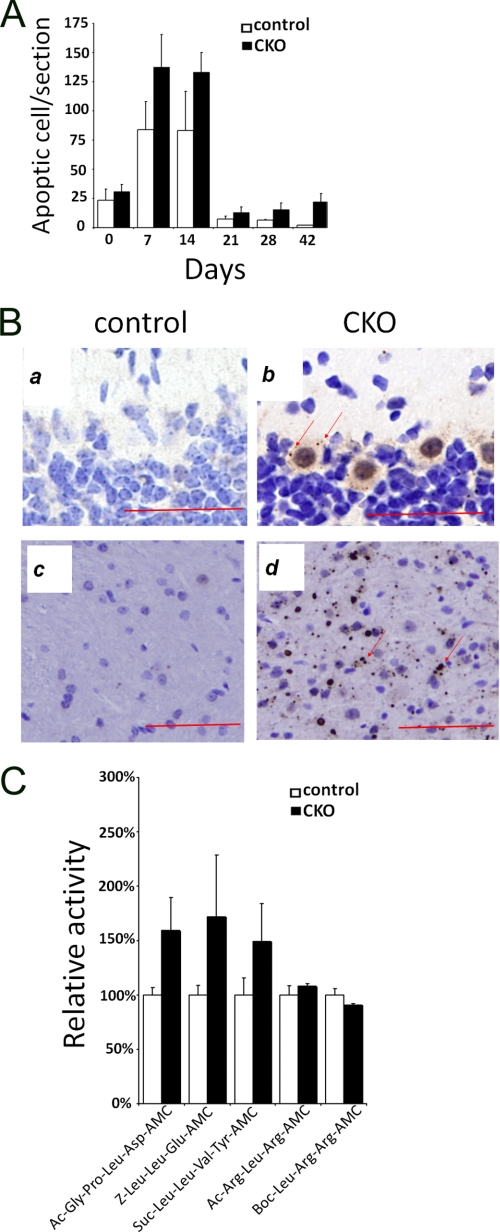
To explore potential mechanisms by which deletion of FIP200 leads to neuron loss and cerebellar degeneration, we examined possible increases in apoptosis in the cerebella of the mutant mice. Indeed, our previous studies showed that deletion of FIP200 resulted in increased apoptosis in the developing heart and liver leading to major defects in these organs and embryonic lethality of KO mice (20). Analysis of cerebellum sections by TUNEL assays showed apoptosis in both control and FIP200f/f;nestin-Cre mice during the first 2 weeks of postnatal development as expected (25), but the mutant mice exhibited an increase in apoptosis compared with control mice at these times (Fig. 4A). As expected, the normal developmental apoptosis in control mice reduced to a minimal level concomitant with the completion of postnatal development of the cerebellum at 3 weeks after birth (2, 3, 25). In contrast, significant apoptosis were still evident in the mutant cerebella at 3 weeks and later after birth. These results suggest that inactivation of FIP200 in the neurons could result in increased apoptosis leading to neural degeneration in the mutant cerebellum.
FIGURE 4.
Analysis of apoptosis, ubiquitinated protein aggregates, and proteasomal activity in cerebellum of FIP200f/f;nestin-Cre mice. A, cerebellum sections from control or FIP200f/f;nestin-Cre (CKO) mice at various postnatal days were analyzed by TUNEL assays. The numbers of apoptotic cells were determined, and the mean ± S.E. from three experiments is shown. B, cerebellum sections from control or FIP200f/f;nestin-CRE mice at 4 weeks of age were analyzed by immunohistochemistry using anti-ubiquitin. Note the ubiquitin-positive aggregates in the Purkinje cells (b) and white matter (d) of FIP200f/f;nestin-CRE mice. Scale bars = 100 μm. C, lysates prepared from cerebellum of control or FIP200f/f;nestin-CRE mice at 2 weeks of age were measured for their proteasome activities using various substrates as indicated. The mean ± S.E. from two independent experiments is shown.
Because many neurodegenerative diseases are associated with accumulation of ubiquitinated protein aggregates correlated with progressive loss of neurons due to apoptosis (26, 27), we next examined the potential accumulation of ubiquitinated protein aggregates by staining of cerebellum sections from FIP200f/f;nestin-Cre and control mice with anti-ubiquitin antibody. Although no aggregates were detected in the control cerebellum (Fig. 4B, panels a and c), large ubiquitin-positive aggregates were found in Purkinje cells and white matter in the mutant cerebellum (Fig. 4B, panels b and d, arrows), suggesting that deletion of FIP200 resulted in abnormal accumulation of ubiquitinated protein aggregates. Because impairment of proteasome functions could lead to abnormal accumulation of ubiquitinated protein aggregates (5–7), we analyzed the proteasome activity in the cerebella of FIP200f/f;nestin-Cre and control mice. The caspase-like, chymotryptic, and tryptic activities of the 20 S proteasomes were measured with Ac-GPLD-AMC (and Z-LLE-AMC), Suc-LLVY-AMC, and Ac-RLR-AMC (and Boc-LRR-AMC) peptides, respectively, as substrates. We found that both the caspase-like and chymotryptic activities were slightly elevated in the cerebella of FIP200f/f;nestin-Cre mice compared with that from control mice at P14 (Fig. 4C) and P28 (data not shown). The tryptic activity was comparable for the mutant and control mice. The increased caspase-like and chymotryptic activities could be due to some feedback mechanism for the cells to attempt to degrade the excess accumulation of the ubiquitinated protein aggregates. Therefore, the abnormal accumulation of ubiquitinated protein aggregates was unlikely a consequence of decreased proteasome activity in the cerebellum upon deletion of FIP200.
Defective Autophagy and Abnormal Mitochondria in FIP200-null Cerebellum
Recent studies have shown that basal autophagy is another important mechanism to remove abnormal protein aggregates (28–31). Suppression of basal autophagy by conditional KO of Atg5 or Atg7 in neuronal cells led to abnormal accumulation of ubiquitinated protein aggregates and increased apoptosis resulting in neurodegeneration in the mutant mice (18, 19). Interestingly, FIP200 has been found recently to form a complex with ULK1 and -2, mammalian orthologs of yeast Atg1, and mammalian Atg13, and to be required for autophagosome formation in MEFs (13, 15–17). To investigate potential deficiency in autophagy that might account for the abnormal accumulation of ubiquitinated protein aggregates upon FIP200 deletion in vivo, we first stained cerebellar sections for p62/SQSTM1 (p62), which regulates protein aggregation and accumulates in autophagy-deficient cells (32–34). As shown in Fig. 5, whereas little p62 staining was detected in the control cerebellum as expected (panel A), a significant amount of p62-positive aggregates was found in the mutant cerebellum (panel B, arrows). These results suggested that, consistent with results in MEFs (20), deletion of FIP200 in neurons resulted in autophagy deficiency in vivo, which could account for the abnormal accumulation of ubiquitinated protein aggregates.
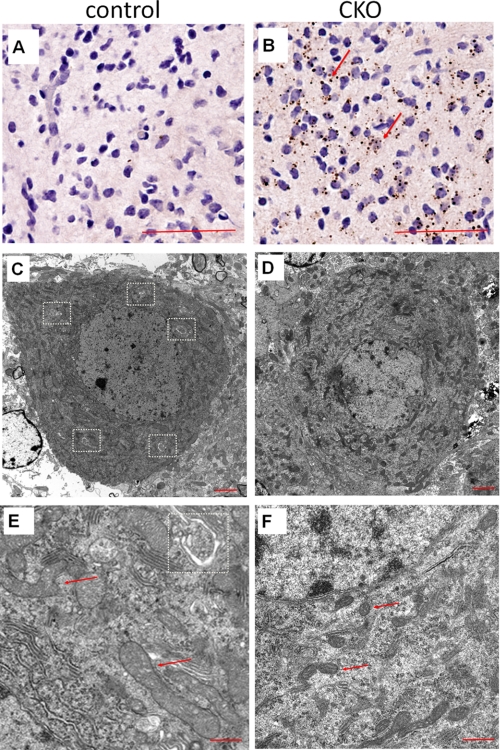
FIGURE 5.
Defective autophagy and mitochondria damage in cerebellum of FIP200f/f;nestin-Cre mice. A and B, cerebellum sections from control (A) or FIP200f/f;nestin-Cre (CKO) (B) mice at 4 weeks of age were analyzed by immunohistochemistry using anti-p62. Note the p62-positive aggregates in the white matter of CKO mice (arrows). Scale bars = 100 μm. C–F, Purkinje cells from control (C and E) and FIP200f/f;nestin-CRE (D and F) mice at 4 weeks of age were examined by transmission electron microscopy. Autophagosomes and mitochondria are marked by dotted rectangles and arrows, respectively. Scale bars = 2 μm (C and D) and 500 nm (E and F), respectively.
To further characterize autophagy deficiency and potentially other cellular abnormalities upon neural specific deletion of FIP200 in vivo, we then examined Purkinje cells in the cerebella of FIP200f/f;nestin-Cre and control mice by transmission electron microscopy. As expected, multiple autophagosomes characterized by the double membrane structures were identified in Purkinje cells from control mice (Fig. 5C, white rectangles). However, no autophagosomes were detected in these cells from FIP200f/f;nestin-Cre mice (Fig. 5D). Furthermore, in contrast to the smooth and extended morphology of mitochondria in control Purkinje cells (Fig. 5E, arrows), mitochondria in the mutant Purkinje cells were smaller with a more condensed contents and sometimes fragmented morphology (Fig. 5F, arrows). These results provide further support that defective autophagy upon deletion of FIP200 is responsible for the accumulation of ubiquitinated protein aggregates. They also suggest that deletion of FIP200 resulted in accumulation of damaged mitochondria in neurons, which could be due to deficiency of autophagy in these cells (35).
Increased Susceptibility to Serum Deprivation-induced Neuronal Death in FIP200-null Cerebellar Neurons
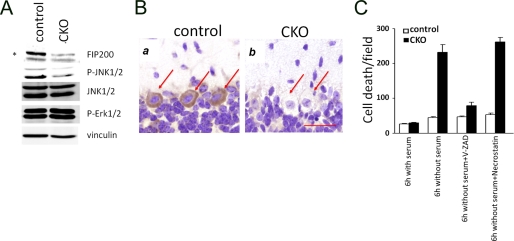
To further explore the mechanisms of apoptosis and neuronal loss upon FIP200 deletion, we prepared primary culture of cerebellar neurons from FIP200f/f;nestin-Cre and control mice and examined them in vitro. Previous studies in MEFs and hepatocytes from FIP200 knock-out embryos suggested that deletion of FIP200 reduced activation of the JNK signaling pathway, leading to increased sensitivity to tumor necrosis factor-α-induced apoptosis in these cells (20). Therefore, we examined activation status of JNK and several other pathways in cerebellar neurons. As shown in Fig. 6A, FIP200 expression is significantly decreased in cerebellar neurons from FIP200f/f;nestin-Cre mice compared with that from control mice. Furthermore, phosphorylation of JNK was also decreased in FIP200f/f;nestin-Cre cerebellar neurons. In contrast, we did not observe any changes in activation of Erk1/2 or Akt upon FIP200 deletion in these neurons. Because the primary cultures of cerebellar neurons are composed of mostly granule cells and Purkinje cells only make up a very small fraction of the isolated neurons, we then examined the activation of JNK in Purkinje cells of FIP200f/f;nestin-Cre by immunohistochemistry. Fig. 6B shows that, consistent with results from granule cells, JNK activation was also significantly reduced in Purkinje cells.
FIGURE 6.
Decreased JNK phosphorylation and increased sensitivity to serum-deprivation induced apoptosis in cerebellar neurons of FIP200f/f;nestin-Cre mice. A, cerebellar neurons were isolated from control or FIP200f/f;nestin-Cre (CKO) mice at P7 and cultured in vitro for 5 days. Lysates were then prepared and then analyzed by Western blotting using various antibodies, as indicated. The asterisk on the left marks a nonspecific band. B, cerebellum sections from control or FIP200f/f;nestin-CRE mice were analyzed by immunohistochemistry with anti-phospho-JNK. Purkinje cells are marked by arrows. Bar = 50 μm. C, cerebellar neurons were isolated and cultured as described in A. They were then treated by serum deprivation in the absence or presence of inhibitors for apoptosis or necroptosis. The neurons were then stained by propidium iodide to detect cell death. The results show the mean ± S.E. from three independent experiments.
Similar to MEFs (20), cerebellar neurons from FIP200f/f; nestin-Cre mice did not exhibit increased cell death compared with that from control mice under normal culture conditions (Fig. 6C). Treatment of the cultured neurons with tumor necrosis factor-α increased cell death in cerebellar neurons from both FIP200f/f;nestin-Cre and control mice, but we did not observe significantly elevated sensitivity upon FIP200 deletion (data not shown), which is unlike that observed in MEFs or hepatocytes previously (20). On the other hand, serum withdraw from growth media dramatically decreased survival of cerebellar neurons from FIP200f/f;nestin-Cre mice while only having a minimal effect on those from control mice (Fig. 6C), suggesting that FIP200 deletion increased susceptibility to cell death induced by deprivation of growth factors and other nutrients in serum. We next examined whether serum deprivation-induced death of FIP200-null cerebellar neurons was through apoptosis or other caspase-independent mechanisms such as necroptosis (36). As shown in Fig. 6C, treatment with V-ZAD, but not necrostatin-1, significantly reduced cell death induced by serum deprivation, suggesting that apoptosis was primarily responsible for the increased cell death in FIP200-null cerebellar neurons. Together, these results suggest that reduced JNK signaling and the increased sensitivity of cerebellar neurons to starvation-induced apoptosis upon FIP200 inactivation and resulting deficiency in autophagy may contribute to the loss of Purkinje cells and granule cells leading to cerebellar ataxia in the mutant mice in vivo.
Spongiosis and Ubiquitinated Protein Aggregates in Other Regions of Brain of FIP200f/f;nestin-Cre Mice
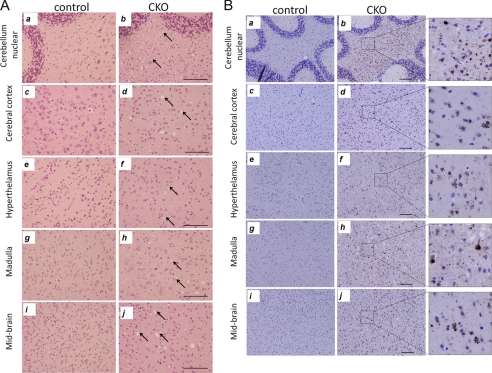
Because FIP200 was also inactivated in other regions of the brain in addition to cerebellum (see Fig. 1A), we examined potentially similar neurodegeneration in other parts of the brain of FIP200f/f;nestin-Cre mice. As shown in Fig. 7A, varying degrees of spongiosis were also observed in several other regions of the brain, including cerebral cortex, hypothalamus, medulla, and midbrain in the FIP200f/f;nestin-Cre mice (arrows). Staining of the sections with anti-ubiquitin antibody also revealed accumulation of ubiquitinated protein aggregates in these regions (Fig. 7B). These results suggest that inactivation of FIP200 in other neurons could also result in accumulation of ubiquitinated protein aggregates due to deficient autophagy, leading to neurodegeneration and spongiosis.
FIGURE 7.
Spongiosis and ubiquitinated protein aggregates in other brain regions of FIP200f/f;nestin-Cre mice. A, sections from cerebellar nuclear (a and b), cerebral cortex (c and d), hypothalamus (e and f), medulla (g and h), and midbrain (i and j) of control (a, c, e, g, and i) and FIP200f/f;nestin-Cre (CKO) (b, d, f, h, and j) mice at 4 weeks of age were stained by hematoxylin and eosin. Arrows mark spongiosis in the various brain regions of mutant mice. Bars = 200 μm. B, sections from cerebellar nuclear (a and b), cerebral cortex (c and d), hypothalamus (e and f), medulla (g and h), and midbrain (i and j) of control (a, c, e, g, and i) and FIP200f/f;nestin-CRE (b, d, f, h, and j) mice at 4 weeks of age were analyzed by immunohistochemistry using anti-ubiquitin. Note the ubiquitin-positive aggregates in different brain regions of FIP200f/f;nestin-CRE mice. The right panels show high magnification view of bracketed areas in b, d, f, h, and j. Scale bars = 200 μm.
DISCUSSION
FIP200 has been implicated in playing important roles in diverse cellular functions through interaction with several cellular proteins and regulation of different signaling pathways (10). Consistent with these observations from in vitro studies, FIP200 knock-out in mice resulted in an embryonic lethal phenotype caused by developmental defects in the heart and liver (20). Although these results indicated a crucial role of FIP200 in cardiac and liver development in vivo, the embryonic lethality prevented analysis of the potential role of FIP200 in different biological and disease processes in adult organisms in vivo. By using a neural-specific conditional KO approach, we present data here suggesting FIP200 as an important regulatory protein in neuronal homeostasis and survival. Deletion of FIP200 in the neural progenitor cells led to several typical neuropathies, including abnormal accumulation of ubiquitinated protein aggregates, progressive axonopathy, and spongiform degeneration, as well as increased neuronal cell death. These cellular defects resulted in growth retardation, development of cerebellar ataxia, and early death of FIP200f/f;nestin-Cre mice.
The defective phenotypes of FIP200f/f;nestin-Cre mice share several characteristics with the recently reported Atg5 or Atg7 conditional KO mice by nestin-Cre (18, 19), such as the increased ubiquitin aggregates without any deficiency in proteasome catalytic functions and loss of Purkinje cells and cerebellar granule cells. Autophagy is an evolutionarily conserved cellular process from yeast to man (37–40), that plays essential roles in the removal of proteins with aberrant structures to maintain cellular homeostasis, which is particularly important in post-mitotic cells such as neurons. Genetic studies in yeast have identified multiple autophagy-related ATG genes and shown a critical importance of the Atg1-Atg13-Atg17 complex for autophagosome initiation (41, 42). Mammalian orthologs for some of the yeast Atg proteins have been identified and characterized to play crucial roles in autophagy in development and diseases processes such as cancer and neurodegeneration (4, 38). Interestingly, several recent studies demonstrated FIP200 interactions with ULK1, mammalian homolog of yeast Atg1, and mammalian Atg13 in a high molecular weight complex, which is required for initiation of starvation-induced autophagy in mammalian cells in vitro (13, 15–17). Consistent with these in vitro data, our results showing similar phenotypes of neural-specific conditional KO of FIP200 and Atg5 or Atg7 provide the first evidence that despite limited homology in their protein sequences FIP200 serves as a mammalian functional counterpart of Atg17 in vivo.
Although they share many of the phenotypes with Atg5 or Atg7 conditional KO, FIP200 conditional KO mice developed many defects such as loss of Purkinje cells as well as movement disorders at an earlier age. As ULK1-Atg13-FIP200 complex is required for autophagosome initiation, whereas Atg5 and Atg7 are involved in the later steps of autophagy (37–40, 43), it is possible that these differences reflect the temporal requirements for autophagy to remove abnormal protein aggregates that were responsible for the various neural degeneration phenotypes. On the other hand, FIP200 has been shown to regulate a number of other signaling pathways and cellular processes besides its interaction with ULK1 and Atg13 in the regulation of autophagy (10). It is therefore possible that perturbations of other signaling pathways may contribute to the earlier and perhaps more severe phenotypes in the FIP200 conditional KO mice.
Our studies also revealed several distinctive features of neural-specific FIP200 conditional KO mice, including the progressive development of spongiosis, that were not observed in the Atg5 or Atg7 conditional KO mice (18, 19) and indeed rarely found in other KO mice models. Signs of spongiform degeneration in the white matter were found as early as 2 weeks of age in FIP200f/f;nestin-Cre mice and precede the loss of Purkinje cells in the mutant mice. Furthermore, we also observed the swelling of segments of Purkinje cell axons both in vivo (see Fig. 3A) and in vitro (data not shown). The blockages in axonal transport resulting from defects in microtubule based motors have been proposed to be responsible for the axonal swelling phenotype (44, 45). Thus, it is possible that ablation of FIP200 could induce the neuronal defects because of abnormality in the axonal transport machinery in Purkinje cells, independent of (or in combination with) the autophagy defects discussed above. Alternatively, such blockages could be caused by the progressive accumulation of ubiquitinated protein aggregates (instead of the transport machinery itself) upon FIP200 deletion but not (or not as severe) in Atg5- or Atg7-deficient neurons. In this regard, it is interesting to note that the aggregation of ubiquitinated proteins was first observed in the white matters correlated with the early signs of spongiform degeneration and anterior to any apparent loss of Purkinje cells. Therefore, the progressive degeneration of Purkinje cell axons could be responsible for the dysfunction of these neurons and their death.
Spongiform degeneration is caused by extensive vacuolization of neuronal cells, which are typically associated with brain damage induced by Prions (46). The underlying mechanism of such vacuolization is not well understood, but mitochondrial dysfunction and increased reactive oxygen species production and/or neuronal sensitivity to reactive oxygen species have been suggested to be responsible for spongiform degeneration in several disease models (47–50). Interestingly, several other mice KO models of neurodegenerative diseases such as those with deletions in mitochondrial protein, Sod2 (51) or PGC1-α (48), also showed spongiform degeneration as prominent features of the neurodegeneration phenotype. Interestingly, unhealthy and fragmented mitochondria were found in Purkinje cells from FIP200f/f;nestin-Cre mice (see Fig. 5F), suggesting a possible role of FIP200 in the regulation of mitochondrial fusion and fission to maintain normal function of mitochondria.
Previous studies showed that total KO of FIP200 caused embryonic lethality resulting from defective heart and liver development as a result of defective tumor necrosis factor-α-JNK and TSC (tuberous sclerosis complex)-mTOR (mammalian target of rapamycin) signaling pathways (20), whereas Atg5−/− and Atg7−/− mice are born at a normal Mendalian ratio and die only shortly after birth because of deficiency in autophagy (18, 19). These results are consistent with the possibility that defects in other signaling pathways in addition to deficient autophagy may also contribute to various neurological phenotypes in FIP200f/f;nestin-Cre mice. Indeed, similar to previous findings in other cells (20), we observed a reduction in JNK activation in cerebellar granule cells and Purkinje cells upon FIP200 deletion. However, the FIP200-null cerebellar neurons showed an increased sensitivity to serum deprivation-induced apoptosis, but not tumor necrosis factor-α-induced cell death as observed in MEFs previously (20), compared with control cells. Future studies will be required to resolve the potential contributions of these and possibly other signaling pathways in mediating neurodegenerative defects in FIP200 conditional KO mice. It will also be interesting to examine potentially other neuronal phenotypes of FIP200f/f;nestin-Cre mice caused by varying degrees of accumulation of ubiquitinated protein aggregates and spongiosis of the other parts of the brain in future studies.
The contents of the abnormally accumulated ubiquitinated protein aggregates in the FIP200 conditional KO mice, or those in the Atg5 or Atg7 conditional KO mice, are unknown. However, it is conceivable that different protein aggregates were accumulated in the FIP200 conditional KO mice versus Atg5 or Atg7 conditional KO mouse, which may also account for the differential phenotype in FIP200 conditional KO mice. In some cases of neurodegenerative mice KO models, the responsible protein aggregates contributing to the neuronal swelling and neurodegeneration are known, such as Prion in TSE (46) and TDP-43 in Sod1−/− mice (52, 53). Future studies will be necessary to identify the proteins in the ubiquitinated aggregates of FIP200 conditional KO mice and to determine whether any of these are responsible for the axonal swelling, spongiform degeneration, and other cerebellar degenerative phenotypes.
In summary, our study suggested a function for FIP200 in the regulation of neuronal cell homeostasis and that its inactivation caused cerebellar degeneration and ataxia accompanied with progressive accumulation of abnormal ubiquitinated protein aggregates, neuronal cell loss, axonal swelling, and spongiosis. These results provide novel insight into the pathogenesis of neurodegenerative diseases and the role of autophagy in the prevention of the diseases. The FIP200 conditional KO mice may provide useful mice models for future studies on the mechanisms of cerebellar degeneration and ataxia as well as promising therapeutic approaches for the diseases in humans.
Acknowledgments
We are grateful to Dr. Yuan Zhu of University of Michigan for discussions, reagents, help throughout this project, critical reading of the manuscript, and helpful comments, Drs. Henry Paulson and Andy Lieberman of University of Michigan for discussions, critical reading of the manuscript, and helpful comments. We also thank members of the Guan laboratory for critical reading of the manuscript and their suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM52890 (to J.-L. G.).
- FIP200
- FAK family-interacting protein of 200 kDa
- KO
- knockout
- PBS
- phosphate-buffered saline
- AMC
- aminomethylcoumarin
- P
- postnatal day
- E
- embryonic day
- MEF
- mouse embryo fibroblast
- MBP
- myelin basic protein
- JNK
- c-Jun N-terminal kinase
- Erk
- extracellular signal-regulated kinase
- Z
- n-benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- Boc
- t-butoxycarbonyl.
REFERENCES
- 1.Goldowitz D., Hamre K. (1998) Trends Neurosci. 21, 375–382 [DOI] [PubMed] [Google Scholar]
- 2.Wang V. Y., Zoghbi H. Y. (2001) Nat. Rev. 2, 484–491 [DOI] [PubMed] [Google Scholar]
- 3.Sillitoe R. V., Joyner A. L. (2007) Annu. Rev. Cell Dev. Biol. 23, 549–577 [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein D. C. (2006) Nature 443, 780–786 [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. (2006) Hematology/the Education Program of the American Society of Hematology, pp. 1–12 and 505–506, American Society of Hematology, Washington, D.C. [DOI] [PubMed] [Google Scholar]
- 6.Berke S. J., Paulson H. L. (2003) Curr. Opin. Genet. Dev. 13, 253–261 [DOI] [PubMed] [Google Scholar]
- 7.Pickart C. M. (2004) Cell 116, 181–190 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N., Klionsky D. J. (2007) Annu. Rev. Nutr. 27, 19–40 [DOI] [PubMed] [Google Scholar]
- 9.Ueda H., Abbi S., Zheng C., Guan J. L. (2000) J. Cell Biol. 149, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan B., Guan J. L. (2008) Cell Signal 20, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chano T., Saji M., Inoue H., Minami K., Kobayashi T., Hino O., Okabe H. (2006) Int. J. Mol. Med. 18, 425–432 [PubMed] [Google Scholar]
- 12.Chano T., Okabe H., Hulette C. M. (2007) Brain Res. 1168, 97–105 [DOI] [PubMed] [Google Scholar]
- 13.Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara T., Mizushima N. (2009) Autophagy 5, 85–87 [DOI] [PubMed] [Google Scholar]
- 15.Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009) J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. (2006) Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. (2006) Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 20.Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. (2006) J. Cell Biol. 175, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei H., Gan B., Wu X., Guan J. L. (2009) J. Biol. Chem. 284, 6004–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dahlstrand J., Lardelli M., Lendahl U. (1995) Brain Res. 84, 109–129 [DOI] [PubMed] [Google Scholar]
- 23.Frappart P. O., Lee Y., Lamont J., McKinnon P. J. (2007) EMBO J. 26, 2732–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates B., Rios M., Trumpp A., Chen C., Fan G., Bishop J. M., Jaenisch R. (1999) Nat. Neurosci. 2, 115–117 [DOI] [PubMed] [Google Scholar]
- 25.Wood K. A., Dipasquale B., Youle R. J. (1993) Neuron 11, 621–632 [DOI] [PubMed] [Google Scholar]
- 26.Ciechanover A., Brundin P. (2003) Neuron 40, 427–446 [DOI] [PubMed] [Google Scholar]
- 27.Bossy-Wetzel E., Schwarzenbacher R., Lipton S. A. (2004) Nat. Med. 10, (suppl.) S2–S9 [DOI] [PubMed] [Google Scholar]
- 28.Ravikumar B., Duden R., Rubinsztein D. C. (2002) Hum. Mol. Genet. 11, 1107–1117 [DOI] [PubMed] [Google Scholar]
- 29.Fortun J., Dunn W. A., Jr., Joy S., Li J., Notterpek L. (2003) J. Neurosci. 23, 10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 31.Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13135–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A, Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 33.Wooten M. W., Geetha T., Babu J. R., Seibenhener M. L., Peng J., Cox N., Diaz-Meco M. T., Moscat J. (2008) J. Biol. Chem. 283, 6783–6789 [DOI] [PubMed] [Google Scholar]
- 34.Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 35.Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G. D., Mitchison T. J., Moskowitz M. A., Yuan J. (2005) Nat. Chem. Biol. 1, 112–119 [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kundu M., Thompson C. B. (2008) Annu. Rev. Pathol. 3, 427–455 [DOI] [PubMed] [Google Scholar]
- 40.Klionsky D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 41.Klionsky D. J. (2005) J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki K., Ohsumi Y. (2007) FEBS Lett. 581, 2156–2161 [DOI] [PubMed] [Google Scholar]
- 43.Simonsen A., Tooze S. A. (2009) J. Cell Biol. 186, 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bendiske J., Bahr B. A. (2003) J. Neuropathol. Exp. Neurol. 62, 451–463 [DOI] [PubMed] [Google Scholar]
- 45.Stokin G. B., Lillo C., Falzone T. L., Brusch R. G., Rockenstein E., Mount S. L., Raman R., Davies P., Masliah E., Williams D. S., Goldstein L. S. (2005) Science 307, 1282–1288 [DOI] [PubMed] [Google Scholar]
- 46.Hooper N. M. (2003) Trends Biotechnol. 21, 144–145 [DOI] [PubMed] [Google Scholar]
- 47.Akassoglou K., Malester B., Xu J., Tessarollo L., Rosenbluth J., Chao M. V. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5075–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J., Wu P. H., Tarr P. T., Lindenberg K. S., St-Pierre J., Zhang C. Y., Mootha V. K., Jäger S., Vianna C. R., Reznick R. M., Cui L., Manieri M., Donovan M. X., Wu Z., Cooper M. P., Fan M. C., Rohas L. M., Zavacki A. M., Cinti S., Shulman G. I., Lowell B. B., Krainc D., Spiegelman B. M. (2004) Cell 119, 121–135 [DOI] [PubMed] [Google Scholar]
- 49.Puccio H., Simon D., Cossée M., Criqui-Filipe P., Tiziano F., Melki J., Hindelang C., Matyas R., Rustin P., Koenig M. (2001) Nat. Genet. 27, 181–186 [DOI] [PubMed] [Google Scholar]
- 50.Matalon R., Rady P. L., Platt K. A., Skinner H. B., Quast M. J., Campbell G. A., Matalon K., Ceci J. D., Tyring S. K., Nehls M., Surendran S., Wei J., Ezell E. L., Szucs S. (2000) J. Gene Med. 2, 165–175 [DOI] [PubMed] [Google Scholar]
- 51.Melov S., Schneider J. A., Day B. J., Hinerfeld D., Coskun P., Mirra S. S., Crapo J. D., Wallace D. C. (1998) Nat. Genet. 18, 159–163 [DOI] [PubMed] [Google Scholar]
- 52.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., Oda T. (2006) Biochem. Biophys. Res. Commun. 351, 602–611 [DOI] [PubMed] [Google Scholar]
- 53.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 54.Craiu A., Gaczynska M., Akopian T., Gramm C. F., Fenteany G., Goldberg A. L., Rock K. L. (1997) J. Biol. Chem. 272, 13437–13445 [DOI] [PubMed] [Google Scholar]